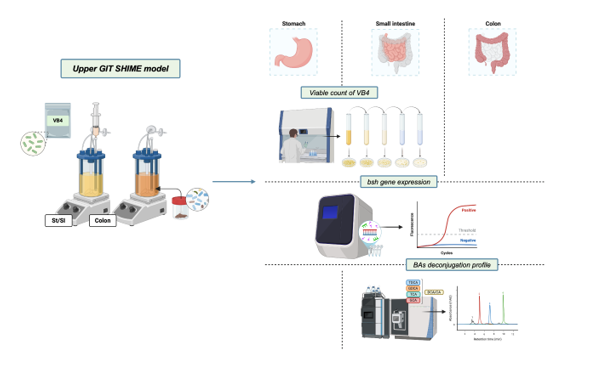
1. Introduction
Lactobacilli, as natural inhabitant of the GIT, have attracted considerable attention for their probiotic trait and for the capacity to synthesize the bile salt hydrolase (BSH), an enzyme closely linked to bile acids (BAs) metabolism [
1]. BSH catalyzes the first reaction of conjugated BAs hydrolysis, necessary for their biotransformation into deconjugated ones. As suggested, BSH ability is directly implicated in different metabolic pathways, and has an essential role on endogenous cholesterol metabolism [
1,
2,
3,
4]. Recently, BSH activity, encoding BSH proteins, is considered a key trait that supports bacterial survivability and colonization into the gastrointestinal tract (GIT) through BAs detoxification [
5,
6]. Additionally, the presence and expression of
bsh genes in lactobacilli genome offering a high perspective in the comprehension of BSH activity and their impact on BAs metabolism [
7]. Taxonomic study on
bsh gene distribution distinguish different phylotype, highlighting different copy number of the gene across
Lactobacillus genera, as well as the synthesis of several BSH variant with different functional properties [
8,
9,
10]. Given the pivotal impact of BSH activity on bacterial survival gut microbiota,
bsh gene detection in lactobacilli, has been recognized as a functional biomarker for probiotic selection [
8].
More recently, research is focused on the use of predictive methods, often integrated with high-throughput technologies, to quantify and characterize cells and molecules involved in the microbial ecosystem. These strategies are applied to detect functional properties arising from the interaction between probiotic supplementation and the gut microbiota. At this regard,
in vitro studies conducted using the Simulator of the Human Microbial Ecosystem (SHIME®, ProDigest, Belgium) are considered a viable strategy to better understand the effect of potential probiotic strains and to make them a more suitable product for the formulation of dietary and/or pharmaceutical supplements [
11]. The SHIME
® is a GIT model, which mimics the dynamic and physiological processes starting from the stomach until the colonic fractions, in response to a probiotic or prebiotic action [
12,
13]. Therefore, in this work, the upper GIT SHIME
® model extended to the colonic fraction was used to assess the survival and BAs deconjugation capacity of the previously BSH-selected
Lacticaseibacillus rhamnosus VB4 strain. In particular, by combining transcriptomic and metabolomic approaches, we sought to better understand the action of BSH by evaluating a dynamic process of BAs colonization and conjugation, monitoring its passage from the upper part of the GIT to the colonic fraction.
2. Material and Methods
2.1. BSH-Positive L. rhamnosus VB4 Strain
The
Lacticaseibacillus rhamnosus VB4 strain, belonging to the microbial collection of ProBioEtna srl, Spin Off of the University of Catania (Italy), was selected for its probiotic properties and high ability to deconjugate BAs, in accordance with the results obtained in the previous work conducted by Agolino and co-workers [
14]. SACCO Srl, Italy. The VB4 strain was freeze-dried and subjected to microbiological analysis by the plate culture method using Lactobacilli MRS agar medium (BD Difco
TM), to ensure the absence of any contaminants. The freeze-dried strain was directly inoculated into the stomach/small intestine reactors and the starting cell density of the VB4 strain was at 9 log
10 CFU/g.
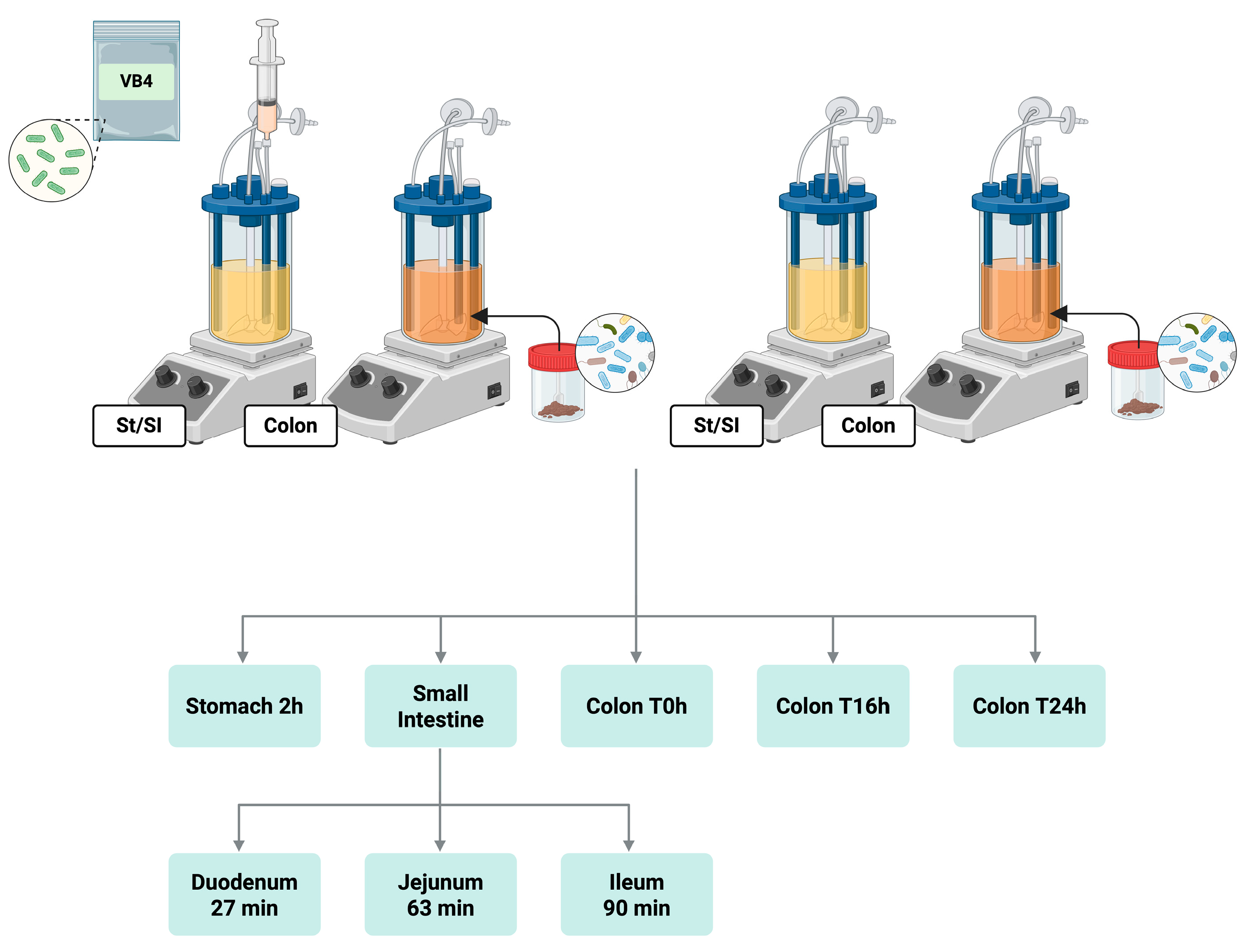
2.2. SHIME® Experiment Set-Up
The dynamic SHIME
® model (ProDigest, Ghent, Belgium) was set up under feeding condition according to the protocols proposed by Marzorati and co-workers [
15] and Jannin and collaborators [
16] to mimic the incubation of stomach, small intestine and colon, using two double-jacketed reactors serially connected, maintained at 37 °C under constant agitation. The lyophilized strain was inoculated with an initial cell density of 9 log
10 CFU/g in the first reactor, simulating the digestion of the stomach and small intestine. In detail, under fed condition, a gastric solution (76 mL), containing SHIME
® nutritional medium (PDNM001B 20.53 g/L, ProDigest, Ghent, Belgium), NaCl (3.63 g/L), KCl (0.65 g/L), 0.4 mL lecithin (13.5 g/L), and 3.6 mL pepsin (40 g/L) at pH 4.6 will be added to the vessel and maintained for 120 min with a sigmoidal pH decrease from 4.6 to 2. After incubation in the stomach, a small intestine phase, comprising duodenum, jejunum, and ileum, will be performed. Specifically, the pH of the small intestine was gradually increased from 2.0 to 6.5 and maintained at this pH over 27 min to simulate the duodenal fraction, followed by jejunal (pH up to 7.5 maintained for 63 min) and ileal (constant pH 7.5 for 90 min) fractions. During the duodenal phase, a pancreatic juice composition (NaHCO
3 7.7 g/L, Oxgall 15 g/L and pancreatin 10 g/L, added with 2.15 mL trypsin 10 g/L and 2.7 mL chymotrypsin 10 g/L) will be added. An increase in pH was achieved by the addition of NaHCO
3 (4.8 g/L) at 60, 90 and 120 min, mimicking the dilution of the intestinal contents. The increase and decrease in pH were automatically controlled, by pH-meter probe (ProSense, Oosterhout, The Netherlands), and adjusted by the dosage of HCl (0.5 M) and NaOH (0.5 M).
2.3. Fecal Slurry Preparation and Colonic Incubation
An extension of the colonic incubation was further proposed. The digested solution from the stomach/small intestine reactor was transferred to the second reactor, simulating colonic conditions, and maintained for 24 hours. A fecal sample from a healthy donor (male, 50y) was chose for the SHIME
® experiment. The collection of data from the use of human fecal sample were approved by the Ethics Committee of the University of Modena and Reggio Emilia (CEAR) on 20.01.2025, and informed consent for the experimentation was obtained from the subject involved in the study. The fecal slurry was obtained according to the protocol proposed by Marzorati and collaborators [
15] and inoculated into the colonic reactor. In detail, a 1:10 (w/v) mixture of fecal sample and anaerobic phosphate buffer (K
2HPO
4 8.8 g/L; KH
2PO
4 6.8 g/L; sodium thioglycolate 0.1 g/L; sodium dithionite 0.015 g/L) will be homogenized for 10 min (BagMixer 400, Interscience, Louvain-LaNeuve, Belgium), and the mixture was centrifuged (2 min, 500 g) and the large particles removed. The obtained fecal slurry (20% v/v) will be added to 160 mL fresh colonic anaerobic medium [KH
2PO
4 (6.6 g/L), K
2HPO
4 (20.5 g/L), NaCl (5 g/L), yeast extract (2 g/L), peptone (2 g/L), glucose (1 g/L), starch (2 g/L), mucin (1 g/L), L-cysteine HCl (0.5 g/L), Tween® 80 (2 mL)], 40 mL of anaerobic PBS [K
2HPO
4 (8.8 g/L), KH
2PO
4 (6.4 g/L), NaCl (8.5 g/L) and L-cysteine HCl (0.5 g/L)]. In the colonic compartment, a fixed pH range between 6.5 and 5.8 was implemented, adjusted automatically, maintaining an anaerobic condition at 37 °C and an agitation of 90 rpm. A reactor without the addition of probiotic strain was used as a control sample, following all of the steps of the digestion, from the stomach to the colon compartments. All the SHIME
® assays were performed in duplicate. In the stomach/small intestine reactor, samples were collected at the end of the incubation phases of stomach, duodenum, jejunum and ileum, and subjected to culture-dependent analysis and culture-independent. While in the colonic fraction, lumen samples were collected after 0, 16 and 24 hours of colonic incubation. All the analysis were assessed in triplicate. A schematic representation of the experimental workflow is shown in
Figure 1.
2.4. Detection of Viability by Culture-Dependent Method
The viability of L. rhamnosus VB4 strain was evaluated during GIT passage, from stomach to ileal phases by plate count. Briefly, the bacterial count was performed by plating serial ten-fold dilution using the anaerobic PBS, containing 8.8 g/L of K2HPO4, 6.4 g/L of KH2PO4, 8.5 g/L of NaCl, and 0.5 g/L of L-cysteine HCl, then cultured on Lactobacilli MRS Agar medium (BD Difco™, Italy) and incubated under anaerobic conditions at 37 °C for 48 hours. Colony counts were performed on plates yielding between 30 and 300 colonies. The plate count assays were performed in triplicate and results were reported as mean values log10 CFU/mL and standard deviation.
2.5. Detection of Gene Expression of BSH-Positive VB4 Strain
2.5.1. RNA Extraction
All samples collected during the simulation with the SHIME model were subjected to RNA extraction. Specifically, 2 mL of stomach/intestinal small intestine and lumen samples and 0.3 g of mucosal samples were centrifuged at 20,000 × g for 10 min, washed twice with anaerobic PBS and subjected to ZymoBIOMICSTM RNA Miniprep Kit (Zymo Research, Orange, CA, USA), with prior mechanical breakage with Precellys Evolution Homogenizer (Bertin Technologies) at 10,000 rpm for 2 min, repeated three times, interspersed with breaks on ice. The concentration, integrity and quality of RNA templates were checked using the QubitTM 4 Fluorometer (Thermo Fisher Scientific, San Jose, CA, USA).
2.5.2. RT-PCR and RT-qPCR Assays
The RNA templates were subjected to reverse transcription PCR (RT-PCR) analysis using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The complementary DNA (cDNA) obtained was subjected to RT-qPCR with a primer pair designed according to Agolino and collaborators [
14]. In detail, the reaction included 10 µL of QuantiNovaTM SYBR Green PCR Kit (Qiagen), 0.7 µM of each primer Bsh_rha_qF2 (GGAATACGGGTGCATACAA) and Bsh_rha_qR2 (CAGGCCAAACATGCCATAAC), 3 µL of cDNA templating and 4.2 µL of Dnase/Rnase-free water. Cycling conditions comprised a holding phase at 95 °C for 2 minutes, followed by 40 cycles at 95 °C for 5 seconds, 60 °C for 10 seconds and 60 °C for 30 seconds. The melting range was set between 60 °C and 95 °C. In addition, RT-qPCR was also performed for the 16S rRNA housekeeping (
HKG) gene to allow for the normalization of gene expression data. The reaction mixture and reagent concentrations included 10 µL of QuantiNovaTM SYBR Green PCR Kit (Qiagen), 0.7 µM of each primer 16S_F1 (GTAGGTGGCAAGCGTTATCC), and 16S_R1 (GATGCGCTTCCTCGGTTAAG). The qPCR cycling parameters including a holding phase at 95 °C for 2 min, followed by 40 cycles at 95 °C for 5 sec and 60 °C for 60 sec. The melting range was set from 60 °C to 95 °C. Specificity and amplification efficiency (E) were performed using a 10x dilution of the microbial gDNA of
L. rhamnosus VB4 as a reference standard. Slope of the regression curve between the logical values of the cDNA concentrations and the mean values of the cycle threshold (Ct) was used to calculate the primer efficiency using the equation: E= 0.5 (10 (-1/slope)) 100. A Rotor Gene Q instrument (Qiagen, Milan, Italy) was used to perform the reaction. Each reaction was repeated at least three times.
2.6. Detection of BAs from SHIME Samples
The semi-quantitative analysis of individual BAs in samples from SHIME
® model (both in control and VB4-inoculated samples) was carried out as reported in Agolino and collaborators [
14] by using a high-resolution mass spectrometer. Chromatographic separation was performed with a UHPLC Ultimate 3000 module (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a C18 column (Acquity UPLC HSS C18 Reversed phase, 2.1 × 100 mm, 1.8 µm particle size, Waters, Milan, Italy). Mass spectrometry analysis was carried out through a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Mobile phases were water with 0.1% formic acid (mobile phase A) and acetonitrile with 0.1% formic acid (mobile phase B) and the flow rate was 0.3 mL/min. An amount of 10 μL of appropriately diluted sample was injected. The chromatographic and mass spectrometry parameters were fully described in Agolino et al. [
14]. The relative amount of bile acids was determined by integrating the area under the peak (AUP). AUP values were quantified from the extracted ion chromatograms (EIC) calculated for each mass-to-charge ratio of a specific compound (tolerance ± 5 ppm) using the Genesis algorithm function in the Thermo Xcalibur Quantitative Browser.
2.7. Statistical Analysis
All the analyses were performed in triplicate, and data are presented as mean values ± standard deviation (SD). Statistical analyses and graph generation were conducted using GraphPad Prism 10 (GraphPad Software, La Jolla, CA, USA). Microbiological counts and gene expression were analyzed using ANOVA One-way, by using Tukey’s post-hoc test. Whereas the BAs profile was assessed using an ANOVA Two-way. Followed by Dunnett’s post-hoc. Statistical significance was set at p-value < 0.05.
3. Results
3.1. L. rhamnosus VB4 Strain Viability in Stomach/Small Intestine
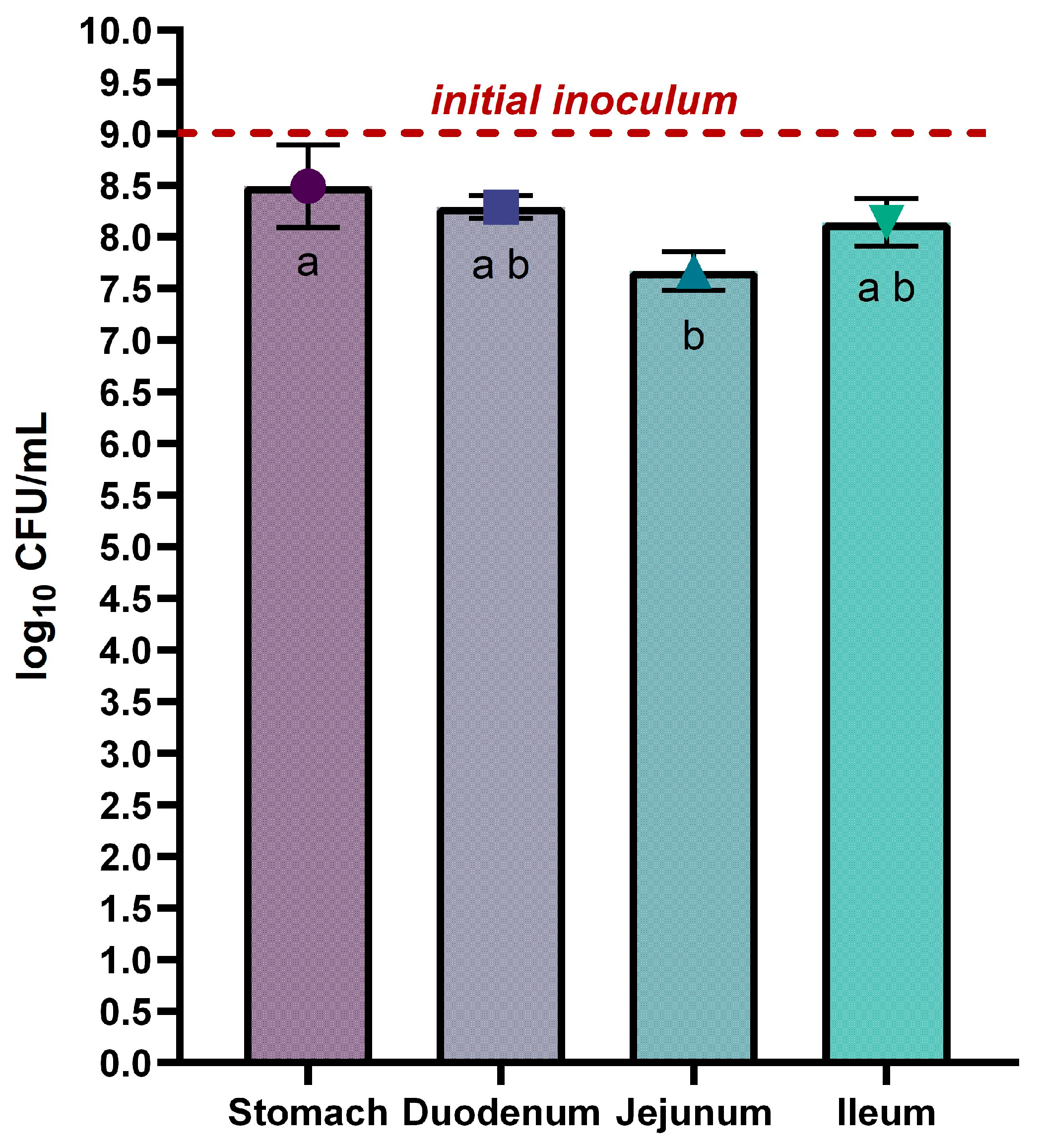
The L. rhamnosus VB4 strain viability in the stomach and small intestine was achieved by plate count. Overall, not substantial differences were detected between the stomach and the small intestine (p > 0.05). In detail, the strain exhibited a slight reduction during the final gastric phase, decreasing from the initial inoculum (9 log10 CFU/mL) to 8.49 ± 0.40 log10 CFU/mL, mainly due to the acidic condition and gastric enzymes of the stomach environment. During the small intestine transit, a progressive decrease of the VB4 cell density was observed from duodenum to jejunum, with values of 8.29 ± 0.11 log10 CFU/mL and 7.67 ± 0.19 log10 CFU/mL, respectively. A slight increase in the ileum with a cell density of 8.14 ± 0.23 log10 CFU/mL was achieved.
Figure 2.
Survival of Lacticaseibacillus rhamnosus VB4 in Stomach/Small Intestine compartments. Viable counts were enumerated on agar plate. Data are expressed as average ± SD of the three independent biological replicates. Results are expressed as mean ± SD of log10 CFU/mL. The horizontal red line indicates the starting cell density of the VB4 strain (9 log10 unit). a–b Different superscript letters indicate significant differences within samples at different GI fractions (at p < 0.05).
Figure 2.
Survival of Lacticaseibacillus rhamnosus VB4 in Stomach/Small Intestine compartments. Viable counts were enumerated on agar plate. Data are expressed as average ± SD of the three independent biological replicates. Results are expressed as mean ± SD of log10 CFU/mL. The horizontal red line indicates the starting cell density of the VB4 strain (9 log10 unit). a–b Different superscript letters indicate significant differences within samples at different GI fractions (at p < 0.05).
3.2. Bsh Gene Detection Through qPCR Analysis
For the evaluation of the transcriptional profile of the
bsh gene in different gastrointestinal compartments, gene expression quantification in both upper gastrointestinal tract and colonic fractions was assessed. The raw qPCR results expressed in ng/µL were converted into absolute copy numbers using an equation based on Avogadro's constant, the genome length of the reference strain (
L. rhamnosus VB4) and the intercept of the standard curve. In addition, the housekeeping (
HKG) gene was also quantified for a more accurate estimate of gene expression capacity.
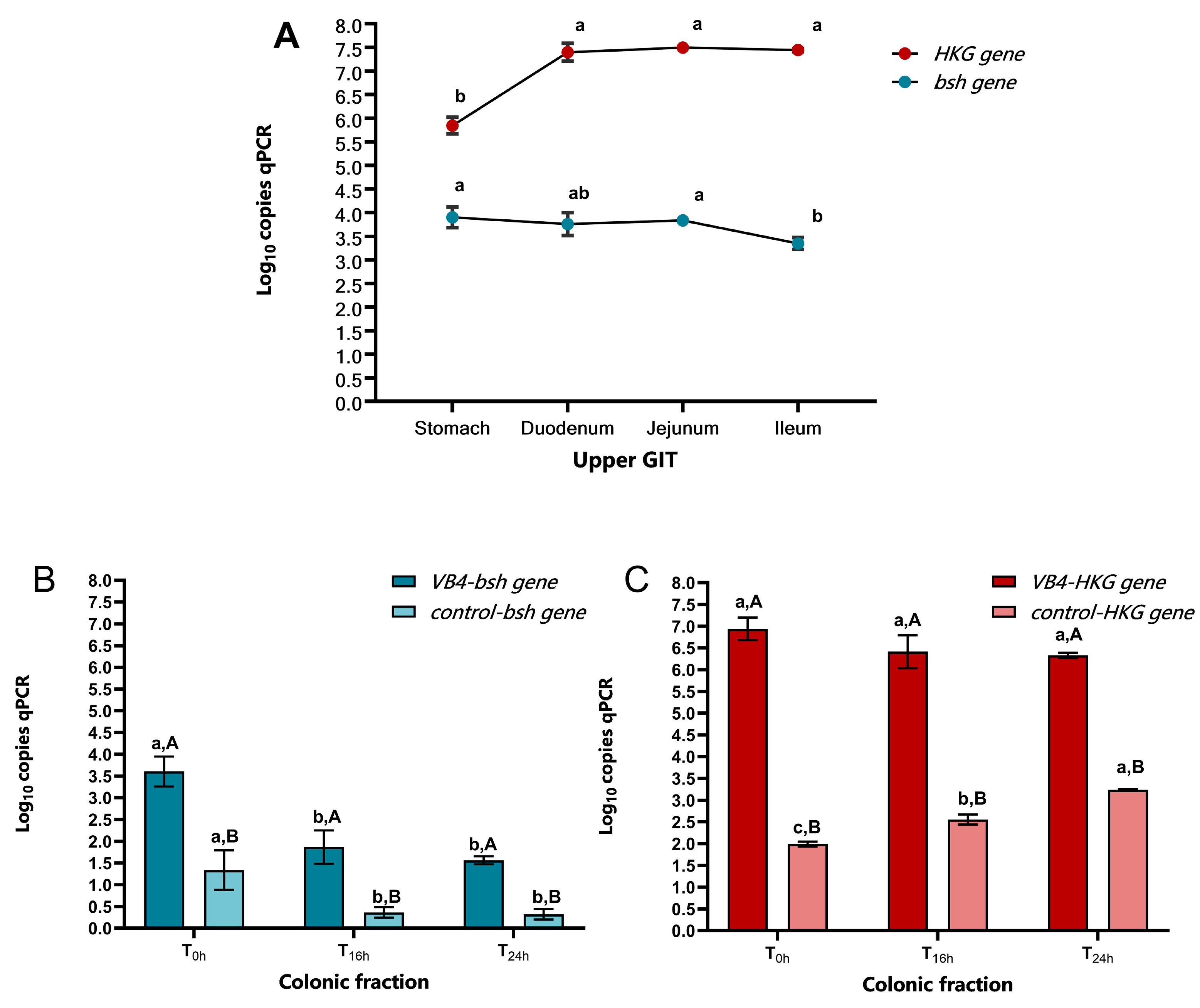
Figure 3 shows the data obtained from the transcriptional test. Specifically,
Figure 3A shows significant differences in the
bsh gene expression profile between the stomach and ileum phases, with values of 3.90 and 3.35 log
10 copies qPCR, respectively, and between the jejunum and ileum phases, with a value of 3.83 log
10 copies qPCR. Conversely, the
HKG gene was lower in the stomach phase with a value of 5.84 log
10 copies qPCR, which was significantly different from the duodenum (7.40 log
10 copies qPCR), jejunum (7.44 log
10 copies qPCR) and ileum (7.50 log
10 qPCR copies), which showed a slight increase towards the end of the small intestine.
In
Figure 3B the transcriptional profile of
bsh gene of VB4-inoculated sample in colonic fraction showed a slight decrease of expression from T
0h to T
24h, with significant differences of T
16h (1.87 log
10 copies qPCR) and T
24h (1.56 log
10 copies qPCR) compared to T
0h (3.61 log
10 copies qPCR). Similar trend was showed in the control sample, where T
0h with value of 1.34 log
10 copies qPCR resulted significant different compared to T
16h and T
24h with 0.36 and 0.32 log
10 copies qPCR, respectively. At the same sampling time,
bsh gene expression resulted more evident in VB4-inoculated sample compared to control one.
For the
HKG gene expression (
Figure 3C), no significant different were found between VB4-inoculated sample at the sampling times, with slightly decrease from T
0h to T
24h, with 6.94 to 6.33 log
10 copies qPCR, respectively. Whereas in the control samples a significant difference was observed at all sampling times, with an increase from T
0h to T
24h, with 1.99 and 3.24 log
10 copies qPCR, respectively. At the same sampling time,
HKG gene expression confirmed the higher transcriptional ability of VB4-inoculated compared to control samples.
3.3. Semi-Quantitative Analysis of BAs in Small Intestine and Colon
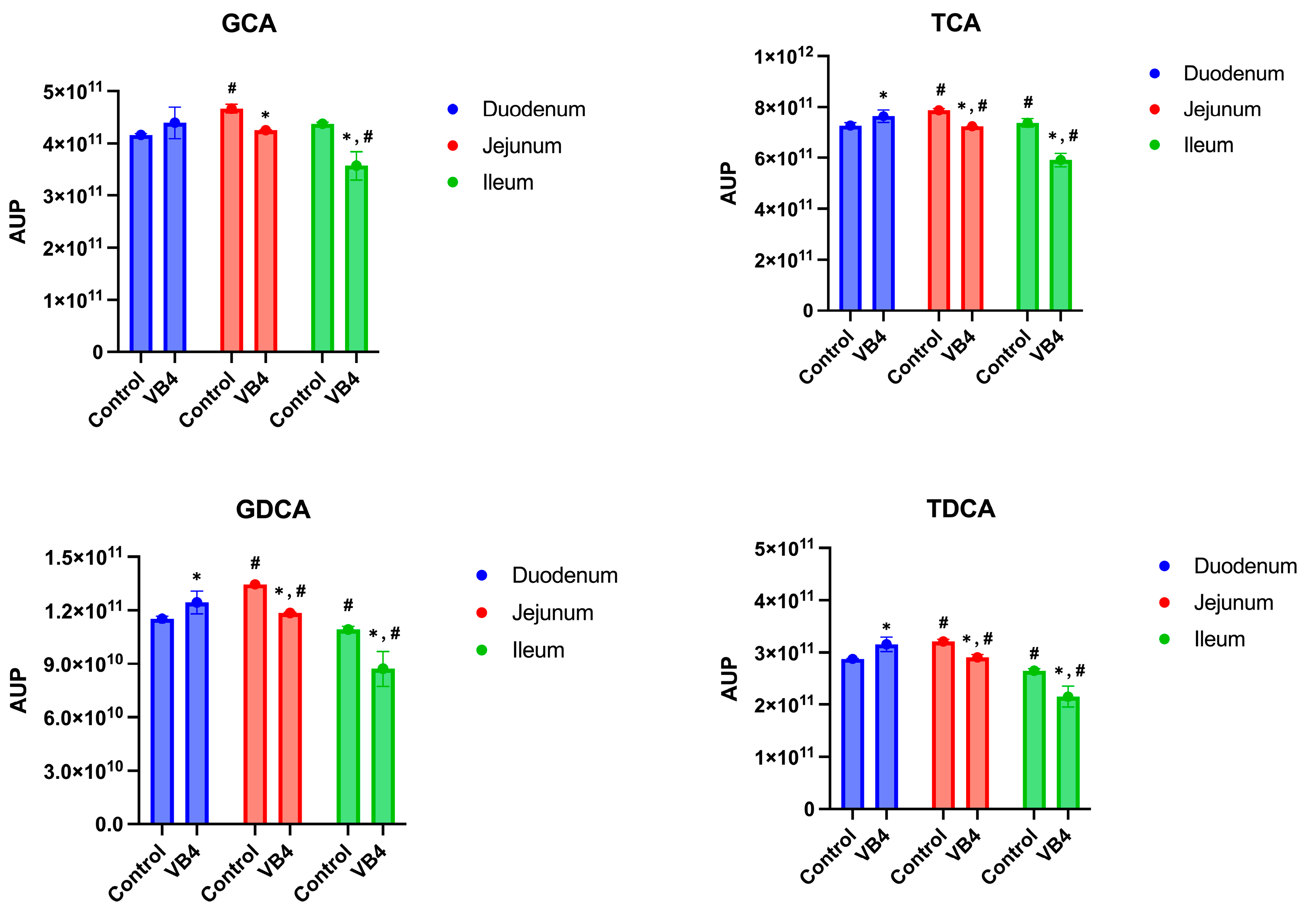
In order to test whether the inoculum of L. rhamnosus VB4 strain changed BAs profile, the amount of four conjugated BAs (taurocholic, glycocholic, taurodeoxycholic, and glycodeoxycholic acids), were detected by high-resolution mass spectrometry during small intestine and colon digestion.
The amount of the conjugated BAs increased from the duodenum to the jejunum in the control vessels (
Figure 4). Differently, a significant decrease was detected among the VB4-inoculated samples (
p < 0.05), with the exception of the GCA sample, which remained stable (
Figure 4). The amount of GCA was not affected after passing into the ileum, whereas a significant (
p < 0.05) decrease in the amount of the other conjugated acids was observed (
Figure 4) in control sample, while significant decrease (
p < 0.05) was showed for the VB4-inoculated sample (
Figure 4).
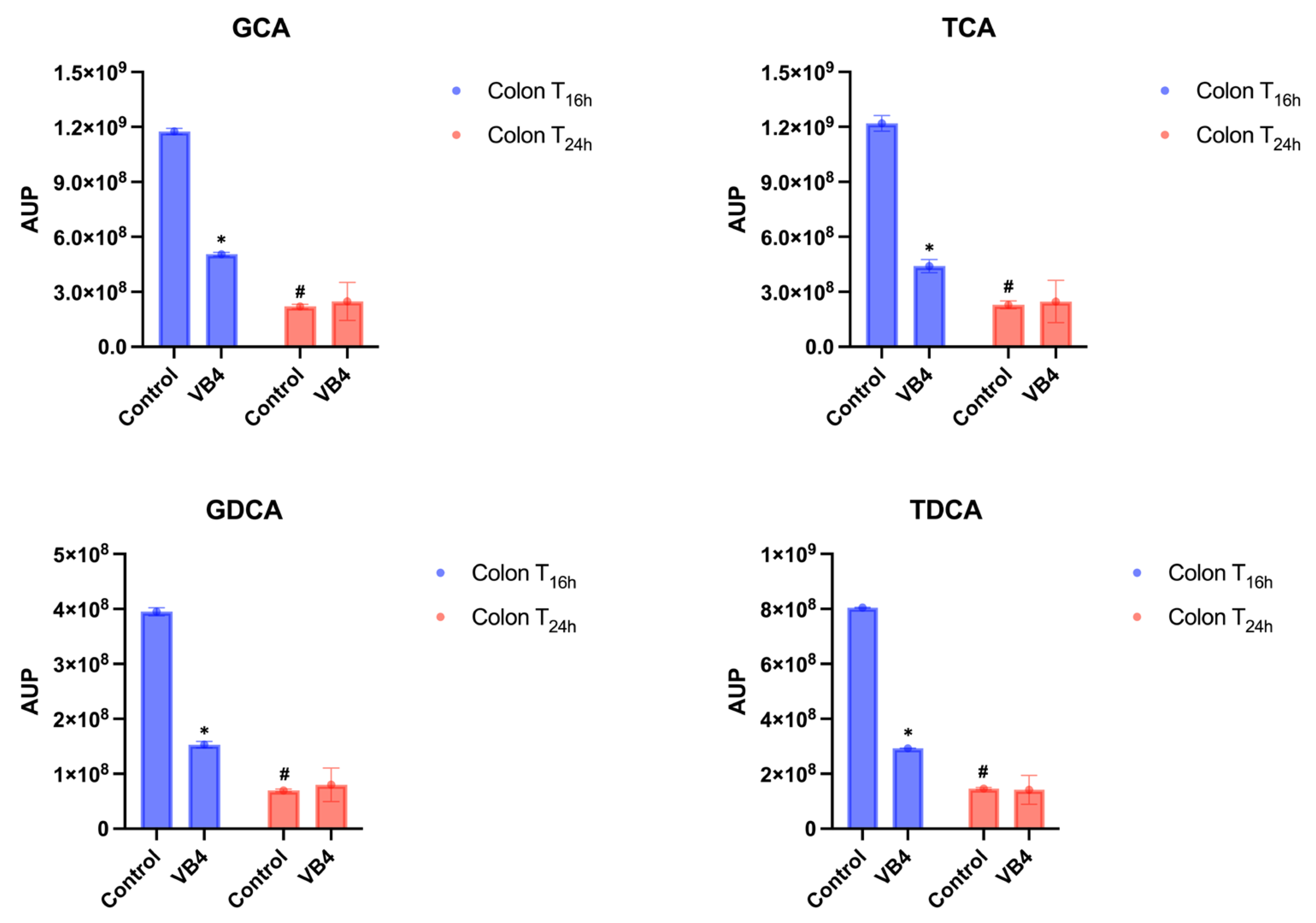
As expected, a decline of 2/3 orders of magnitude from ileum to colonic fraction were observed in both control and VB4-inoculates samples, probably due to the physiological dilution of digested during the transit in the colonic fraction, reaching a value of approximately of 10
9-10
8 and AUP in the colon fraction, after 16 h (T
16h) of incubation (
Figure 5). It is interesting to highlight that a more pronounced decrease of Bas was revealed in the VB4-inoculated samples, compared to the control. In the T
24h colon sample, no significant differences (
p < 0.05) between the control and the inoculated conjugated BAs were found (
Figure 5). Otherwise, in VB4-inoculated reactor, the amount of conjugated BAs significantly decreased (
p < 0.05) in T
24h colon samples compared with T
16h (
Figure 5).
4. Discussion
Bile salt hydrolase (BSH) activity is a key probiotic function, particularly in
Lactobacillus species, as it promotes gastrointestinal survival, modulates bile acid metabolism, and contributes to cholesterol reduction and cardiovascular health [
17,
18]. In the present study, BSH activity of the
L. rhamnosus VB4 strain was assessed using the upper GIT SHIME® model, under FED condition, providing a more realistic physiological state in which BSH activity can be optimally expressed as a probiotic feature. Furthermore, to elucidate the behavior of the VB4 administration under the influence of the gut microbial community, which represents a natural reservoir of BSH-positive bacteria in healthy individual [
19], the experiment was extended to colonic fraction.
The strain survivability trough the GIT passage was revealed by culture-dependent analysis and
bsh gene expression profile was detected by qPCR. Across the simulated GI transit, the VB4 strain exhibited high resilience, with a modest loss of around 1 log unit, from the stomach to the ileum-end in FED condition. A similar upper GIT SHIME experiment was conducted by Govaert and collaborators [
20], in which a freeze-dried probiotic formulation, inoculated at log
10 CFU/g under FASTED (probiotic administration before a meal) condition, preserved high viability from the stomach to the ileal phases, showing around 2 log reduction in cell density at the final stage. According to that, previous studies have highlighted that exposure to gastric juice can drastically reduce probiotic survival. These findings, figure out the ability of the probiotic strain to adapt to gastric pH and enzymes, as well as to bile salts and pancreatic enzyme, which represent an additional critical factor to be overcome.
According to transcriptional data, bsh gene expression showed significant variations in several compartments of the gastrointestinal tract. In particular, when inoculated, L. rhamnosus VB4 showed higher gene expression levels in the stomach and a gradual decline towards the duodenal, jejunal and ileal phases.
Conversely as observed in literature [
21], that
bsh gene expression is inducible in conditions of bile richness, in this study it is clear to see that its maximum expression is already present and detectable in the gastric compartment and then attenuates with the decrease in bile salt concentrations along the small intestine. This could confirm the constitutively expression of the
bsh gene. In addition,
bsh gene was further detected in the colon compartment up to 24 h of incubation. Despite the
bsh expression related to endogenous commensal microbiota, VB4-inoculated samples displayed sustained
bsh expression compared to the control sample, suggesting an overtime contribution of VB4 strain in enhancing deconjugation of BAs compared to the control.
Scientific evidence has revealed a close relationship between BAs deconjugation ability and
bsh gene expression [
9,
22]. According to that, in this study the impact of the BAs profile after VB4 administration by detecting the main human glyco- and tauro-conjugated acids was demonstrated.
In physiological condition, BAs deconjugation starting in the small intestine with maximal activity occurring in the ileum and in the proximal colon [
23,
24]. On this basis, our study showed a progressive decrease in each detected conjugated BAs from the duodenum to the jejunum phases after VB4 administration, followed by a substantial decrease after the ileal phase. These results could confirm the highest deconjugation potential at the end of the small intestine. VB4 administration, in the colonic compartment after 16 h of incubation (T
16h), significantly affected the deconjugation of each conjugated BAs, compared to the control, suggesting an enhanced BSH potential of the microbiota, induced by strain supplementation. However, at T
24h, although the further reduction, VB4 did not alter BAs profile in the colonic fraction, except for a slight increase in GDCA (compared to the control). These finding suggests that, at this stage, VB4 starting to loss their BSH potential, mainly expressed in the proximal colon stages, as described to Ridlon and co-worker [
25] in their review article which described the BAs metabolism.
5. Conclusions
This study provides novel insight into the dynamic interplay between the BSH-positve L. rhamnosus VB4 strain and BAs metabolism during GIT. The obtained results demonstrate that VB4 exhibit robust gastrointestinal survivability and maintain transcriptional activity of the bsh gene, indicating its resilience during the harsh condition of the digestive process. The modulation of the BAs profile, characterized by a marked reduction of conjugated BAs in the ileal phase, supports the strain’s active deconjugation potential. These finding contribute to the growing body of evidence to support the use of BSH-positive probiotics shaping host BAs pools. Combining untargeted metabolomics with dynamic gut models offers a valuable approach to study the probiotic influence in BAs biotransformation’s, posing a new possibility for developing a personalized probiotic treatment in restoring gut microbiota and BAs dysregulation. These approaches will enable future studies to be conducted focusing on the effect of a daily administration of BSH-positive strains on the gut microbiota, using hypercholesterolemic subjects, in order to clarify the positive and negative effects of BSH probiotic activity on host physiology.
Author Contributions
Writing—original draft preparation, A.V, G.A., and A.C.; writing—review and editing, A.P., D.T., L.S., C.C. and C.L.R.; data curation, A.V., G.A., A.P., M.C., and A.C.; formal analysis, A.V, G.A., M.C., D.T., and A.C..; investigation, A.P., D.T., C.C., L.S. and C.L.R.; methodology, A.P., D.T., L.S. and C.L.R.; conceptualization, L.S., C.C., and C.L.R.; visualization, A.V, A.P, D.T., A.C., and C.L.R; and funding acquisition, A.P., L.S. and C.L.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Fecal samples of the donor were collected according to the ethical approval of approved by the Ethics Committee of the University of Modena and Reggio Emilia (CEAR) on 20.01.2025 related to the project “LEVANTE - Analisi probiogenomica e caratterizzazione funzionale di nuovi candidate lattobacillari probiotici con attività idrolasica degli acidi biliari (BSH)” (num. Prot. 332033).
Acknowledgments
The authors acknowledge the project PIAno di inCEntivi per la RIcerca di Ateneo 2020/2022—Linea di Intervento 3 “Starting Grant”. Project Title “Selezione di probiotici con attività idrolasica dei sali biliari (BSH) per la salute umana”, ProBSH. The project was partially supported by FAR2024 (University of Modena and Reggio Emilia) and by the NRRP, Mission 4 Component 2 Investment 1.4 – Call for tender No. 3138 of 16 December 2021, rectified by Decree n. 3175 of 18 December 2021 of the Italian Ministry of University and Research funded by the European Union – NextGenerationEU. Project Code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, CUP E93C22001090001, “National Biodiversity Future Center – NBFC’’.
Conflicts of Interest
The authors Alessandra Pino, Cinzia Caggia and Cinzia Lucia Randazzo are founder of ProBioEtna a spinoff of the University of Catania. They have two affiliations but the employer is the University of Catania. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Kumar, M.; Nagpal, R.; Kumar, R.; Hemalatha, R.; Verma, V.; Kumar, A.; Chakraborty, C.; Singh, B.; Marotta, F.; Jain, S.; Yadav, H. Cholesterol-Lowering Probiotics as Potential Biotherapeutics for Metabolic Diseases. Exp. Diabetes Res. 2012, 2012, 902917. [Google Scholar] [CrossRef]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G. Regulation of Host Weight Gain and Lipid Metabolism by Bacterial Bile Acid Modification in the Gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef]
- Agolino, G.; Pino, A.; Vaccalluzzo, A.; Cristofolini, M.; Solieri, L.; Caggia, C.; Randazzo, C.L. Bile Salt Hydrolase: The Complexity behind Its Mechanism in Relation to Lowering-Cholesterol Lactobacilli Probiotics. J. Funct. Foods 2024, 120, 106357. [Google Scholar] [CrossRef]
- Deng, C.; Pan, J.; Zhu, H.; Chen, Z.Y. Effect of Gut Microbiota on Blood Cholesterol: A Review on Mechanisms. Foods 2023, 12, 4308. [Google Scholar] [CrossRef] [PubMed]
- De Boever, P.; Wouters, R.; Verschaeve, L.; Berckmans, P.; Schoeters, G.; Verstraete, W. Protective Effect of the Bile Salt Hydrolase-Active Lactobacillus reuteri against Bile Salt Cytotoxicity. Appl. Microbiol. Biotechnol. 2000, 53, 709–714. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y. Bile Acids as Metabolic Regulators. Curr. Opin. Gastroenterol. 2015, 31, 159–165. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Briner Crawley, A.; Theriot, C.M.; Barrangou, R. The Lactobacillus Bile Salt Hydrolase Repertoire Reveals Niche-Specific Adaptation. mSphere 2018, 3, e00108-18. [Google Scholar] [CrossRef]
- Kusada, H.; Morinaga, K.; Tamaki, H. Identification of Bile Salt Hydrolase and Bile Salt Resistance in a Probiotic Bacterium Lactobacillus gasseri JCM1131T. Microorganisms 2021, 9, 1011. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Li, J. Taxonomic Profiling and Populational Patterns of Bacterial Bile Salt Hydrolase (BSH) Genes Based on Worldwide Human Gut Microbiome. Microbiome 2019, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Feng, S.; Zhou, X.; Song, Z.; Li, J.; Li, P. Taxonomic Identification of Bile Salt Hydrolase-Encoding Lactobacilli: Modulation of the Enterohepatic Bile Acid Profile. iMeta 2023, 2, e128. [Google Scholar] [CrossRef]
- Marzorati, M. SHIME®: An Advanced In Vitro Technology Platform for Studying the Mode-of-Action of Probiotics in the Gastrointestinal Tract. Nutrafoods 2018, 17, 213–217. [Google Scholar]
- Rudzka, A.; Patloka, O.N.D.; Plecha, M.; Królikowski, T.; Oczkowski, M.; Zborowski, M.; Kołożyn-Krajewska, D.; Zielinska, D. Changes in the Microbiome of a Human and in the Simulator of Human Intestinal Microbial Ecosystem (SHIME®) in Response to a Diet and Probiotic Supplementation. Żywność Nauka Technologia Jakość 2023, 30, 121–136. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Mackie, T.A., Eds.; Springer: Cham, Switzerland, 2015. [Google Scholar] [CrossRef]
- Agolino, G.; Cristofolini, M.; Vaccalluzzo, A.; Tagliazucchi, D.; Cattivelli, A.; Pino, A.; Caggia, C.; Solieri, L.; Randazzo, C.L. Genome Mining and Characterization of Two Novel Lacticaseibacillus rhamnosus Probiotic Candidates with Bile Salt Hydrolase Activity. Biomolecules 2025, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Marzorati, M.; Calatayud, M.; Rotsaert, C.; Van Mele, M.; Duysburgh, C.; Durkee, S.; White, T.; Fowler, K.; Jannin, V.; Bellamine, A. Comparison of Protection and Release Behavior of Different Capsule Polymer Combinations Based on L. acidophilus Survivability and Function and Caffeine Release. Int. J. Pharm. 2021, 607, 120977. [Google Scholar] [CrossRef] [PubMed]
- Jannin, V.; Duysburgh, C.; Gonzalez, V.; Govaert, M.; Agisson, M.; Marzorati, M.; Madit, N. In Vitro Evaluation of the Gastrointestinal Delivery of Acid-Sensitive Pancrelipase in a Next Generation Enteric Capsule Using an Exocrine Pancreatic Insufficiency Disease Model. Int. J. Pharm. 2023, 630, 122441. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Allen, G.; Rivera, A.J.; Stewart, A.K.; Barrangou, R.; Theriot, C.M. Lactobacillus Bile Salt Hydrolase Substrate Specificity Governs Bacterial Fitness and Host Colonization. Proc. Natl. Acad. Sci. USA 2021, 118, e2017709118. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.; Zhou, S.; Huang, L.; Chen, Y.; Huan, H. Bile Salt Hydrolase Can Improve Lactobacillus plantarum Survival in Gastrointestinal Tract by Enhancing Their Adhesion Ability. FEMS Microbiol. Lett. 2019, 366, fnz100. [Google Scholar] [CrossRef]
- Bourgin, M.; Kriaa, A.; Mkaouar, H.; Mariaule, V.; Jablaoui, A.; Maguin, E.; Rhimi, M. Bile Salt Hydrolases: At the Crossroads of Microbiota and Human Health. Microorganisms 2021, 9, 1122. [Google Scholar] [CrossRef]
- Govaert, M.; Rotsaert, C.; Vannieuwenhuyse, C.; Duysburgh, C.; Medlin, S.; Marzorati, M.; Jarrett, H. Survival of Probiotic Bacterial Cells in the Upper Gastrointestinal Tract and the Effect of the Surviving Population on the Colonic Microbial Community Activity and Composition. Nutrients 2024, 16, 2791. [Google Scholar] [CrossRef]
- Duary, R.B.; Shriram, V.; Batish, V.K.; Grover, S. Bile-Salt Hydrolase Is an Inducible Activity in Lactobacillus, and bsh Gene Expression in L. plantarum Increased Sixfold after Exposure to 2% Bile. Microb. Cell Fact. 2012, 11, 153. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, W.; Dong, W.; Chen, G.; Sun, Y.; Zeng, X. Anti-Diabetic Effect of Dicaffeoylquinic Acids Is Associated with the Modulation of Gut Microbiota and Bile Acid Metabolism. J. Adv. Res. 2025, 72, 17–35. [Google Scholar] [CrossRef]
- Deyaert, S.; Moens, F.; Pirovano, W.; van den Bogert, B.; Klaassens, E.S.; Marzorati, M.; Van den Abbeele, P. Development of a Reproducible Small Intestinal Microbiota Model and Its Integration into the SHIME®-System, a Dynamic In Vitro Gut Model. Front. Microbiol. 2023, 13, 1054061. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, R.M.; Cariello, M.; Crudele, L.; Moschetta, A. Bile Salt Hydrolase-Competent Probiotics in the Management of IBD: Unlocking the “Bile Acid Code”. Nutrients 2022, 14, 3212. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).