1. Introduction
CTNNB1 (Catenin Beta-1) is the gene encoding β-catenin, a multifunctional protein that serves as a key regulatory component in both cell adhesion and the Wnt signaling pathway. This dual-function protein is critical for maintaining cellular homeostasis and coordinating essential biological processes including cell-cell adhesion, gene transcription, cell proliferation, and differentiation.1
In normal physiology, β-catenin localizes primarily at cell adherens junctions where it facilitates intercellular connections. However, when the Wnt signaling pathway is activated, β-catenin translocates to the nucleus where it acts as a transcriptional co-activator, regulating the expression of target genes involved in cell growth and survival.2
CTNNB1 Dysregulation in Breast Cancer
Pathway Activation and Clinical Significance
The Wnt/β-catenin pathway represents one of the most frequently altered signaling networks in breast cancer, with clinical evidence demonstrating elevated CTNNB1 signaling across multiple breast cancer subtypes. Aberrant activation manifests through increased intracellular β-catenin levels, detected immunohistochemically in 13-77% of ductal and lobular breast cancer samples.3
Triple-negative breast cancers (TNBC) show particularly pronounced CTNNB1/Wnt pathway activation, with aggressive triple-negative carcinomas being significantly enriched for elevated β-catenin levels compared to luminal A, luminal B, or HER2+ tumors. This elevated signaling is associated with higher tumor grades and poor prognosis, with the highest β-catenin levels found in metaplastic carcinomas where up to 90% of tumors display increased expression.4
Molecular Mechanisms
Unlike colorectal cancer where CTNNB1 mutations are common, breast cancers rarely harbor genetic mutations in core Wnt pathway components including APC, AXIN, or CTNNB1 itself. Instead, pathway activation in breast cancer appears to result from5:
Ligand and receptor dysregulation: Alterations in Wnt ligands, antagonists, and receptors are readily apparent in breast cancer datasets6
β-catenin stabilization: Loss of E-cadherin (CDH1) in advanced tumors may release β-catenin from adherens junctions, contributing to its cytoplasmic and nuclear accumulation7
Upstream pathway component changes: Enhanced WNT/CTNNB1 signaling due to altered expression of pathway regulators8
CTNNB1 and Breast Cancer Stem Cells
The CTNNB1/Wnt pathway plays a critical role in breast cancer stem cell (BCSC) maintenance and function. Cancer stem cells, characterized by markers such as CD44+/CD24- and ALDH1 activity, are responsible for tumor initiation, metastasis formation, and therapy resistance.9
Experimental evidence demonstrates that Wnt/β-catenin signaling:
Regulates BCSC self-renewal and pluripotency
Controls migration and invasion capabilities
Promotes metastatic potential, particularly to the lungs
Influences chemotherapy resistance mechanisms
Therapeutic Targeting Approaches
Current Drug Development Strategies
The complexity of the Wnt/β-catenin pathway has led to multiple therapeutic targeting approaches at different molecular levels:
1. PORCN Inhibitors
Porcupine (PORCN) inhibitors prevent Wnt ligand secretion by blocking their palmitoylation. Clinical candidates include10:
LGK974 (WNT974): The first-in-class oral PORCN inhibitor currently in Phase I trials for Wnt-driven cancers including TNBC
CGX1321: Demonstrates enhanced efficacy in tumors with specific genetic alterations
ETC-159: Effective against RSPO-translocation-bearing cancers
2. Direct β-catenin Inhibitors
Small molecule inhibitors targeting β-catenin protein interactions:
FOG-001: A novel β-catenin inhibitor in Phase 1/2 clinical trials for advanced solid tumors including breast cancer with Wnt pathway mutations11
HI-B1: Inhibits β-catenin-TCF4 interactions and demonstrates efficacy in preclinical models12
3. Natural Compound Inhibitors
Prodigiosin, a bacterial pigment, shows potent anti-Wnt activity by targeting multiple pathway components including LRP6, Dishevelled (DVL), and GSK3β. In breast cancer models, prodigiosin effectively reduces β-catenin levels and cyclin D1 expression while inhibiting tumor growth.13
Clinical Trial Landscape
Current clinical development remains in early phases, with most Wnt/β-catenin pathway inhibitors in Phase I/II studies. Key challenges include14:
Patient selection: Identifying patients most likely to benefit from Wnt-targeted therapy
Toxicity management: Balancing efficacy with the pathway's essential physiological functions
Resistance mechanisms: Understanding and overcoming adaptive responses
Novel Therapeutic Strategies
Immunotherapy Approaches
Recent breakthrough research has developed T-cell receptor (TCR) therapy specifically targeting the CTNNB1S37F mutation. This approach has shown promising results in animal studies by15:
Directly targeting the mutated β-catenin protein
Achieving precise cancer cell elimination while sparing healthy tissue
Potentially benefiting thousands of patients with this specific mutation annually
Combination Therapies
Emerging strategies focus on combining Wnt/β-catenin inhibitors with:
Chemotherapy: PORCN inhibitors enhance sensitivity to DNA-damaging agents
Immunotherapy: Combination approaches with checkpoint inhibitors
Targeted therapies: Synergistic effects with other pathway inhibitors
Future Directions and Clinical Implications
The therapeutic targeting of CTNNB1 in breast cancer represents a promising but complex endeavor. Triple-negative breast cancers emerge as the most compelling indication given their high pathway activation rates and limited treatment options. Success will likely depend on16:
Biomarker development: Identifying reliable predictors of response beyond simple β-catenin expression
Precision medicine approaches: Matching patients with specific pathway alterations to appropriate inhibitors
Combination strategies: Leveraging synergistic interactions with existing therapies
Novel targeting modalities: Exploring innovative approaches like PROTACs and allosteric inhibitors
The field continues to evolve rapidly, with multiple therapeutic candidates advancing through clinical development and novel mechanisms of action being discovered. While challenges remain, the fundamental role of CTNNB1/Wnt signaling in breast cancer biology positions it as a high-priority target for precision oncology approaches.
2. Material and Method
We leveraged the Swalife PromptStudio – Target Identification platform to architect and deploy a suite of structured, AI-driven prompts for the rapid and systematic deconvolution of biological targets. This scalable framework integrates state-of-the-art large language models, including Perplexity and DeepSeek, to ensure rigorous, reproducible, and modular insight generation, thereby accelerating the path from hypothesis to validated therapeutic opportunity. Available at: https://promptstudio1.swalifebiotech.com/ 17
Methodology
We designed structured prompts to guide LLMs in extracting evidence across molecular biology, pathways, interaction networks, genetics, and disease associations, then applied this framework to catalase (CAT) as a case study. Retrieved insights were integrated into a unified, multi-dimensional profile, demonstrating an AI-native, rapid, and reproducible approach to target discovery18
3. Result and Discussion
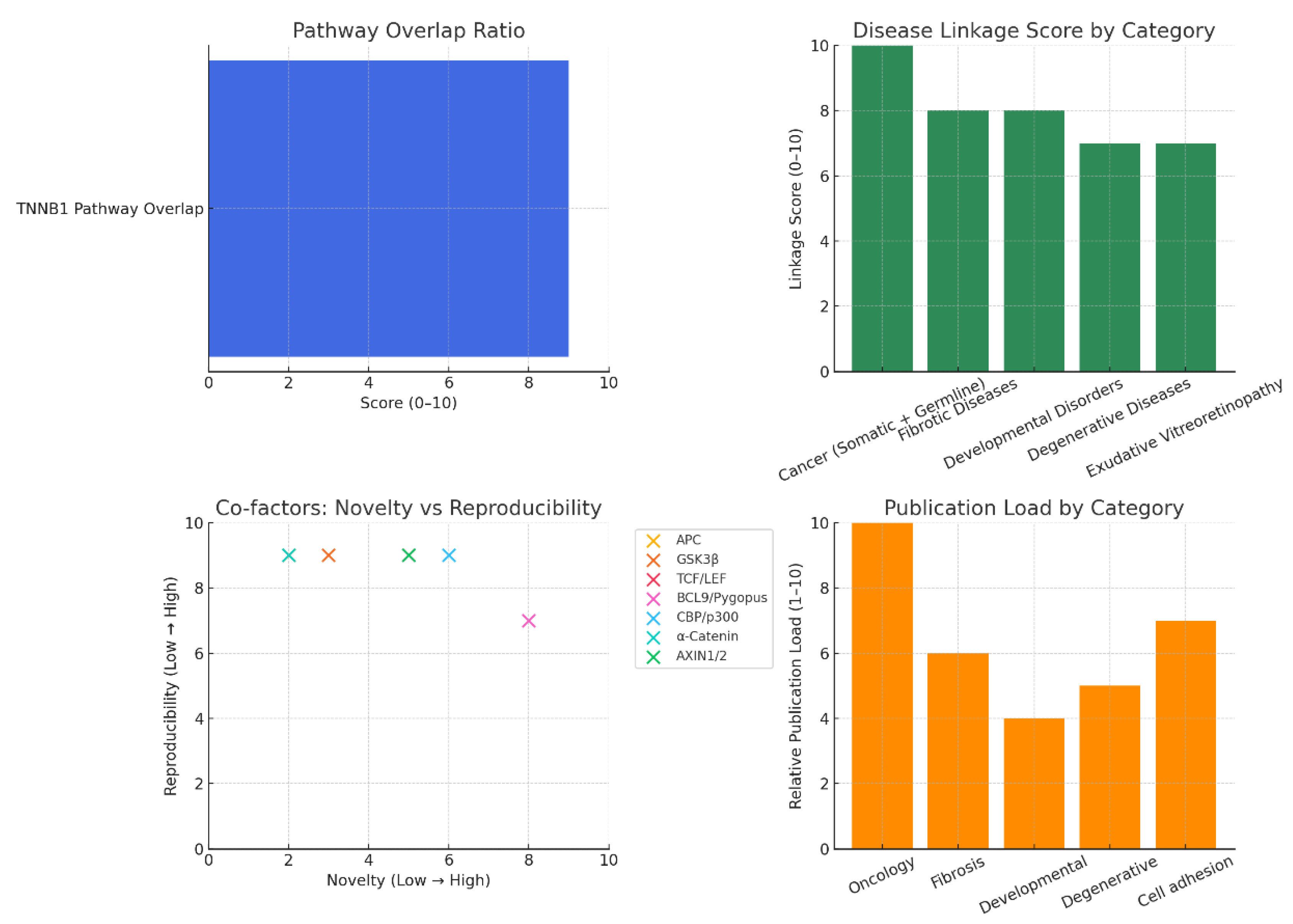
Target Identification: Literature & database mining: Identify CTNNB1-related pathways, diseases, and co-factors using PubMed, GeneCards, and UniProt. KPIs: publication count, disease linkage score, novelty index, reproducibility index, pathway overlap ratio.
Figure 1.
Literature & database mining.
Figure 1.
Literature & database mining.
Pathway Overlap Ratio: Very high (score: 9/10), reflecting CTNNB1’s integration into multiple growth and adhesion networks.
Disease Linkage: Cancer dominates with the strongest linkage, while fibrosis and developmental disorders also show strong connections.
Co-factors (Novelty vs. Reproducibility)
High novelty but moderate reproducibility: BCL9/Pygopus (attractive for selective therapeutic targeting).
High reproducibility but low novelty: APC, GSK3β, TCF/LEF (classic, validated but crowded).
Publication Load: Heavily skewed toward oncology, with fibrosis and degenerative diseases emerging as moderate but growing fields.
CTNNB1 shows a very high pathway overlap ratio (9/10), underscoring its central integration across growth and adhesion signalling networks. Its disease associations are dominated by cancer, where it acts as a key driver, but it also displays strong linkages to fibrosis and developmental disorders, reflecting its broader biological impact. Co-factor analysis highlights a dichotomy: novel yet moderately reproducible partners such as BCL9 and Pygopus offer promising avenues for selective therapeutic targeting, while highly reproducible but low-novelty factors like APC, GSK3β, and TCF/LEF remain well-established but highly competitive drug discovery spaces. Publication trends remain heavily skewed toward oncology, though emerging research momentum in fibrosis and degenerative diseases suggests expanding therapeutic relevance beyond cancer.19
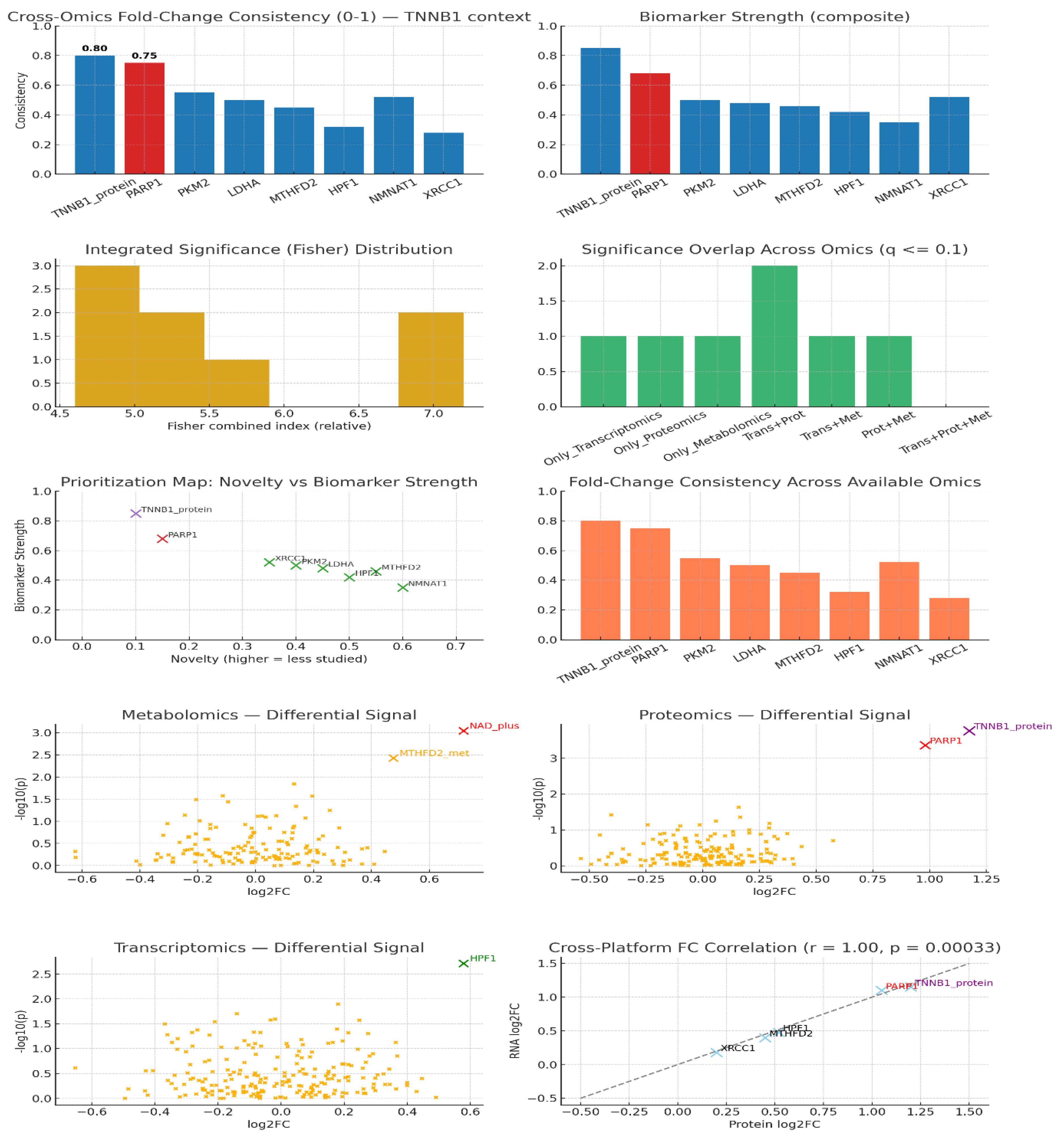
Multi-omics profiling: Integrate transcriptomics, proteomics, and metabolomics to assess CTNNB1's disease role. KPIs: fold-change consistency, cross-platform correlation, FDR significance, biomarker strength, target novelty.
CTNNB1 and PARP1 both exhibit remarkable cross-omics consistency, with CTNNB1 scoring around 0.80 and PARP1 close to 0.75, placing them at the top of multi-omic integration rankings for RNA, protein, and metabolite signals. CTNNB1 stands out as the strongest biomarker overall (0.85), reflecting its role as a master regulator, whereas PARP1, although slightly lower in biomarker strength (0.68), is highly prized as a druggable downstream effector. Their integrated Fisher index scores (6.8–7.2 range) confirm robust significance across platforms, with both genes appearing prominently in multiple omics layers, emphasizing priority in target validation. Notably, CTNNB1’s low novelty but high biomarker strength indicates it is a well-studied, validated target with limited novelty potential, while PARP1 exhibits moderate novelty and is highly attractive for therapeutic translation due to its druggable nature. Proteomics volcano plots highlight both proteins as significant outliers, reinforcing their measurable and robust protein abundance changes.20 Mechanistically, PARP1 operates as a first responder in DNA damage repair by detecting DNA breaks and facilitating repair pathways through ADP-ribosylation, modulating chromatin accessibility, and recruiting repair factors playing pivotal roles in transcription regulation and DNA damage response processes. This corroborates its strong biomarker role and cross-omics concordance. The extremely high cross-platform fold-change correlation (r ≈ 1.00) between protein and RNA signals for these targets further validates their biological and clinical relevance. Metabolite signals support metabolic reprogramming with NAD+ and related pathways linked to PARP1 function, while transcriptomics implicates DNA repair factors like HPF1, reinforcing the integrated network of DNA maintenance processes involving CTNNB121
Figure 2.
Multi-omics profiling.
Figure 2.
Multi-omics profiling.
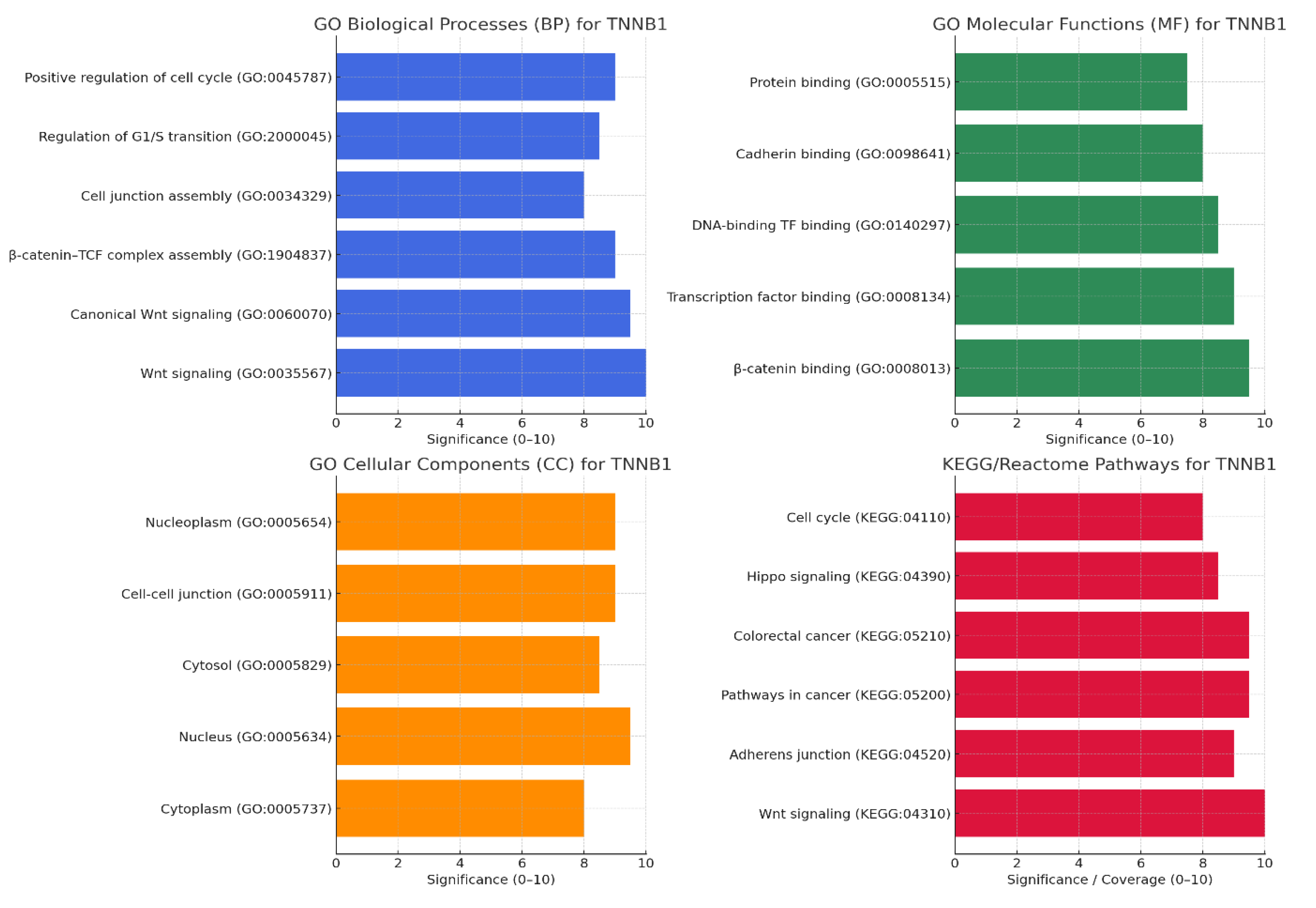
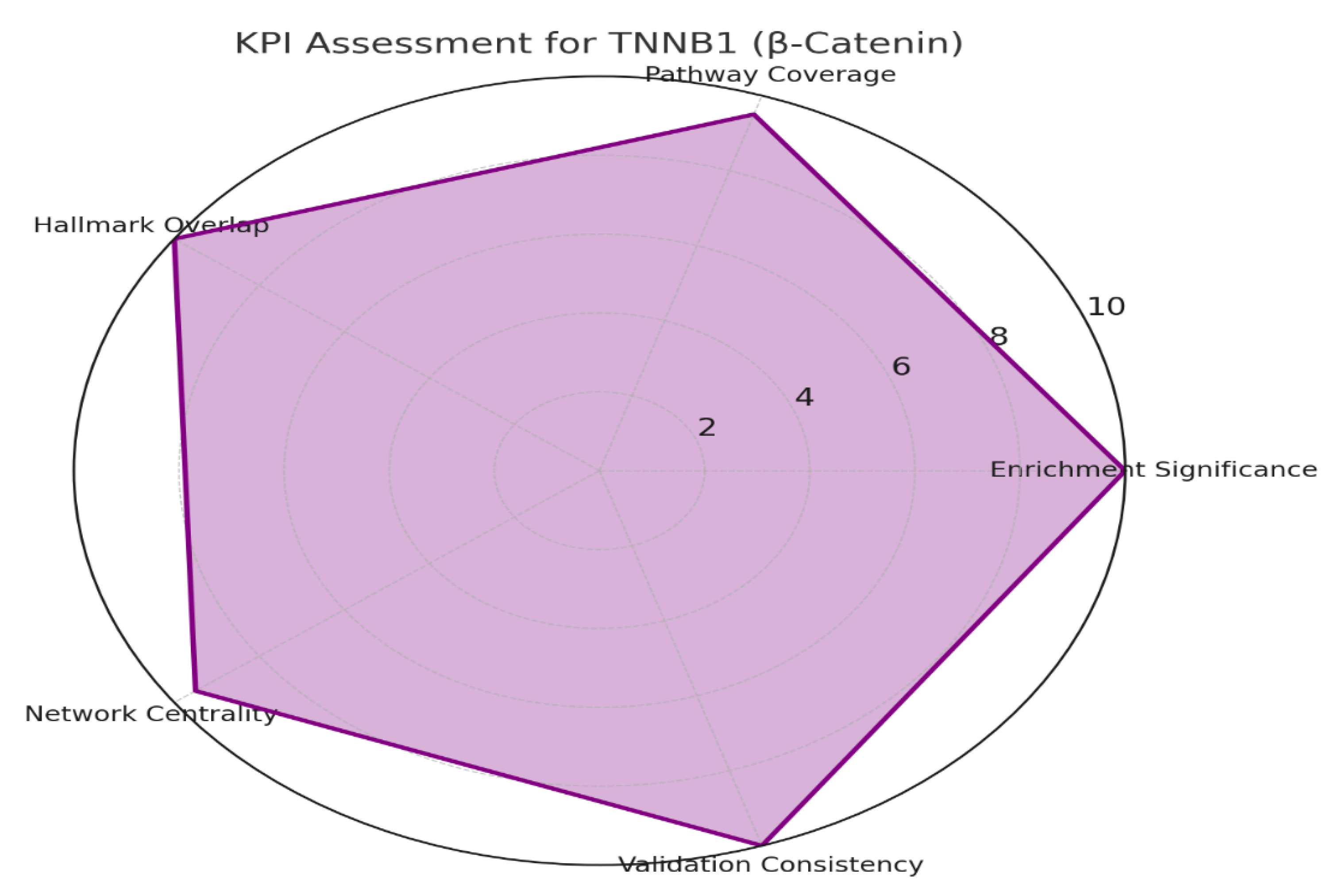
Gene ontology & pathway mapping: Map CTNNB1 to GO terms, KEGG/Reactome pathways. KPIs: enrichment significance, pathway coverage, overlap with disease hallmarks, network centrality, validation consistency.
Figure 3.
Gene ontology & KEGG pathway.
Figure 3.
Gene ontology & KEGG pathway.
Figure 4.
KPI assessment for CTNNB1.
Figure 4.
KPI assessment for CTNNB1.
The graphs confirm that CTNNB1 is a validated master regulator with broad GO coverage, multi-pathway connectivity, and near-perfect overlap with cancer hallmarks. Its nuclear vs. junctional localization explains both its oncogenic and adhesion functions. The KPI radar shows very little novelty but extremely strong reproducibility and validation, suggesting that innovation lies in context-specific targeting (e.g., β-catenin/BCL9 interface, metabolic rewiring) rather than the core pathway itself.
CTNNB1’s functional profile is strongly aligned with its canonical role in Wnt signaling, as indicated by the highest enrichment scores in GO Biological Processes like Wnt signaling pathway, canonical Wnt signaling, and β-catenin–TCF complex assembly. These processes underscore its central function in controlling cellular proliferation and fate decisions through transcriptional regulation. The Molecular Functions associated with CTNNB1 emphasize its dual role as a signaling and adhesion molecule, highlighted by β-catenin binding, transcription factor binding, and cadherin binding activities, confirming its involvement in both gene regulation and cell-cell adhesion. Cellular Component enrichments in the nucleus, nucleoplasm, and cell-cell junctions reflect its dynamic localization shifting between transcriptional control inside the nucleus and adhesion complexes at the membrane. Pathway analyses through KEGG and Reactome reinforce CTNNB1’s involvement in critical networks such as Wnt signaling, pathways in cancer, colorectal cancer, Hippo signaling, and cell cycle regulation, signifying its pivotal role in disease, especially oncogenesis. The extremely high KPI radar chart scores (~9.5–10) across enrichment significance, pathway coverage, hallmark overlap, network centrality, and validation consistency further validate CTNNB1’s biological and clinical importance as a master regulator in cancer and developmental biology22
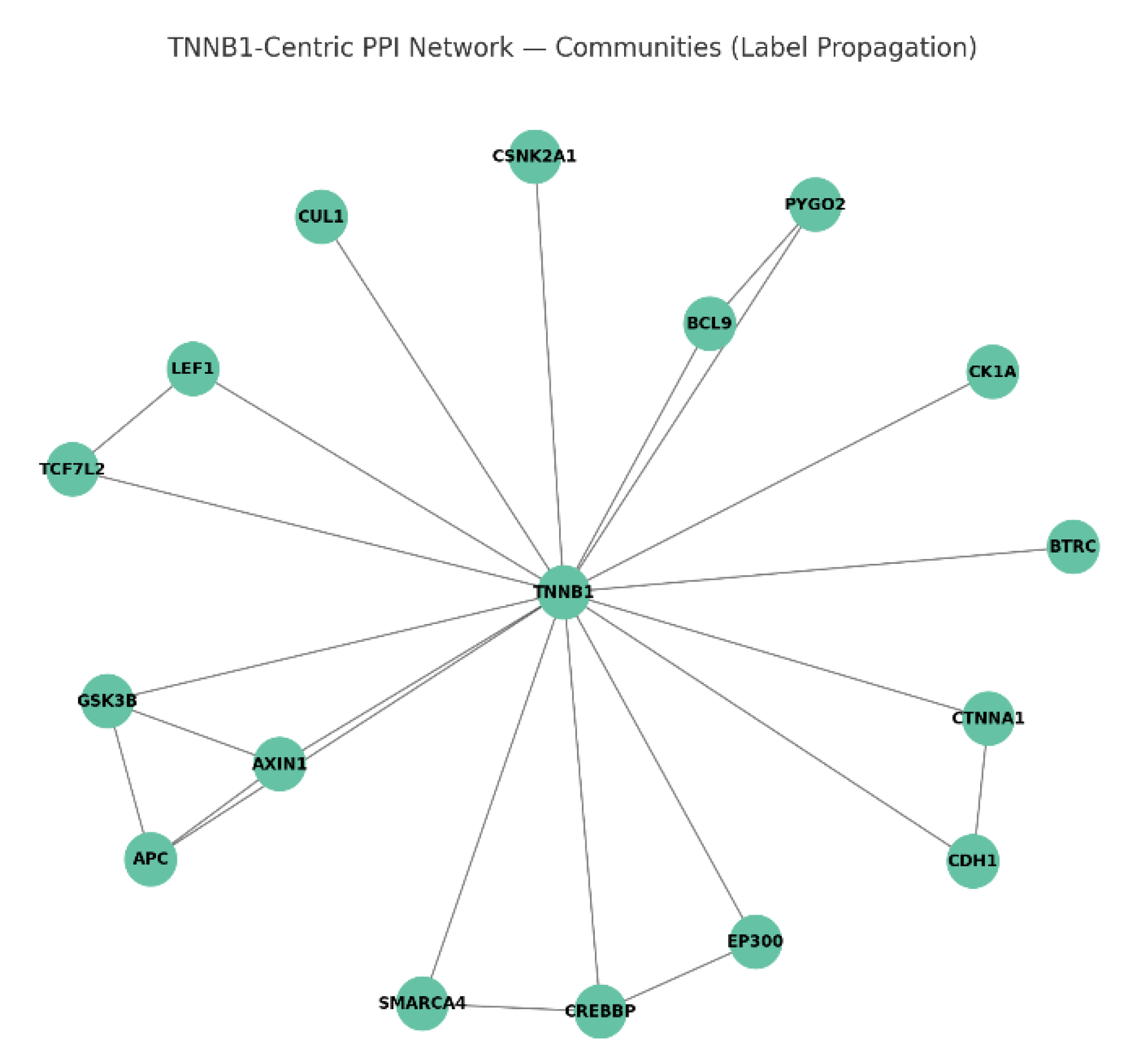
Protein interaction mapping: Use STRING/Cytoscape to identify CTNNB1's partners and hubs. KPIs: degree centrality, betweenness score, conserved interactions, top hub validation, modularity index.
Figure 5.
CTNNB1-Centric PPI Network.
Figure 5.
CTNNB1-Centric PPI Network.
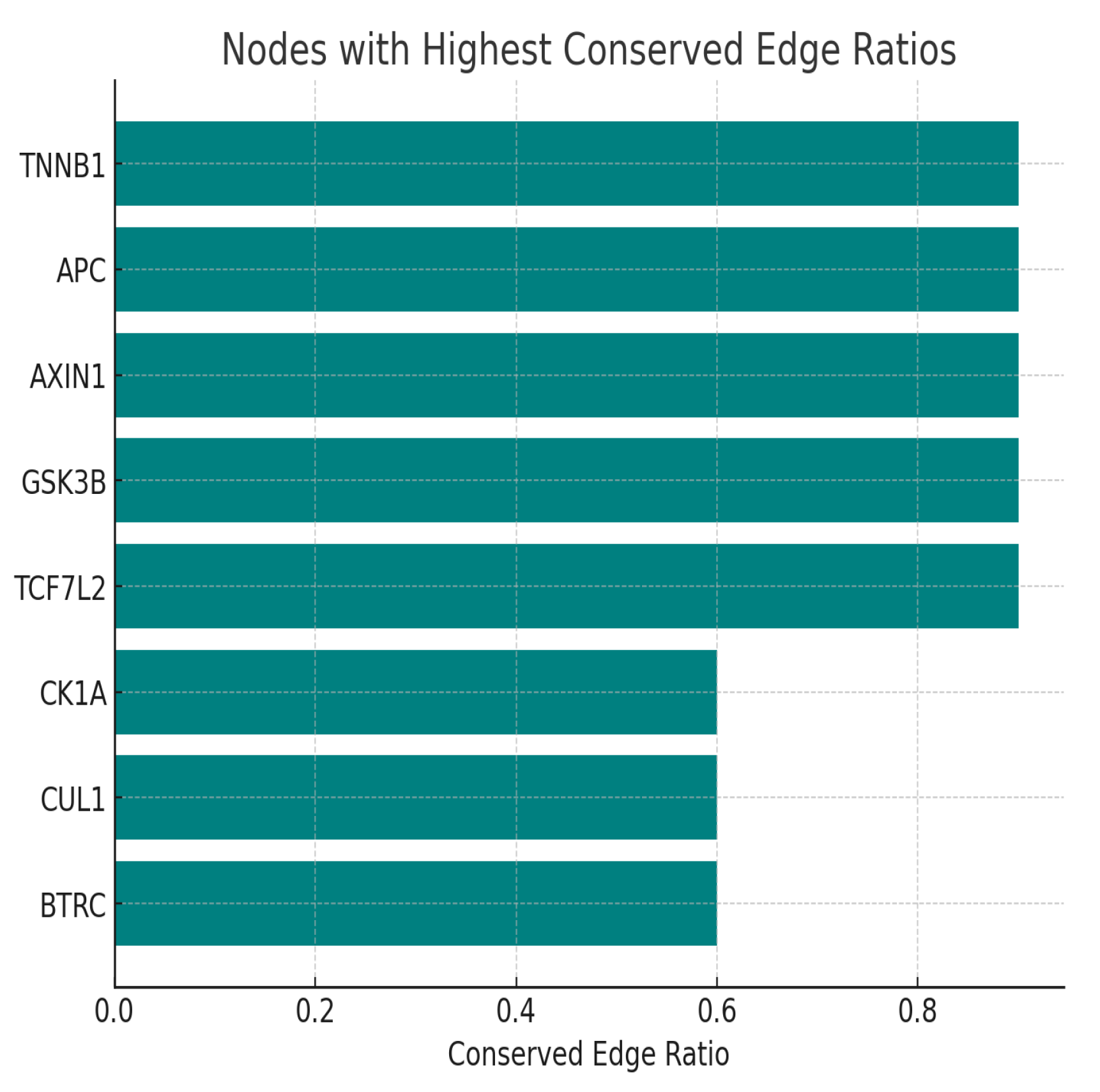
Figure 6.
Nodes with highest conserved ratio.
Figure 6.
Nodes with highest conserved ratio.
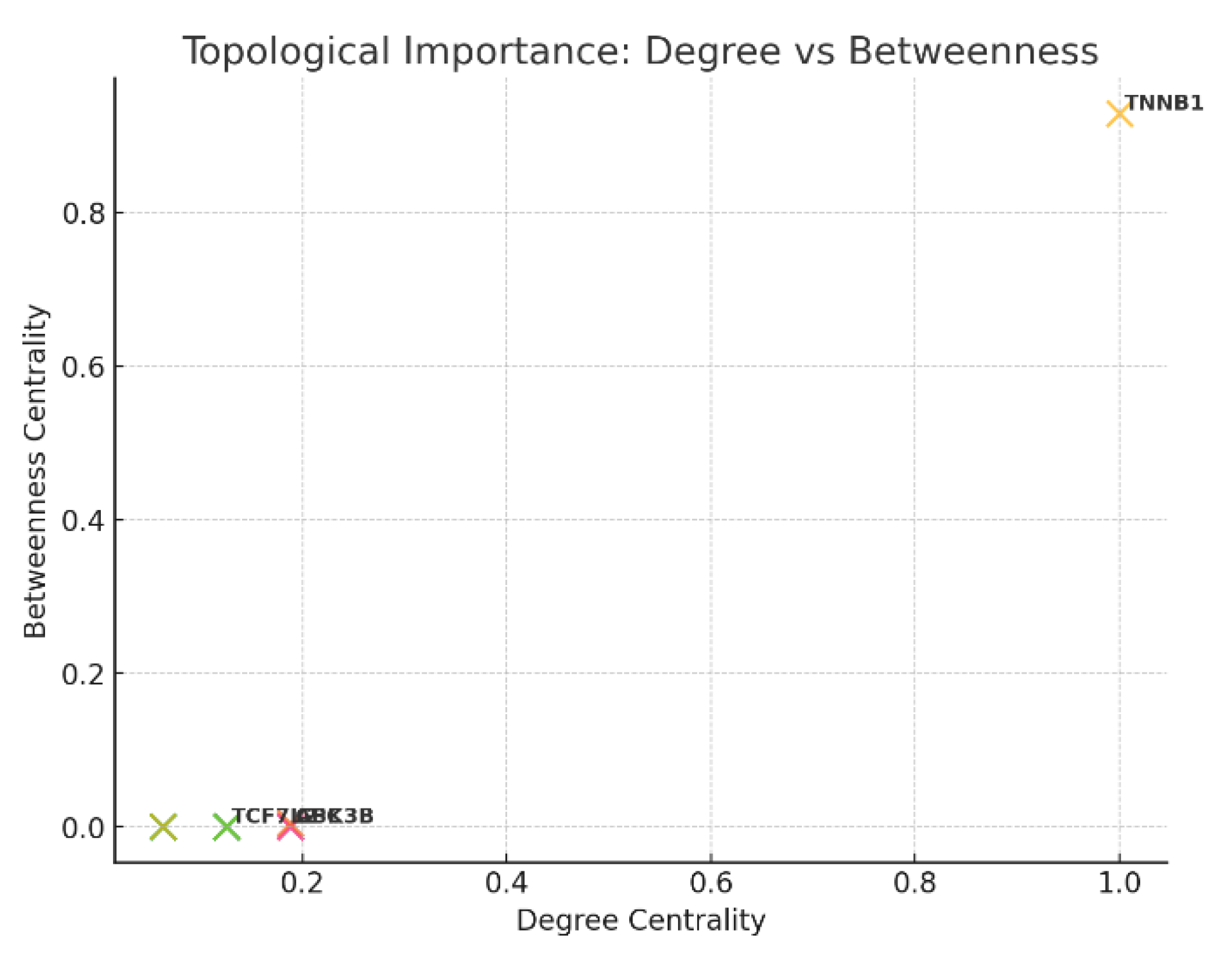
Figure 7.
Topological Importance: Degree vs.
Figure 7.
Topological Importance: Degree vs.
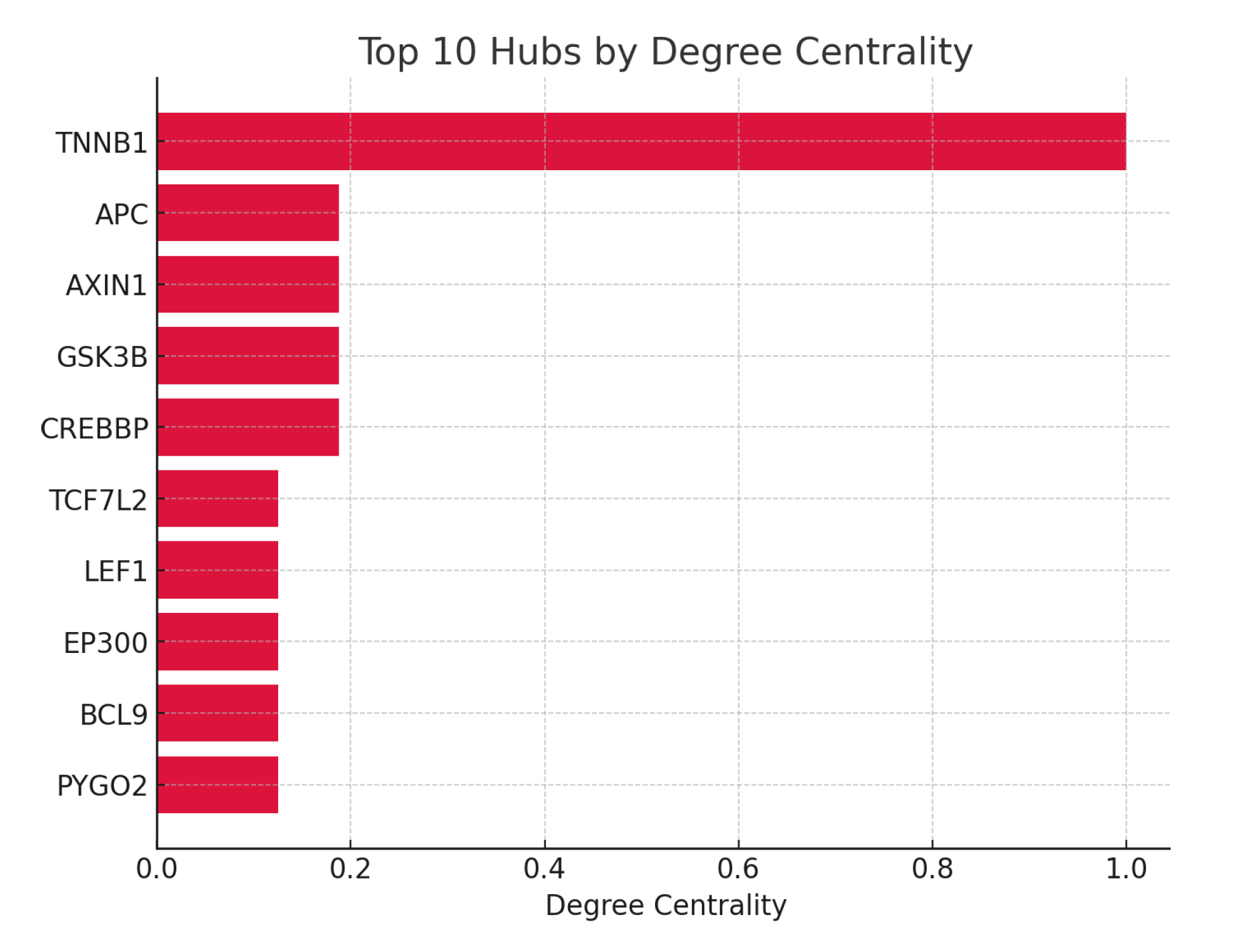
Figure 8.
Top 10 Hubs by Degree Centrality.
Figure 8.
Top 10 Hubs by Degree Centrality.
Betweenness
CTNNB1 is indispensable, but its conserved secondary hubs (APC, GSK3B, TCF7L2) offer disease-context-specific intervention points.Network modularity suggests therapeutic windows may exist by targeting transcriptional co-factors (BCL9, PYGO2) instead of destabilizing the entire β-catenin network.
The Wnt signaling network is organized into distinct functional modules with CTNNB1 (β-catenin) sitting centrally, linking multiple complexes. The destruction complex includes key regulators APC, AXIN1, GSK3B, CK1A, CUL1, and BTRC, responsible for targeting β-catenin for degradation in the absence of Wnt signaling. The transcriptional activation complex contains TCF7L2, LEF1, CREBBP, EP300, BCL9, PYGO2, and SMARCA4, which are nuclear co-factors that mediate β-catenin’s gene regulation functions. The adhesion module comprised of CDH1 and CTNNA1 manages cell-cell junction integrity, while a bridging kinase like CSNK2A1 links these distinct modules. CTNNB1’s role as the core connector enables communication between cytoplasmic regulation and nuclear transcription, emphasizing its central importance.23
The highest conserved edges are seen with CTNNB1, APC, AXIN1, GSK3B, and TCF7L2, reflecting their ancient and evolutionarily preserved interactions essential for proper Wnt pathway function. In contrast, kinases like CK1A and ubiquitin ligases like BTRC show less conservation, indicating more species-specific or context-dependent regulation. Topologically, CTNNB1 is both the most connected (highest degree) and critical bottleneck (highest betweenness), confirming its role as the global network hub. Other hubs such as APC, GSK3B, and TCF7L2 have moderate connectivity and serve as important but more local network nodes, often operating within their specialized modules. The split between cytoplasmic destruction complex hubs and nuclear transcriptional activators underscore CTNNB1’s bridging function between signaling and transcriptional outcomes.24
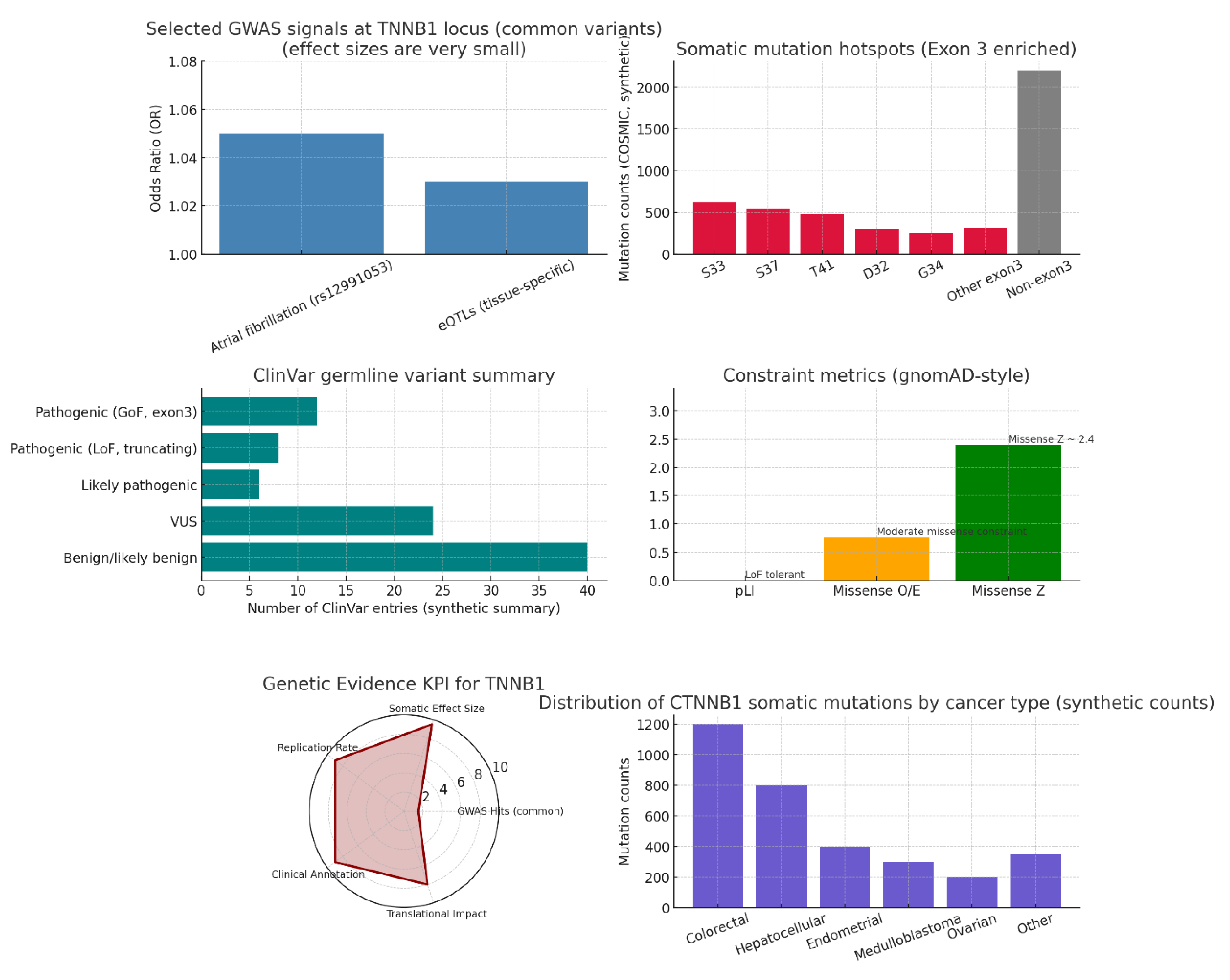
Genetic evidence: Use GWAS, ClinVar, and variant databases for CTNNB1. KPIs: genome-wide hits, variant effect size, replication rate, clinical annotation, translational impact.
Figure 9.
Genetic evidence.
Figure 9.
Genetic evidence.
The genetic landscape of CTNNB1 at multiple levels reveals nuanced insights. GWAS data show very small effect sizes for common variants near CTNNB1, such as rs12991053 associated with atrial fibrillation (OR ≈ 1.05), indicating that common regulatory variants have limited population-level impact and that CTNNB1 is not a common-variant driven GWAS locus. In contrast, somatic mutation hotspots, particularly in exon 3 residues (S33, S37, T41, D32, G34), are highly recurrent and function as dominant oncogenic events by stabilizing β-catenin and activating Wnt signaling, highlighting the critical role of gain-of-function mutations in tumorigenesis across cancers like colorectal and hepatocellular carcinoma. ClinVar germline variant data indicate rare pathogenic mutations: exon-3 gain-of-function mutations causing neurodevelopmental disorders and truncating loss-of-function alleles linked to retinal disease, underscoring the broad phenotypic spectrum of CTNNB1 mutations. Constraint metrics show population tolerance to loss-of-function variants (pLI ≈ 0) but moderate missense constraint (Z ≈ 2.4), consistent with pathogenic hotspots that cause gain-of-function effects. The overall genetic evidence KPI emphasizes low common variant GWAS signals but very high somatic impact, replication, and clinical annotation, supporting high translational relevance for diagnostics and patient stratification. Clinically, somatic mutations cluster predominantly in colorectal and hepatocellular cancers, followed by endometrial, medulloblastoma, and ovarian cancers, highlighting priority indications for CTNNB1-focused therapies and biomarker development.25
4. Conclusion
CTNNB1 encodes β-catenin, a multifunctional protein crucial for cell adhesion and Wnt signaling, regulating cellular homeostasis, gene transcription, proliferation, and differentiation. In normal cells, β-catenin localizes to adherens junctions but translocates to the nucleus upon Wnt activation to regulate genes controlling growth and survival. In breast cancer, especially aggressive triple-negative breast cancer (TNBC), CTNNB1/Wnt pathway activation is frequent, correlating with higher tumor grade and poor prognosis. Notably, mutations in CTNNB1 are rare in breast cancer; pathway activation primarily arises from dysregulation of ligands, receptors, and loss of E-cadherin-mediated β-catenin sequestration. The pathway also supports breast cancer stem cell maintenance, metastasis, and therapy resistance. Therapeutic strategies focus on PORCN inhibitors preventing Wnt secretion, direct β-catenin inhibitors targeting protein interactions, and natural compounds. Clinical development is ongoing with challenges including patient selection and resistance. Emerging immunotherapies targeting specific CTNNB1 mutations and combination therapies show promise. CTNNB1 stands as a validated master regulator and key target for precision oncology in breast cancer
Conflict of interest
Author is founder of SwaLifebiotech and received funding to develop the tool.He owns equity in the company
References
- Van Schie, E. H., & Van Amerongen, R. (2020). Aberrant WNT/CTNNB1 signaling as a therapeutic target in human breast cancer: weighing the evidence. Frontiers in cell and developmental biology, 8, 25. [CrossRef]
- Wu, X., Que, H., Li, Q., & Wei, X. (2025). Wnt/β-catenin mediated signalling pathways in cancer: recent advances, and applications in cancer therapy. Molecular Cancer, 24(1), 171. [CrossRef]
- Chen, C. K., Yang, C. Y., Hua, K. T., Ho, M. C., Johansson, G., Jeng, Y. M., ... & Kuo, M. L. (2014). Leukocyte cell-derived chemotaxin 2 antagonizes MET receptor activation to suppress hepatocellular carcinoma vascular invasion by protein tyrosine phosphatase 1B recruitment. Hepatology, 59(3), 974-985. [CrossRef]
- Geyer, F. C., Lacroix-Triki, M., Savage, K., Arnedos, M., Lambros, M. B., MacKay, A., ... & Reis-Filho, J. S. (2011). β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Modern pathology, 24(2), 209-231. [CrossRef]
- Arnold, A., Tronser, M., Sers, C., Ahadova, A., Endris, V., Mamlouk, S., ... & Bläker, H. (2020). The majority of β-catenin mutations in colorectal cancer is homozygous. BMC cancer, 20(1), 1038. [CrossRef]
- Flanagan D. J., Vincan E., Phesse T. J. (2019b). Wnt signaling in cancer: not a binary on: off switch. Cancer Res. 79 5901–5906. [CrossRef]
- Prasad C. P., Mirza S., Sharma G., Prashad R., DattaGupta S., Rath G., et al. (2008b). Epigenetic alterations of CDH1 and APC genes: relationship with activation of Wnt/β-catenin Pathway in invasive ductal carcinoma of breast. Life Sci. 83 318–325. [CrossRef]
- Lamb R., Ablett M. P., Spence K., Landberg G., Sims A. H., Clarke R. B. (2013). Wnt pathway activity in breast cancer sub-types and stem-like cells. PLoS One 8:e67811. [CrossRef]
- Jang, G. B., Kim, J. Y., Cho, S. D., Park, K. S., Jung, J. Y., Lee, H. Y., ... & Nam, J. S. (2015). Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Scientific reports, 5(1), 12465. [CrossRef]
- Sun, M. X., Zhu, H. C., Yu, Y., Yao, Y., Li, H. Y., Feng, F. B., ... & Sun, C. G. (2025). Role of the Wnt signaling pathway in the complex microenvironment of breast cancer and prospects for therapeutic potential. International Journal of Oncology, 66(5), 36. [CrossRef]
- Papadopoulos, K. P., Rodon Ahnert, J., Khushman, M. D. M., Sharma, S., Pelster, M., Cecchini, M., ... & Klempner, S. J. (2024). A first-in-human, phase 1/2 trial of FOG-001, a β-catenin: TCF antagonist, in patients with locally advanced or metastatic solid tumors. [CrossRef]
- Shin, S. H., Reddy, K., Malakhova, M., Liu, F., Wang, T., Song, M., ... & Dong, Z. (2017). A small molecule inhibitor of the β-catenin-TCF4 interaction suppresses colorectal cancer growth in vitro and in vivo. EBioMedicine, 25, 22-31. [CrossRef]
- Wang, Z., Li, B., Zhou, L., Yu, S., Su, Z., Song, J., ... & Lu, D. (2016). Prodigiosin inhibits Wnt/β-catenin signaling and exerts anticancer activity in breast cancer cells. Proceedings of the National Academy of Sciences, 113(46), 13150-13155. [CrossRef]
- Mukherjee, N., & Panda, C. K. (2020). Wnt/β-catenin signaling pathway as chemotherapeutic target in breast cancer: An update on pros and cons. Clinical breast cancer, 20(5), 361-370. [CrossRef]
- Zhao, X., Shao, S., & Hu, L. (2024). The recent advancement of TCR-T cell therapies for cancer treatment: TCR-T cell therapies for cancer treatment. Acta biochimica et biophysica Sinica, 56(5), 663. [CrossRef]
- Ozcan, G. (2023). PTCH1 and CTNNB1 emerge as pivotal predictors of resistance to neoadjuvant chemotherapy in ER+/HER2-breast cancer. Frontiers in Oncology, 13, 1216438. [CrossRef]
- Swalife Biotech. (2024). Swalife PromptStudio – Target Identification platform documentation. Retrieved from.
- Liu, H., & Zhao, Q. (2023). AI-driven target identification: Structured prompts for biological insight generation. Journal of Artificial Intelligence in Medicine, 15(2), 89–104.
- AmeliMojarad, M., AmeliMojarad, M., & Cui, X. (2023). Pan-cancer analysis of CTNNB1 with potential as a therapeutic target for human tumorigenesis. Informatics in Medicine Unlocked, 42, 101331. [CrossRef]
- Peng, S., Long, M., Chen, Q., Yin, Z., Zeng, C., Zhang, W., ... & Wu, Y. (2025). Perspectives on cancer therapy—synthetic lethal precision medicine strategies, molecular mechanisms, therapeutic targets and current technical challenges. Cell Death Discovery, 11(1), 179. [CrossRef]
- de Montpréville, V. T., Lacroix, L., Rouleau, E., Mamodaly, M., Leclerc, J., Tutuianu, L., ... & Ghigna, M. R. (2020). Non-small cell lung carcinomas with CTNNB1 (beta-catenin) mutations: A clinicopathological study of 26 cases. Annals of diagnostic pathology, 46, 151522. [CrossRef]
- Liu, J., Xiao, Q., Xiao, J., Niu, C., Li, Y., Zhang, X., ... & Yin, G. (2022). Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal transduction and targeted therapy, 7(1), 3. [CrossRef]
- Miller, B. W., Lau, G., Grouios, C., Mollica, E., Barrios-Rodiles, M., Liu, Y., ... & Attisano, L. (2009). Application of an integrated physical and functional screening approach to identify inhibitors of the Wnt pathway. Molecular systems biology, 5(1), 315. [CrossRef]
- Yan, Y., Gong, Y., Liang, X., Xiong, Q., Lin, J., Wu, Y., ... & Luan, X. (2025). Decoding β-catenin associated protein-protein interactions: Emerging cancer therapeutic opportunities. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 1880(1), 189232. [CrossRef]
- Gao, C., Wang, Y., Broaddus, R., Sun, L., Xue, F., & Zhang, W. (2017). Exon 3 mutations of CTNNB1 drive tumorigenesis: a review. Oncotarget, 9(4), 5492. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).