Submitted:
07 December 2023
Posted:
07 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
- Bacterial strain and culture conditions
- 2.
- Genome Sequencing and Annotation
- 3.
- Evolutionary analysis
- 4.
- Nucleotide sequence accession number
- 5.
- In-vitro evaluation of probiotic properties of MCC0200
- 6.
- Antioxidant activity
- 7.
- In-vitro Evaluation of the Anti-hypercholesterolemic Effect of MCC0200
- 8.
- Screening of MCC0200 for beta-galactosidase production
- 9.
- Safety Assessment
3. Results and Discussion
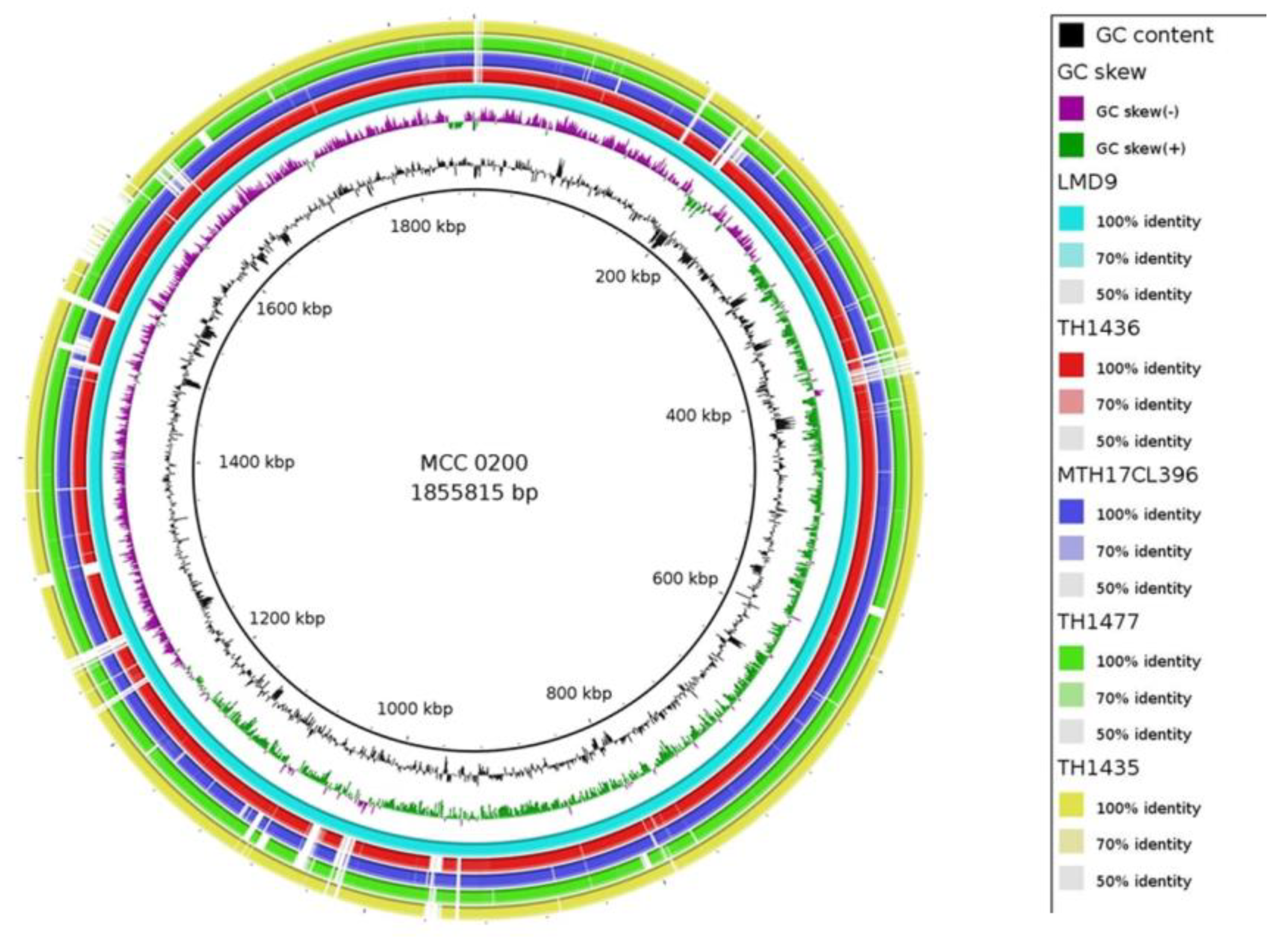
3.1. Genome attributes of S. thermophilus MCC0200:
3.2. Evolutionary analysis and comparison of MCC0200 with other S. thermophilus strains:
3.3. Assessment of Probiotic Properties
3.3.1. Resistance to gastric conditions
- F1–F0 ATPases/ ATP synthases: This multi-subunit enzyme actively pumps protons out of cells to maintain a relatively neutral intracellular pH. The F1 protein is responsible for catalyzing intracellular ATP hydrolysis or synthesis, while the F0 protein plays a crucial role in proton translocation. Bacteria employ these mechanisms to regulate cytoplasmic pH efficiently, utilizing ATP hydrolysis to pump H+ out of cells. This process helps maintain pH homeostasis, protecting cells from damage induced by acidic environments. Studies on S. thermophilus LMD-9 have revealed the involvement of proton translocating F0F1-ATPase system in response mechanism to acid stress [36].
- Na+/H+ antiporters: These membrane proteins contribute to cytoplasmic pH homeostasis by actively effluxing Na+ using the electrochemical gradient of protons generated across the cell membrane by specific transporters, such as ion-pumping ATPases [37].
- Urease system: This system is commonly employed by LAB under acid stress conditions. The urease system produces NH3 and CO2 from urea, providing protection against acid stress. This system has been extensively studied in S. thermophilus, and S. salivarius [38]. In congruence with the above study, genes encoding ureI, structural (ureABC) and accessory (ureEFGD) genes were detected in MCC0200 genome, indicating a probable mechanism for acid tolerance.
- Protection of macromolecules: Specific proteins induced by acid stress play a crucial role in protecting or repairing macromolecules like DNA and proteins.
- Chaperones, including DnaK, DnaJ, GrpE, HrcA, GroEL, GroES, Clp proteases, and EF-Tu, are known to facilitate protein repair during acid stress.
- recA participates in DNA recombinational repair alongside RecN and AddAB (exonuclease V). The nucleotide excision repair system functions on damaged DNA resulting from base modification, single-strand breaks, and abasic sites, making it a critical DNA repair mechanism.
- UvrABCD, DNA polymerase, and DNA ligase: These components actively support the repair of acid-induced DNA damage by performing functions such as damage recognition, base excision, and gap filling.
3.3.2. Adhesion potential of MCC0200
3.3.2.1. Supplemental assays for evaluating bacterial adhesion
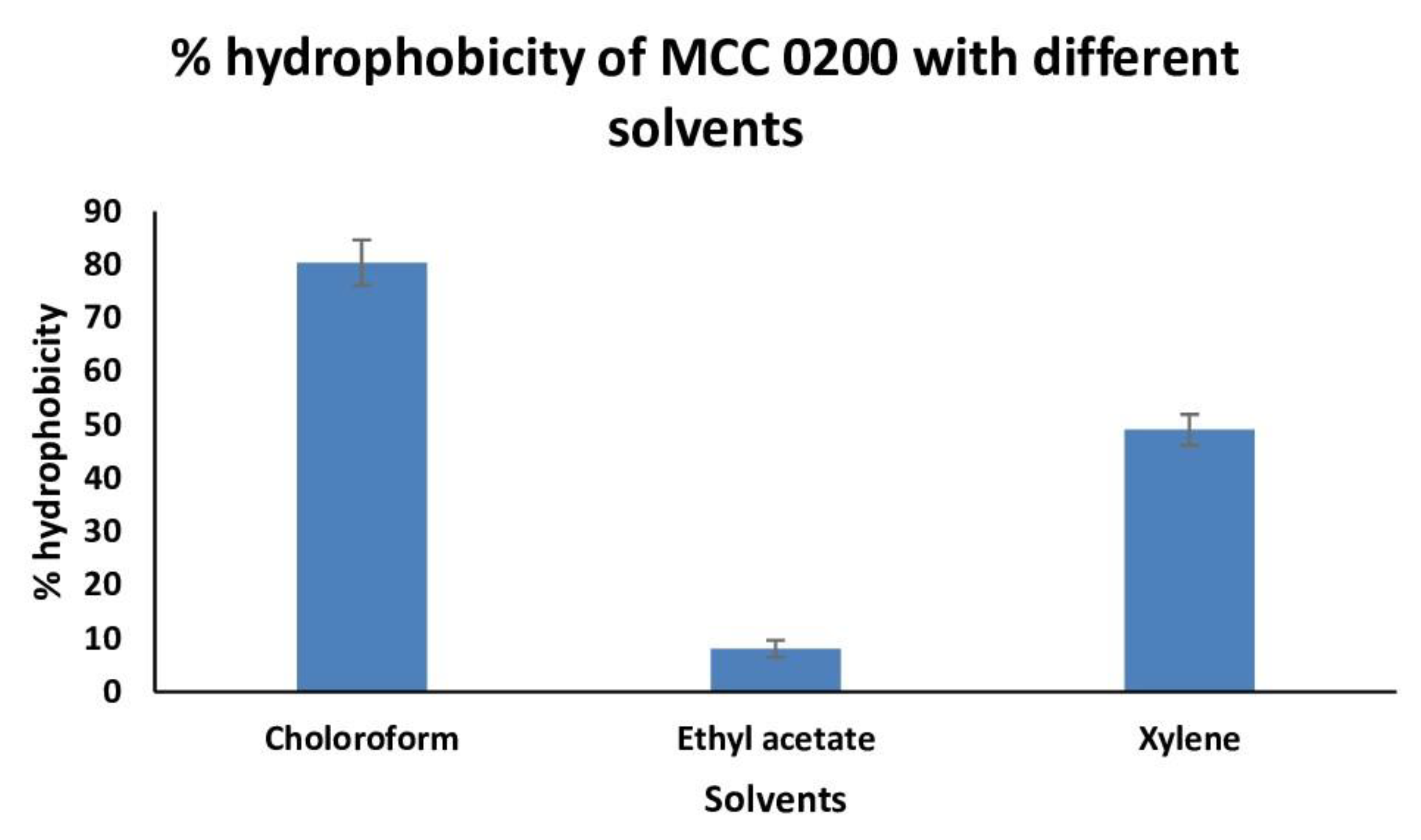
3.3.2.1.1. Cell surface Hydrophobicity of MCC0200
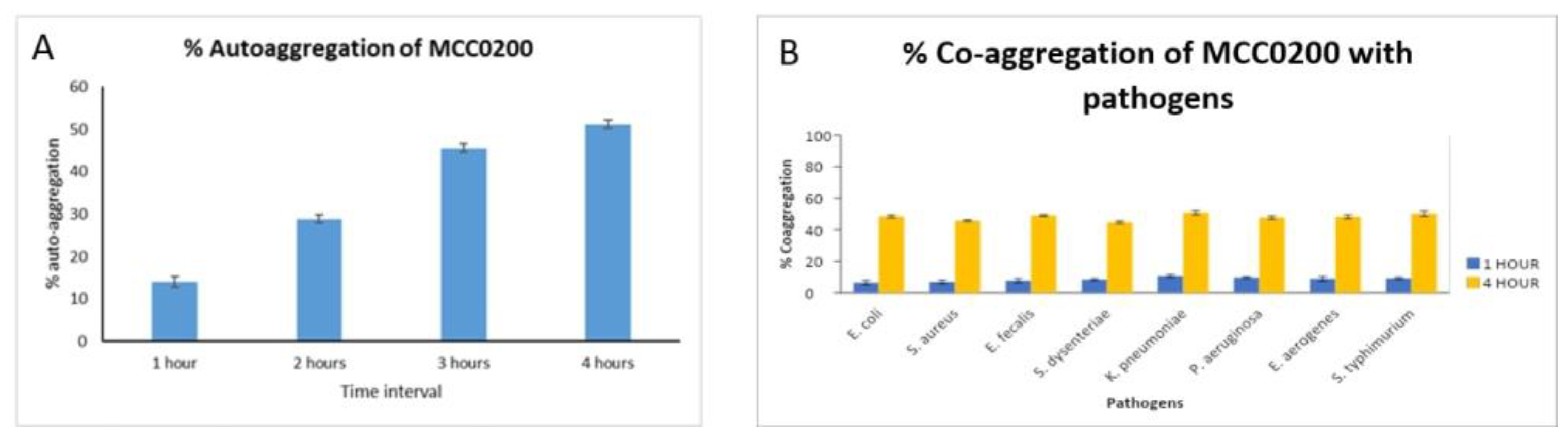
3.3.2.1.2. Aggregation ability of MCC0200
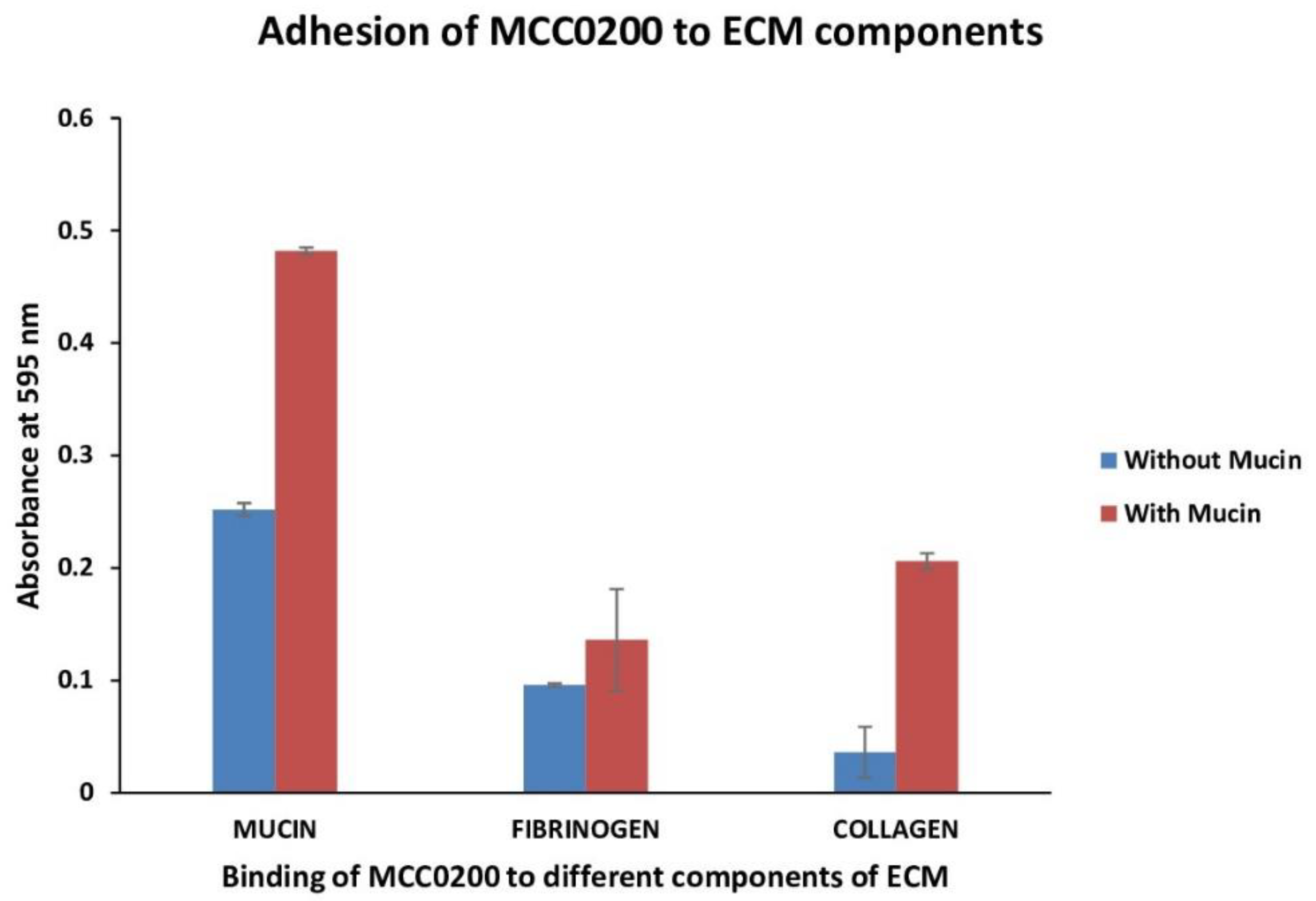
3.3.2.1.3. Adhesion to mucin, fibrinogen and collagen
3.3.2.2. Adhesion of MCC0200 to HT-29 cell line
3.3.3. Antioxidant activity
3.3.3.1. The redox system in MCC0200
3.3.4. MCC0200 as nutrient factory: Biosynthetic capabilities
3.3.5. Beta galactosidase production
3.3.6. In-vitro Evaluation of the Anti-hypercholesterolemic Effect of MCC0200
3.4. Safety Assessment of MCC0200 as a probiotic
3.4.1. Genome based safety evaluation of MCC0200
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Alexandraki, V. , Kazou, M., Blom, J., Pot, B., Papadimitriou, K. and Tsakalidou, E., 2019. Comparative genomics of Streptococcus thermophilus support important traits concerning the evolution, biology and technological properties of the species. Frontiers in microbiology, 10, p.2916.
- Cui, Y. , Xu, T., Qu, X., Hu, T., Jiang, X. and Zhao, C., 2016. New insights into various production characteristics of Streptococcus thermophilus strains. International journal of molecular sciences, 17(10), p.1701.
- Martinović, A. , Cocuzzi, R., Arioli, S. and Mora, D., 2020. Streptococcus thermophilus: to survive, or not to survive the gastrointestinal tract, that is the question!. Nutrients, 12(8), p.2175.
- Uriot, O. , Denis, S., Junjua, M., Roussel, Y., Dary-Mourot, A. and Blanquet-Diot, S., 2017. Streptococcus thermophilus: from yogurt starter to a new promising probiotic candidate?. Journal of Functional Foods, 37, pp.74-89.
- Hu, T. , Cui, Y., Zhang, Y., Qu, X. and Zhao, C., 2020. Genome analysis and physiological characterization of four Streptococcus thermophilus strains isolated from Chinese traditional fermented milk. Frontiers in Microbiology, 11, p.184.
- Roux, E. , Nicolas, A., Valence, F., Siekaniec, G., Chuat, V., Nicolas, J., Le Loir, Y. and Guédon, E., 2022. The genomic basis of the Streptococcus thermophilus health-promoting properties. BMC genomics, 23(1), p.210.
- Cui, Y. , Jiang, X., Hao, M., Qu, X. and Hu, T., 2017. New advances in exopolysaccharides production of Streptococcus thermophilus. Archives of microbiology, 199, pp.799-809.
- Xiong, Z.Q. , Kong, L.H., Lai, P.F.H., Xia, Y.J., Liu, J.C., Li, Q.Y. and Ai, L.Z., 2019. Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3. Journal of dairy science, 102(6), pp.4925-4934.
- Iyer, R. , Tomar, S.K., Maheswari, T.U. and Singh, R., 2010. Streptococcus thermophilus strains: Multifunctional lactic acid bacteria. International Dairy Journal, 20(3), pp. 133-141.
- Hao, M. , Cui, Y. and Qu, X., 2018. Analysis of CRISPR-Cas system in Streptococcus thermophilus and its application. Frontiers in Microbiology, 9, p.257.
- Tian, H. , Muhammad, Z., Evivie, S.E., Gu, C.T. and Huo, G.C., 2018. Exact identification of six starter-strain candidates of Streptococcus thermophilus by analysing genotypic and industrial properties. International Journal of Dairy Technology, 71, pp.11-21.
- Prajapati, J.B. , Nathani, N.M., Patel, A.K., Senan, S. and Joshi, C.G., 2013. Genomic analysis of dairy starter culture Streptococcus thermophilus MTCC 5461. J Microbiol Biotechnol, 23(4), pp.459-466.
- Rasmussen, T.B. , Danielsen, M., Valina, O., Garrigues, C., Johansen, E. and Pedersen, M.B., 2008. Streptococcus thermophilus core genome: comparative genome hybridization study of 47 strains.
- Vendramin, V. , Treu, L., Campanaro, S., Lombardi, A., Corich, V. and Giacomini, A., 2017. Genome comparison and physiological characterization of eight Streptococcus thermophilus strains isolated from Italian dairy products. Food microbiology, 63, pp.47-57.
- Parks, D.H. , Imelfort, M., Skennerton, C.T., Hugenholtz, P. and Tyson, G.W., 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome research, 25(7), pp.1043-1055.
- Aziz, R.K. , Bartels, D., Best, A.A., DeJongh, M., Disz, T., Edwards, R.A., Formsma, K., Gerdes, S., Glass, E.M., Kubal, M. and Meyer, F., 2008. The RAST Server: rapid annotations using subsystems technology. BMC genomics, 9(1), pp.1-15.
- Kanehisa, M. , Sato, Y. and Morishima, K., 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of molecular biology, 428(4), pp.726-731.
- Vecchione, A. , Celandroni, F., Mazzantini, D., Senesi, S., Lupetti, A. and Ghelardi, E., 2018. Compositional quality and potential gastrointestinal behavior of probiotic products commercialized in Italy. Frontiers in medicine, 5, p.59.
- Prokopovich, P. and Perni, S., 2009. An investigation of microbial adhesion to natural and synthetic polysaccharide-based films and its relationship with the surface energy components. Journal of Materials Science: Materials in Medicine, 20(1), pp.195-202.
- Farniya, F. , Jamalli, A. and Dadgar, T., 2019. Physicochemical surface characteristics in different pathogenic bacteria. Cogent Biology, 5(1), p.1638572.
- Sharma, S. and Kanwar, S.S., 2017. Adherence potential of indigenous lactic acid bacterial isolates obtained from fermented foods of Western Himalayas to intestinal epithelial Caco-2 and HT-29 cell lines. Journal of food science and technology, 54, pp.3504-3511.
- Inturri, R. , Stivala, A., Sinatra, F., Morrone, R. and Blandino, G., 2014. Scanning electron microscopy observation of adhesion properties of Bifidobacterium longum W11 and chromatographic analysis of its exopolysaccaride. Food and Nutrition Sciences, 5(18), p.1787.
- Mu, G. , Gao, Y., Tuo, Y., Li, H., Zhang, Y., Qian, F. and Jiang, S., 2018. Assessing and comparing antioxidant activities of lactobacilli strains by using different chemical and cellular antioxidant methods. Journal of dairy science, 101(12), pp.10792-10806.
- Yan, F. , Yu, X. and Jing, Y., 2018. Optimized preparation, characterization, and antioxidant activity of chitooligosaccharide–glycine Maillard reaction products. Journal of food science and technology, 55, pp.712-720.
- Tomaro-Duchesneau, C. , Jones, M.L., Shah, D., Jain, P., Saha, S. and Prakash, S., 2014. Cholesterol assimilation by Lactobacillus probiotic bacteria: an in-vitro investigation. BioMed research international, 2014.
- Rudel, L.L. and Morris, M.D., 1973. Determination of cholesterol using o-phthalaldehyde. Journal of Lipid Research, 14(3), pp.364-366.
- EFSA Panel on Biological Hazards (BIOHAZ), Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., Fernández Escámez, P.S., Girones, R., Koutsoumanis, K., Lindqvist, R. and Nørrung, B., 2018. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 8: suitability of taxonomic units notified to EFSA until March 2018. EFSA Journal, 16(7), p.e05315.
- Alcock, B.P. , Raphenya, A.R., Lau, T.T., Tsang, K.K., Bouchard, M., Edalatmand, A., Huynh, W., Nguyen, A.L.V., Cheng, A.A., Liu, S. and Min, S.Y., 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic acids research, 48(D1), pp.D517-D525.
- Bortolaia, V. , Kaas, R.S., Ruppe, E., Roberts, M.C., Schwarz, S., Cattoir, V., Philippon, A., Allesoe, R.L., Rebelo, A.R., Florensa, A.F. and Fagelhauer, L., 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. Journal of Antimicrobial Chemotherapy, 75(12), pp.3491-3500.
- Arndt, D. , Grant, J.R., Marcu, A., Sajed, T., Pon, A., Liang, Y. and Wishart, D.S., 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic acids research, 44(W1), pp.W16-W21.
- Couvin, D. , Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., Rocha, E.P., Vergnaud, G., Gautheret, D. and Pourcel, C., 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic acids research, 46(W1), pp.W246-W251.
- Carattoli, A. , Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., Møller Aarestrup, F. and Hasman, H., 2014. In-silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial agents and chemotherapy, 58(7), pp.3895-3903.
- Auch, A.F. , von Jan, M., Klenk, H.P. and Göker, M., 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Standards in genomic sciences, 2, pp.117-134.
- Lee, I. , Ouk Kim, Y., Park, S.C. and Chun, J., 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. International journal of systematic and evolutionary microbiology, 66(2), pp.1100-1103.
- Alikhan, N.F. , Petty, N.K., Ben Zakour, N.L. and Beatson, S.A., 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC genomics, 12(1), pp.1-10.
- Goh, Y.J. , Goin, C., O’Flaherty, S., Altermann, E. and Hutkins, R., 2011, December. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. In Microbial cell factories (Vol. 10, No. 1, pp. 1-17). BioMed Central.
- Padan, E. , 2008. The enlightening encounter between structure and function in the NhaA Na+–H+ antiporter. Trends in biochemical sciences, 33(9), pp.435-443.
- Mora, D. , Monnet, C., Parini, C., Guglielmetti, S., Mariani, A., Pintus, P., Molinari, F., Daffonchio, D. and Manachini, P.L., 2005. Urease biogenesis in Streptococcus thermophilus. Research in Microbiology, 156(9), pp.897-903.
- Bustos, A.Y. , de Valdez, G.F., Fadda, S. and Taranto, M.P., 2018. New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health. Food Research International, 112, pp.250-262.
- Tuncer, B.O. and Tuncer, Y., 2014. Exopolysaccharide producer Streptococcus thermophilus ST8. 01 strain; a potential probiotic culture. GIDA, 39(4), pp.195-202.
- Arias, C.A. and Murray, B.E., 2009. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. New England Journal of Medicine, 360(5), pp.439-443.
- Ruiz, L. , Margolles, A. and Sánchez, B., 2013. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Frontiers in microbiology, 4, p.396.
- Kebouchi, M. , Galia, W., Genay, M., Soligot, C., Lecomte, X., Awussi, A.A., Perrin, C., Roux, E., Dary-Mourot, A. and Le Roux, Y., 2016. Implication of sortase-dependent proteins of Streptococcus thermophilus in adhesion to human intestinal epithelial cell lines and bile salt tolerance. Applied microbiology and biotechnology, 100, pp.3667-3679.
- Pfeiler, E.A. , Azcarate-Peril, M.A. and Klaenhammer, T.R., 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. Journal of bacteriology, 189(13), pp.4624-4634.
- Weiss, G. and Jespersen, L., 2010. Transcriptional analysis of genes associated with stress and adhesion in Lactobacillus acidophilus NCFM during the passage through an in-vitro gastrointestinal tract model. Microbial Physiology, 18(4), pp.206-214.
- Duary, R.K., Bhausaheb, M.A., Batish, V.K. and Grover, S., 2012. Anti-inflammatory and immunomodulatory efficacy of indigenous probiotic Lactobacillus plantarum Lp91 in colitis mouse model. Molecular biology reports, 39, pp.4765-4775.
- Lebeer, S. , Vanderleyden, J. and De Keersmaecker, S.C., 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiology and Molecular Biology Reviews, 72(4), pp.728-764.
- Tuo, Y. , Yu, H., Ai, L., Wu, Z., Guo, B. and Chen, W., 2013. Aggregation and adhesion properties of 22 Lactobacillus strains. Journal of dairy science, 96(7), pp.4252-4257.
- Wang, L.Q. , Meng, X.C., Zhang, B.R., Wang, Y. and Shang, Y.L., 2010. Influence of cell surface properties on adhesion ability of bifidobacteria. World Journal of Microbiology and Biotechnology, 26, pp.1999-2007.
- Taj, R. , Masud, T., Sohail, A., Sammi, S., Naz, R., Sharma Khanal, B.K. and Nawaz, M.A., 2022. In-vitro screening of EPS-producing Streptococcus thermophilus strains for their probiotic potential from Dahi. Food Science & Nutrition, 10(7), pp.2347-2359.
- Collado, M.C. , Isolauri, E. and Salminen, S., 2008. Specific probiotic strains and their combinations counteract adhesion of Enterobacter sakazakii to intestinal mucus. FEMS microbiology letters, 285(1), pp.58-64.
- Ayyash, M. , Abushelaibi, A., Al-Mahadin, S., Enan, M., El-Tarabily, K. and Shah, N., 2018. In-vitro investigation into probiotic characterisation of Streptococcus and Enterococcus isolated from camel milk. LWT, 87, pp.478-487.
- Nishiyama, K. , Sugiyama, M. and Mukai, T., 2016. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms, 4(3), p.34.
- Jonsson, H. , Ström, E. and Roos, S., 2001. Addition of mucin to the growth medium triggers mucus-binding activity in different strains of Lactobacillus reuteri in-vitro. FEMS Microbiology Letters, 204(1), pp.19-22.
- Fernandez, N. , Wrzosek, L., Radziwill-Bienkowska, J.M., Ringot-Destrez, B., Duviau, M.P., Noordine, M.L., Laroute, V., Robert, V., Cherbuy, C., Daveran-Mingot, M.L. and Cocaign-Bousquet, M., 2018. Characterization of mucus-related properties of Streptococcus thermophilus: from adhesion to induction. Frontiers in Physiology, 9, p.980.
- Haeri, A. , Khodaii, Z., Ghaderian, S.M.H., Tabatabaei Panah, A.S. and Akbarzadeh Najar, R., 2012. Comparison of adherence patterns of a selection of probiotic bacteria to Caco-2, HEp-2, and T84 cell lines. Annals of microbiology, 62(1), pp.339-344.
- Kapczynski, D.R. , Meinersmann, R.J. and Lee, M.D., 2000. Adherence of Lactobacillus to intestinal 407 cells in culture correlates with fibronectin binding. Current microbiology, 41, pp.136-141.
- Kainulainen, V. and Korhonen, T.K., 2014. Dancing to another tune—adhesive moonlighting proteins in bacteria. Biology, 3(1), pp.178-204.
- Wang, Y. , Wu, Y., Wang, Y., Xu, H., Mei, X., Yu, D., Wang, Y. and Li, W., 2017. Antioxidant properties of probiotic bacteria. Nutrients, 9(5), p.521.
- Lobo, R.E. , Gómez, M.I., de Valdez, G.F. and Torino, M.I., 2019. Physicochemical and antioxidant properties of a gastroprotective exopolysaccharide produced by Streptococcus thermophilus CRL1190. Food Hydrocolloids, 96, pp.625-633.
- Kang, C.H. , Kim, J.S., Park, H.M., Kim, S. and Paek, N.S., 2021. Antioxidant activity and short-chain fatty acid production of lactic acid bacteria isolated from Korean individuals and fermented foods. 3 Biotech, 11(5), p.217.
- Feng, T. and Wang, J., 2020. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes, 12(1), p.1801944.
- Agbas, A. and Moskovitz, J., 2009. The role of methionine oxidation/reduction in the regulation of immune response. Current signal transduction therapy, 4(1), pp.46-50.
- Saubade, F. , Hemery, Y.M., Guyot, J.P. and Humblot, C., 2017. Lactic acid fermentation as a tool for increasing the folate content of foods. Critical Reviews in Food Science and Nutrition, 57(18), pp.3894-3910.
- Juers, D.H. , Matthews, B.W. and Huber, R.E., 2012. LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Science, 21(12), pp.1792-1807.
- Yañez-Ñeco, C.V. , Cervantes, F.V., Amaya-Delgado, L., Ballesteros, A.O., Plou, F.J. and Arrizon, J., 2021. Synthesis of β (1→ 3) and β (1→ 6) galactooligosaccharides from lactose and whey using a recombinant β-galactosidase from Pantoea anthophila. Electronic Journal of Biotechnology, 49, pp.14-21.
- Kumar, M. , Nagpal, R., Kumar, R., Hemalatha, R., Verma, V., Kumar, A., Chakraborty, C., Singh, B., Marotta, F., Jain, S. and Yadav, H., 2012. Cholesterol-lowering probiotics as potential biotherapeutics for metabolic diseases. Journal of Diabetes Research, 2012.
- Ziarno, M. , 2010. Viability and cholesterol uptake by Streptococcus thermophilus cultures in artificial GIT fluids. Acta Scientiarum Polonorum Technologia Alimentaria, 9(1), pp.83-94.
- Noh, D.O. , Kim, S.H. and Gilliland, S.E., 1997. Incorporation of cholesterol into the cellular membrane of Lactobacillus acidophilus ATCC 43121. Journal of dairy science, 80(12), pp.3107-3113.
- Lee, J. , Kim, Y., Yun, H.S., Kim, J.G., Oh, S. and Kim, S.H., 2010. Genetic and proteomic analysis of factors affecting serum cholesterol reduction by Lactobacillus acidophilus A4. Applied and Environmental Microbiology, 76(14), pp.4829-4835.
- Sharma, P. , Tomar, S.K., Goswami, P., Sangwan, V. and Singh, R., 2014. Antibiotic resistance among commercially available probiotics. Food research international, 57, pp.176-195.
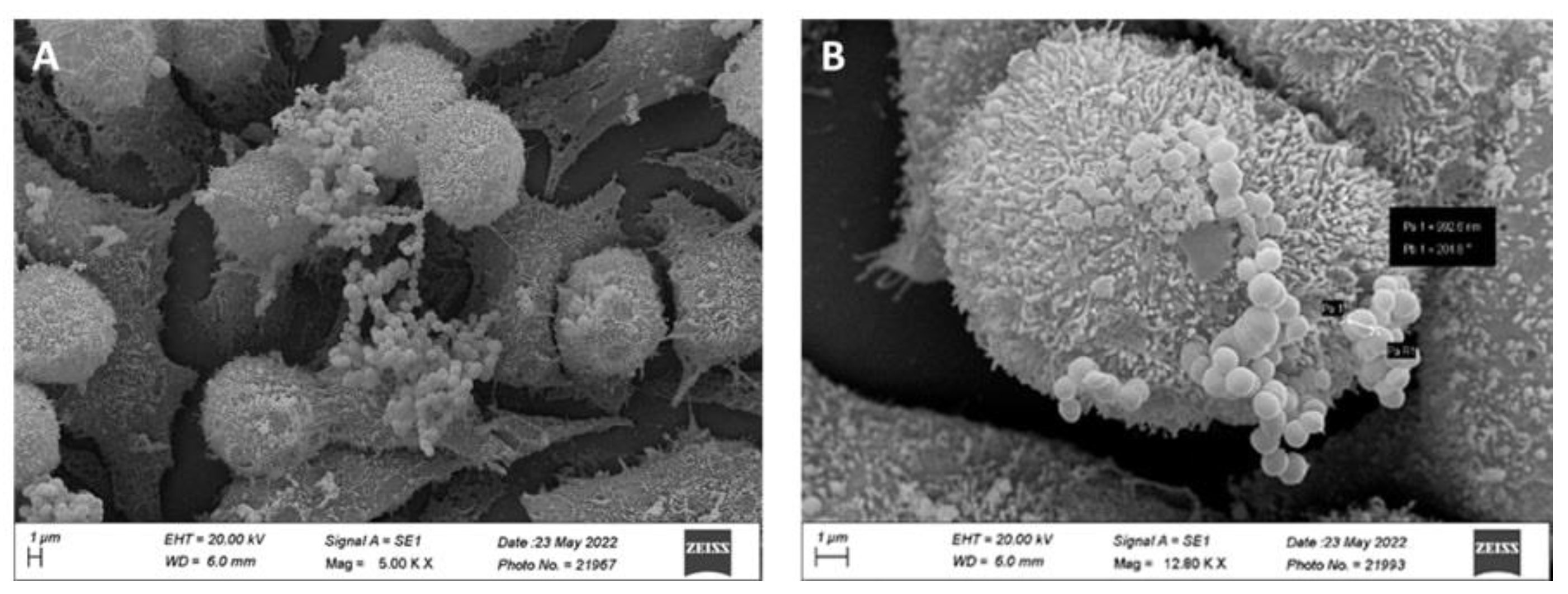
| Genome attributes | Values | |
|---|---|---|
| Genome size (bp) | 1,855,815 | |
| GC content % | 39.1 | |
| Number of contigs | 6 | |
| Protein coding genes (CDS) | 2239 | |
| Subsystems | 219 | |
| RNA encoding genes | 83 | |
| Reference strain | % ANI | % DDH |
|---|---|---|
| Streptococcus thermophilus LMD-9 | 99.93 | 99.70 |
| Streptococcus thermophilus TH1477 | 98.96 | 91.70 |
| Streptococcus thermophilus MTH17CL396 | 99.01 | 92.20 |
| Streptococcus thermophilus TH1436 | 99.25 | 94.00 |
| Streptococcus thermophilus TH1435 | 99.31 | 93.90 |
| Genes detected in MCC0200 | FigFam no. | Predicted function |
|---|---|---|
| ATP synthase subunit a | fig|6666666.935801.peg.921 | |
| ATP synthase subunit b | fig|6666666.935801.peg.922 | Acid tolerance by maintaining pH homeostasis |
| ATP synthase subunit c | fig|6666666.935801.peg.920 | |
| ATP synthase alpha chain | fig|6666666.935801.peg.924 | |
| ATP synthase Beta chain | fig|6666666.935801.peg.926 | |
| ATP synthase Gamma chain | fig|6666666.935801.peg.925 | |
| ATP synthase Epsilon chain | fig|6666666.935801.peg.927 | |
| ATP synthase delta chain | fig|6666666.935801.peg.923 | |
| Na+/H+ antiporter | fig|6666666.935801.peg.2147 | |
|
Urease system Urease cluster protein Alpha Beta Gamma Accessory proteins: Urease accessory protein UreD Urease accessory protein UreE Urease accessory protein UreF Urease accessory protein UreG |
fig|6666666.935801.peg.706 fig|6666666.935801.peg.707 fig|6666666.935801.peg.711 fig|6666666.935801.peg.710 fig|6666666.935801.peg.709 fig|6666666.935801.peg.715 fig|6666666.935801.peg.712 fig|6666666.935801.peg.713 fig|6666666.935801.peg.714 |
Acid tolerance by Alkali production |
| Ffh DnaK DnaJ GrpE HrcA GroEL GroES Clp proteases EF-Tu recA recN exonuclease V UvrABCD DNA polymerase DNA ligase |
fig|6666666.935801.peg.1370 fig|6666666.935801.peg.487 fig|6666666.935801.peg.488 fig|6666666.935801.peg.485 fig|6666666.935801.peg.484 fig|6666666.935801.peg.603 fig|6666666.935801.peg.601 fig|6666666.935801.peg.801 fig|6666666.935801.peg.929 fig|6666666.935801.peg.410 fig|6666666.935801.peg.1668 fig|6666666.935801.peg.1681 fig|6666666.935801.peg.31 fig|6666666.935801.peg.1985 fig|6666666.935801.peg.1783 fig|6666666.935801.peg.1458 fig|6666666.935801.peg.46 fig|6666666.935801.peg.2048 |
Proteins involved in protection and repair of molecules under acid stress |
| Sortase A Sortase-dependent proteins HtrA DnaJ GroEL |
fig|6666666.935801.peg.1755 fig|6666666.935801.peg.992 fig|6666666.935801.peg.1848 fig|6666666.935801.peg.349 fig|6666666.935801.peg.488 fig|6666666.935801.peg.603 |
Proteins involved in bile salt tolerance |
| Genes detected in MCC0200 |
Predicted function | FigFam no. |
|---|---|---|
| Fibronectin/fibrinogen-binding protein | Binds to fibronectin | fig|6666666.935801.peg.1423 |
| Sortase A, LPXTG specific | Cell surface localization and peptidoglycan interaction | fig|6666666.935801.peg.1755 |
| Moonlighting proteins | ||
| Enolase | Binding to plasmin(ogen), fibronectin, laminin, albumin, collagen, salivary mucin, intestinal epithelial cells, | fig|6666666.935801.peg.1108 |
| EF-Tu | Binding to plasmin(ogen), plasma Factor H and Factor H-related protein 1 (FHR-1), intestinal epithelial cells and HT-MTX-derived mucus, salivary mucin, fibronectin | fig|6666666.935801.peg.929 |
| EF-G | Binding to salivary mucin | fig|6666666.935801.peg.75 |
| Triosephosphate isomerase | Binding to plasmin(ogen), intestinal epithelial cells, | fig|6666666.935801.peg.930 |
| GroEL | Binding to intestinal HT-29 cells and mucus | fig|6666666.935801.peg.603 |
| DnaK | Binding to plasmin(ogen) | fig|6666666.935801.peg.486 |
| Pyruvate kinase | Binding to salivary mucin | fig|6666666.935801.peg.1651 |
| Inosine 5′-monophosphate dehydrogenase (IMPDH) | Binding to plasmin(ogen) | fig|6666666.935801.peg.342 |
| Glutamine synthetase |
Binding to plasmin(ogen), laminin, collagen I, fibronectin | fig|6666666.935801.peg.61 |
| Glucose-6-phosphate isomerase (GPI) | Binding to collagen | fig|6666666.935801.peg.585 |
| Gene detected in MCC0200 genome | FigFam no. | Predicted function |
|---|---|---|
| Thiol peroxidase, Tpx-type (EC 1.11.1.15) NADH peroxidase |
fig|6666666.935801.peg.1462 fig|6666666.935801.peg.1758 |
H2O2-degrading enzymes |
| Superoxide dismutase [Mn] (EC 1.15.1.1) | fig|6666666.935801.peg.1192 | Hydroperoxide radical detoxification |
| Thioredoxin reductase (EC 1.8.1.9) Thioredoxin |
fig|6666666.935801.peg.1905 fig|6666666.935801.peg.88 |
Redox homeostasis |
| Peptide-methionine (S)-S-oxide reductase MsrA/MrsB |
fig|6666666.935801.peg.1824 fig|6666666.935801.peg.2133 |
Resistance to oxidative stress |
| recA | fig|6666666.935801.peg.410 | Induces DNA repair mechanism |
| GroES/EL, clp proteases, CtsR, HrcA | fig|6666666.935801.peg.602 fig|6666666.935801.peg.603 fig|6666666.935801.peg.801 fig|6666666.935801.peg.425 fig|6666666.935801.peg.484 |
Targeting and degradation of misfolded proteins. |
| HtrA | fig|6666666.935801.peg.349 | Proteolysis of abnormal proteins |
| GrpE | fig|6666666.935801.peg.485 | Proper protein folding |
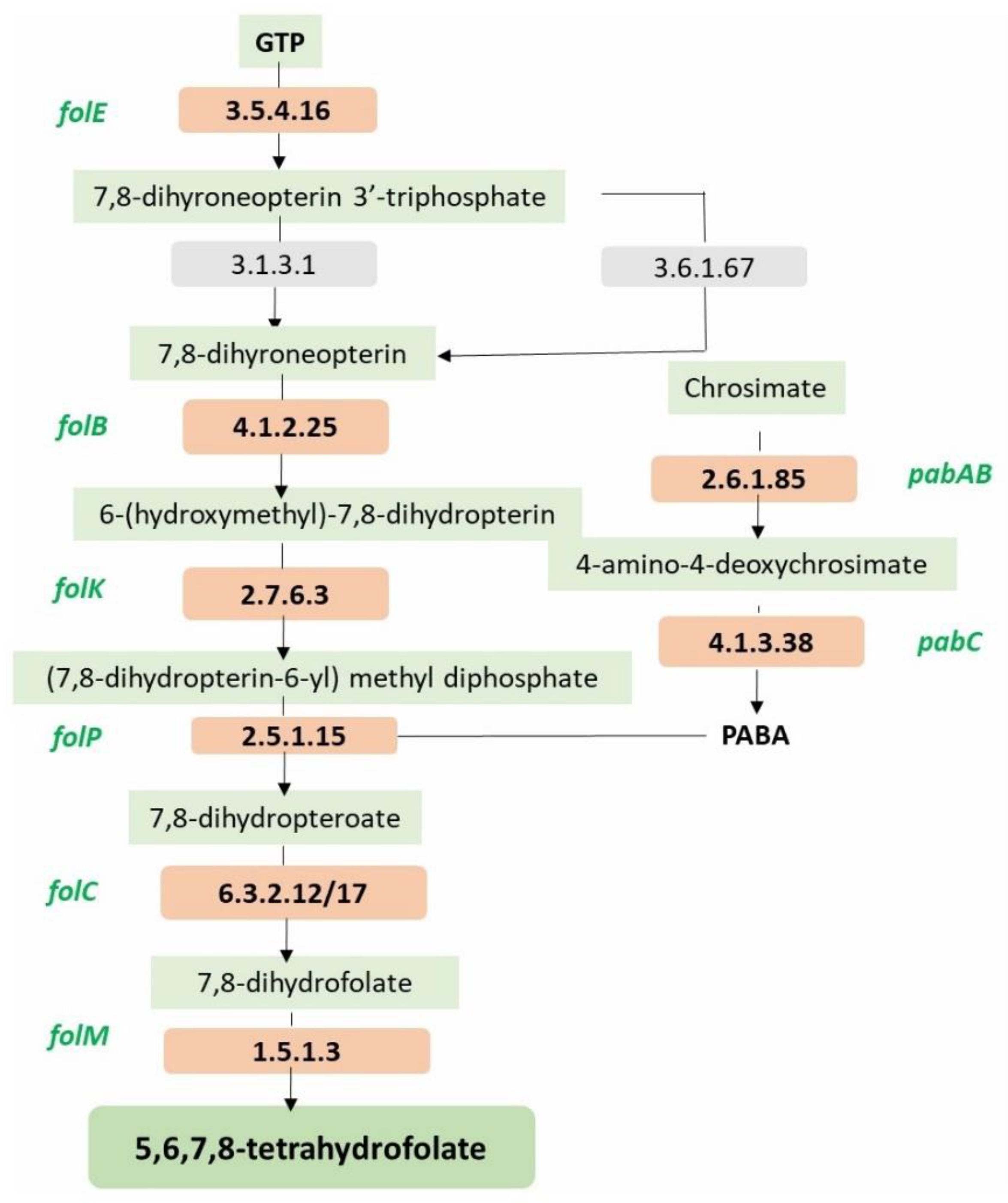
| Folate biosynthesis Protein/gene/system detected in the MCC0200 | FigFam no. |
|---|---|
| FolE, GTP cyclohydrolase I (EC 3.5.4.16) type 1 | fig|6666666.935801.peg.2035 |
| FolB, dihydroneopterin aldolase | fig|6666666.935801.peg.2032 |
| FolK,2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase (EC 2.7.6.3) | fig|6666666.935801.peg.2031 |
| FolP, Dihydropteroate synthase (EC 2.5.1.15) | fig|6666666.935801.peg.2034 |
| FolC1, Dihydrofolate synthase (EC 6.3.2.12) | fig|6666666.935801.peg.846 |
| FolC2, Dihydrofolate synthase (EC 6.3.2.12) | fig|6666666.935801.peg.2038 |
| FolM, FolA, Dihydrofolate reductase (EC 1.5.1.3) | fig|6666666.935801.peg.1044 |
| PabC, Aminodeoxychorismate lyase | fig|6666666.935801.peg.1232 |
| PabAB, Para-aminobenzoate synthase, aminase component (EC 2.6.1.85) | fig|6666666.935801.peg.1232 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





