Submitted:
03 September 2025
Posted:
08 September 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Results
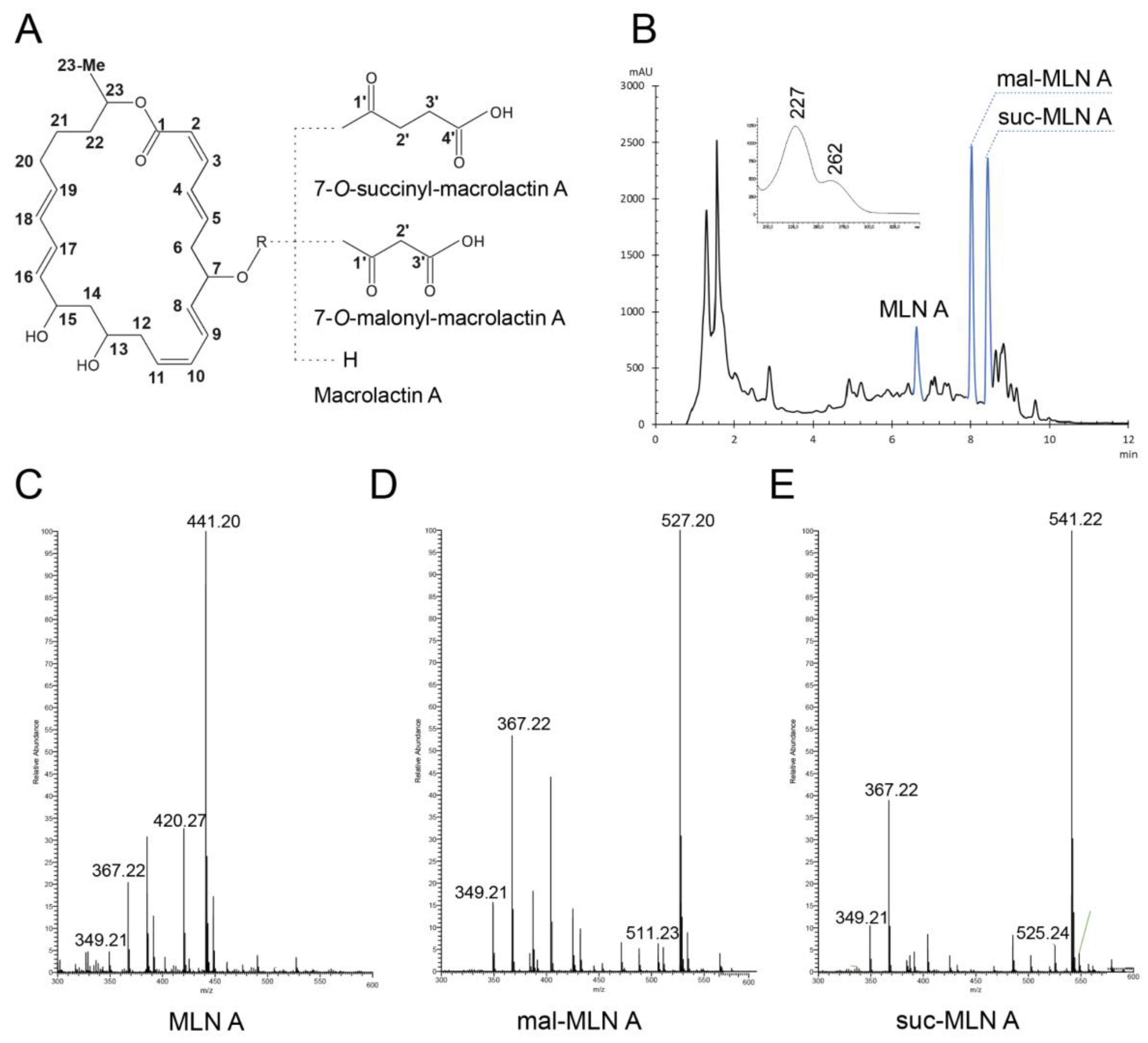
Cultivation of the Strain, Isolation and Identification of the Metabolites with Antibiotic Activity
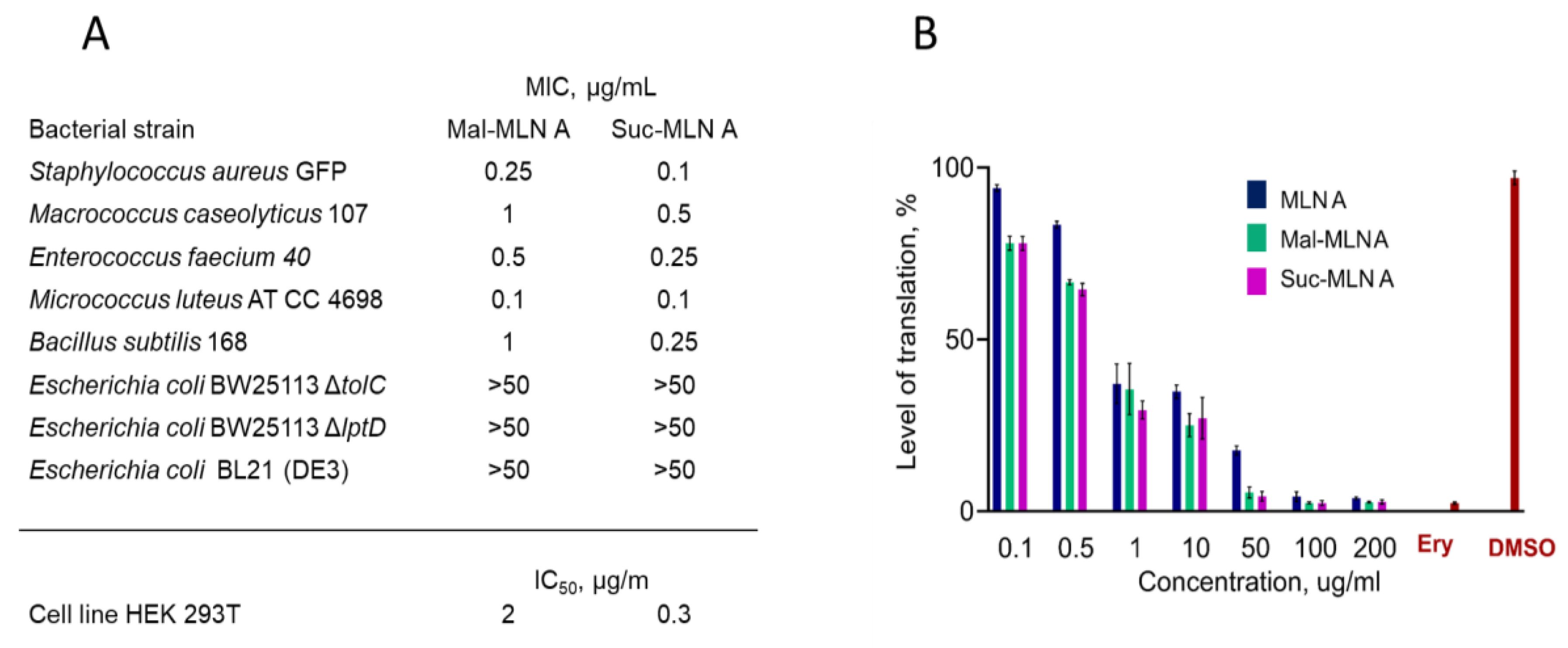
Bacterial Susceptibility and Cytotoxicity of the Macrolactins
Inhibition of Bacterial Protein Synthesis In Vitro by the Macrolactins
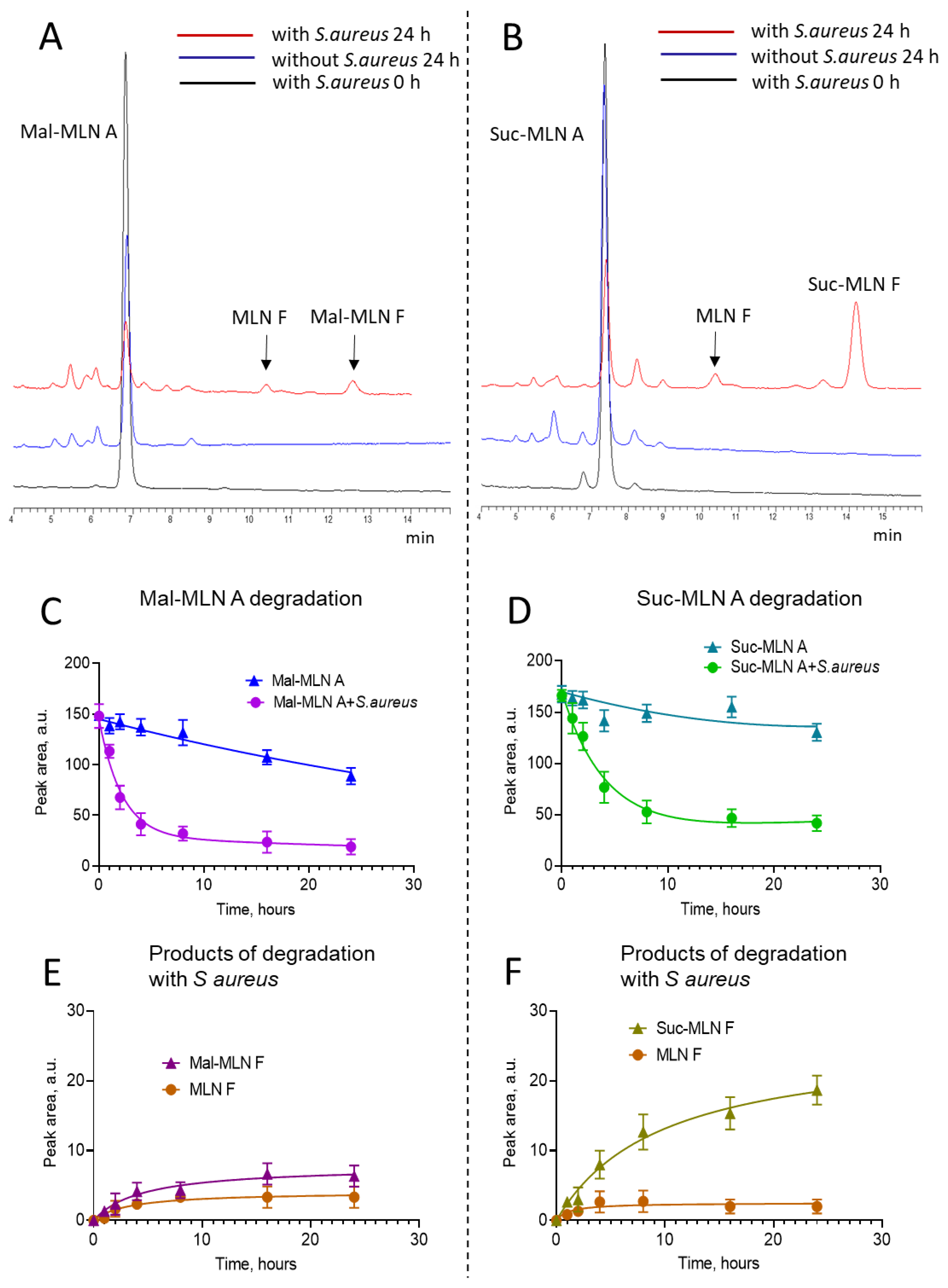
Transformation of Acylated Macrolactins in S. aureus Cells
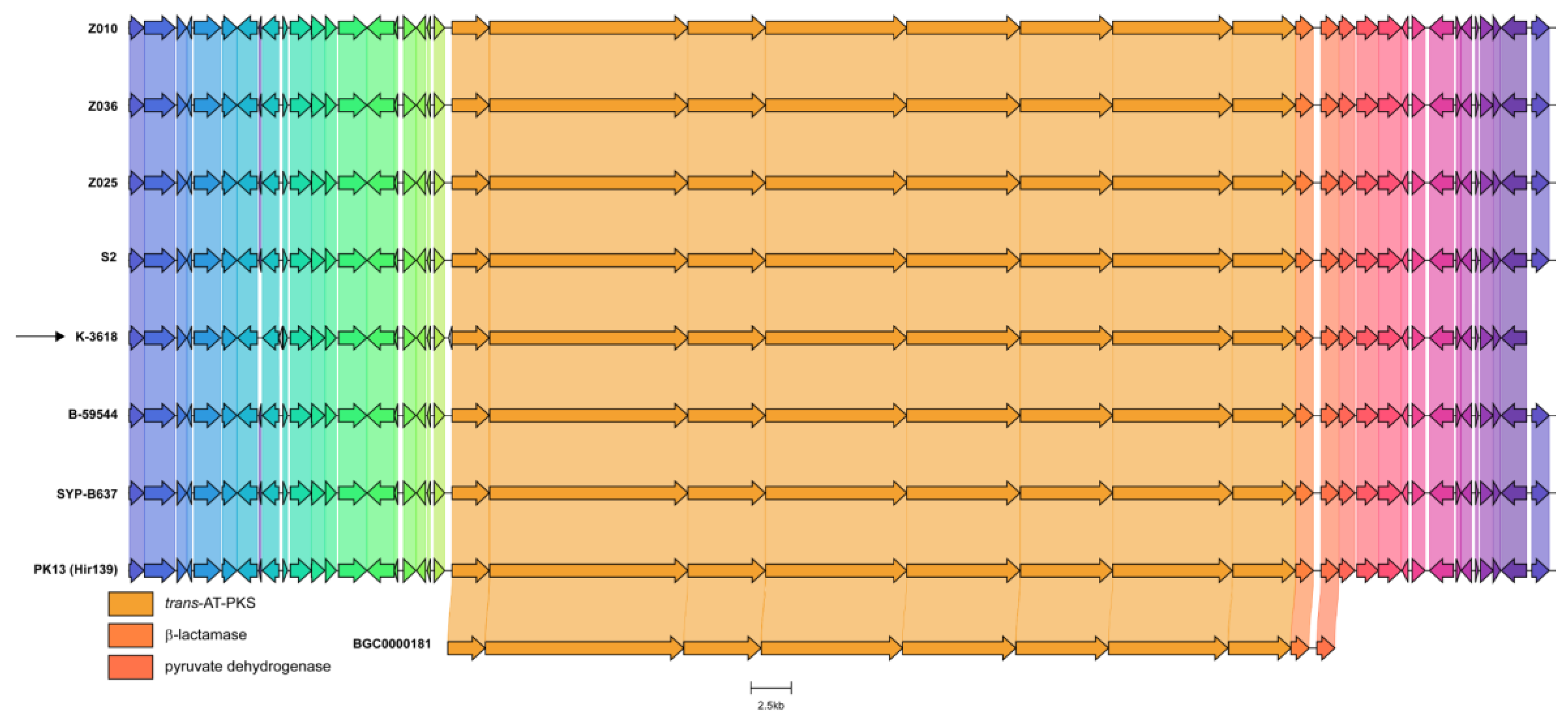
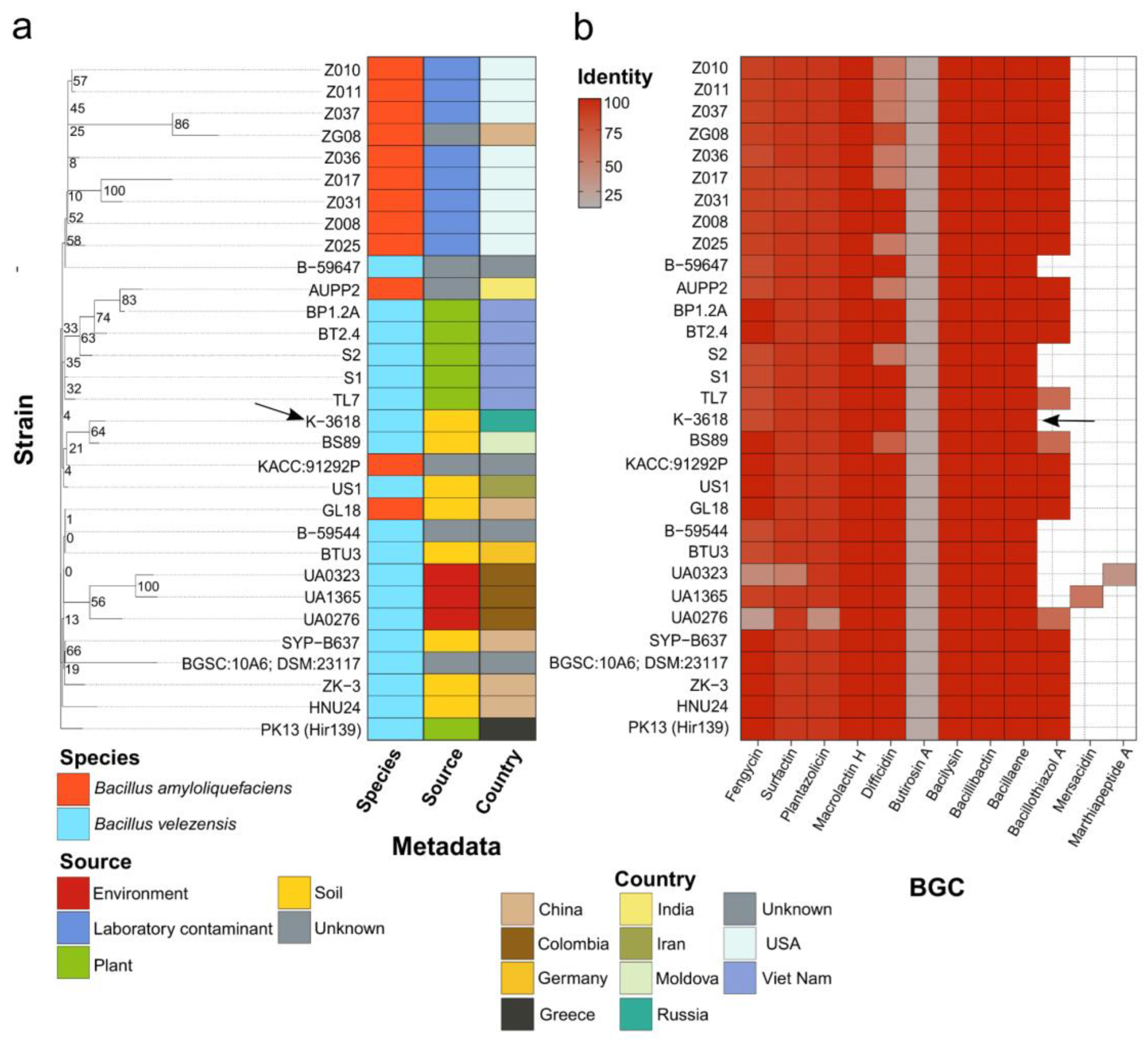
MLN Biosynthetic Gene Cluster is Highly Abundant in B. velezensis and B. amyloliquefaciens Species
Discussion
Materials and Methods
Bacterial Strains and Cell Lines
Genome Sequencing, Assembly and Annotation
Phylogeny Reconstruction
Cultivation, Isolation and Purification of Macrolactins
LC-MS Analysis
NMR
Antibacterial Activity Assessment
Cytotoxicity Assessment
Inhibition Assays in Cell-Free Translation System
Biotransformation of with suc-MLN A and mal-MLN A by S.aureus
Analysis of Reaction Broth, Isolation and Purification of Target Compounds After Incubation of the Macrolactins with S.aureus Cells
Conclusions
Supplementary Materials
Data Availability Statement
Acknowledgments
References
- Baker, B.P.; Green, T.A.; Loker, A.J. Biological Control and Integrated Pest Management in Organic and Conventional Systems. Biol. Control 2020, 140, 104095. [CrossRef]
- Zhong, X.; Jin, Y.; Ren, H.; Hong, T.; Zheng, J.; Fan, W.; Hong, J.; Chen, Z.; Wang, A.; Lu, H.; et al. Research Progress of Bacillus Velezensis in Plant Disease Resistance and Growth Promotion. Front. Ind. Microbiol. 2024, 2, 1442980. [CrossRef]
- Rabbee, M.F.; Hwang, B.-S.; Baek, K.-H. Bacillus Velezensis: A Beneficial Biocontrol Agent or Facultative Phytopathogen for Sustainable Agriculture. Agronomy 2023, 13, 840. [CrossRef]
- Tran, C.; Cock, I.E.; Chen, X.; Feng, Y. Antimicrobial Bacillus: Metabolites and Their Mode of Action. Antibiotics 2022, 11, 88. [CrossRef]
- Chen, L.; Wang, X.; Liu, Y. Contribution of Macrolactin in Bacillus Velezensis CLA178 to the Antagonistic Activities against Agrobacterium Tumefaciens C58. Arch. Microbiol. 2021, 203, 1743–1752. [CrossRef]
- Yuan, J.; Li, B.; Zhang, N.; Waseem, R.; Shen, Q.; Huang, Q. Production of Bacillomycin- and Macrolactin-Type Antibiotics by Bacillus Amyloliquefaciens NJN-6 for Suppressing Soilborne Plant Pathogens. J. Agric. Food Chem. 2012, 60, 2976–2981. [CrossRef]
- Wang, J.; Peng, Y.; Xie, S.; Yu, X.; Bian, C.; Wu, H.; Wang, Y.; Ding, T. Biocontrol and Molecular Characterization of Bacillus Velezensis D against Tobacco Bacterial Wilt. Phytopathol. Res. 2023, 5, 50. [CrossRef]
- Yousfi, S.; Krier, F.; Deracinois, B.; Steels, S.; Coutte, F.; Frikha-Gargouri, O. Characterization of Bacillus Velezensis 32a Metabolites and Their Synergistic Bioactivity against Crown Gall Disease. Microbiol. Res. 2024, 280, 127569. [CrossRef]
- Lu, X.; Xu, Q.; Liu, X.; Cao, X.; Ni, K.; Jiao, B. Marine Drugs – Macrolactins. Chem. Biodivers. 2008, 5, 1669–1674. [CrossRef]
- Wu, T.; Xiao, F.; Li, W. Macrolactins: Biological Activity and Biosynthesis. Mar. Life Sci. Technol. 2021, 3, 62–68. [CrossRef]
- Gustafson, K.; Roman, M.; Fenical, W. The Macrolactins, a Novel Class of Antiviral and Cytotoxic Macrolides from a Deep-Sea Marine Bacterium. J. Am. Chem. Soc. 1989, 111, 7519–7524. [CrossRef]
- Xu, Y.; Song, Y.; Ning, Y.; Li, S.; Qu, Y.; Jiao, B.; Lu, X. Macrolactin XY, a Macrolactin Antibiotic from Marine-Derived Bacillus Subtilis Sp. 18. Mar. Drugs 2024, 22, 331. [CrossRef]
- Jaruchoktaweechai, C.; Suwanborirux, K.; Tanasupawatt, S.; Kittakoop, P.; Menasveta, P. New Macrolactins from a Marine Bacillus Sp. Sc026. J. Nat. Prod. 2000, 63, 984–986. [CrossRef]
- Romero-Tabarez, M.; Jansen, R.; Sylla, M.; Lünsdorf, H.; Häußler, S.; Santosa, D.A.; Timmis, K.N.; Molinari, G. 7- O -Malonyl Macrolactin A, a New Macrolactin Antibiotic from Bacillus Subtilis Active against Methicillin-Resistant Staphylococcus Aureus , Vancomycin-Resistant Enterococci, and a Small-Colony Variant of Burkholderia Cepacia. Antimicrob. Agents Chemother. 2006, 50, 1701–1709. [CrossRef]
- Schneider, K.; Chen, X.-H.; Vater, J.; Franke, P.; Nicholson, G.; Borriss, R.; Süssmuth, R.D. Macrolactin Is the Polyketide Biosynthesis Product of the Pks2 Cluster of Bacillus Amyloliquefaciens FZB42. J. Nat. Prod. 2007, 70, 1417–1423. [CrossRef]
- Xiao, F.; Dong, S.; Liu, Y.; Feng, Y.; Li, H.; Yun, C.-H.; Cui, Q.; Li, W. Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase. J. Am. Chem. Soc. 2020, 142, 16031–16038. [CrossRef]
- Bae, S.H.; Kwon, M.J.; Park, J.B.; Kim, D.; Kim, D.-H.; Kang, J.-S.; Kim, C.-G.; Oh, E.; Bae, S.K. Metabolic Drug-Drug Interaction Potential of Macrolactin A and 7- O -Succinyl Macrolactin A Assessed by Evaluating Cytochrome P450 Inhibition and Induction and UDP-Glucuronosyltransferase Inhibition In Vitro. Antimicrob. Agents Chemother. 2014, 58, 5036–5046. [CrossRef]
- Kim, J.M.; Jung, J.W.; Kim, D.H.; Kang, J.S.; Kim, C.G.; Kang, H.E. A Simple and Sensitive HPLC-UV Determination of 7- O -succinyl Macrolactin A in Rat Plasma and Urine and Its Application to a Pharmacokinetic Study. Biomed. Chromatogr. 2013, 27, 273–279. [CrossRef]
- Jung, J.W.; Kim, J.M.; Kwon, M.H.; Kim, D.H.; Kang, H.E. Pharmacokinetics of Macrolactin A and 7- O -Succinyl Macrolactin A in Mice. Xenobiotica 2014, 44, 547–554. [CrossRef]
- Salazar, F.; Ortiz, A.; Sansinenea, E. A Strong Antifungal Activity of 7-O-Succinyl Macrolactin A vs Macrolactin A from Bacillus Amyloliquefaciens ELI149. Curr. Microbiol. 2020, 77, 3409–3413. [CrossRef]
- Kim, D.H.; Kim, H.K.; Kim, K.M.; Kim, C.K.; Jeong, M.H.; Ko, C.Y.; Moon, K.H.; Kang, J.S. Antibacterial Activities of Macrolactin a and 7-O-Succinyl Macrolactin a from Bacillus Polyfermenticus KJS-2 against Vancomycin-Resistant Enterococci and Methicillin-Resistant Staphylococcus Aureus. Arch. Pharm. Res. 2011, 34, 147–152. [CrossRef]
- Nagao, T.; Adachi, K.; Sakai, M.; Nishijima, M.; Sano, H. Novel Macrolactins as Antibiotic Lactones from a Marine Bacterium. J. Antibiot. (Tokyo) 2001, 54, 333–339. [CrossRef]
- Vasilchenko, A.S.; Lukyanov, D.A.; Dilbaryan, D.S.; Usachev, K.S.; Poshvina, D.V.; Taldaev, A.K.; Nikandrova, A.A.; Imamutdinova, A.N.; Garaeva, N.S.; Bikmullin, A.G.; et al. Macrolactin a Is an Inhibitor of Protein Biosynthesis in Bacteria. Biochimie 2025, 232, 25–34. [CrossRef]
- Yoo, J.-S.; Zheng, C.-J.; Lee, S.; Kwak, J.-H.; Kim, W.-G. Macrolactin N, a New Peptide Deformylase Inhibitor Produced by Bacillus Subtilis. Bioorg. Med. Chem. Lett. 2006, 16, 4889–4892. [CrossRef]
- Sohn, M.-J.; Zheng, C.-J.; Kim, W.-G. Macrolactin S, a New Antibacterial Agent with Fab G-Inhibitory Activity from Bacillus Sp. AT28. J. Antibiot. (Tokyo) 2008, 61, 687–691. [CrossRef]
- Kalai, K.; Rufus, A.C.; Manz, A.M.; Elangovan, E. Unveiling the Antimicrobial Potential of 7-O-Succinyl Macrolactin F from Bacillus Subtilis Group against HtsA Siderophore Receptor of Staphylococcus Aureus: A Computational Exploration. Biomed. Biotechnol. Res. J. 2024, 8, 92–99. [CrossRef]
- Terekhov, S.S.; Smirnov, I.V.; Malakhova, M.V.; Samoilov, A.E.; Manolov, A.I.; Nazarov, A.S.; Danilov, D.V.; Dubiley, S.A.; Osterman, I.A.; Rubtsova, M.P.; et al. Ultrahigh-Throughput Functional Profiling of Microbiota Communities. Proc. Natl. Acad. Sci. 2018, 115, 9551–9556. [CrossRef]
- Duddeck, H. E. Pretsch, P. Bühlmann, C. Affolter. Structure Determination of Organic Compounds—Tables of Spectra Data. Springer, Berlin, 2000. 421 Pp. plus CD-ROM. Price £ 40.39, DM 79.00. ISBN 3 540 67815 8. Magn. Reson. Chem. 2002, 40, 247–247. [CrossRef]
- Bacillus Subtilis CH13: A Highly Effective Biocontrol Agent for the Integrated Management of Plant Diseases. IOBC-WPRS.
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [CrossRef]
- Zdouc, M.M.; Blin, K.; Louwen, N.L.L.; Navarro, J.; Loureiro, C.; Bader, C.D.; Bailey, C.B.; Barra, L.; Booth, T.J.; Bozhüyük, K.A.J.; et al. MIBiG 4.0: Advancing Biosynthetic Gene Cluster Curation through Global Collaboration. Nucleic Acids Res. 2025, 53, D678–D690. [CrossRef]
- Chen, X.-H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and Functional Characterization of Three Polyketide Synthase Gene Clusters in Bacillus Amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative Analysis of the Complete Genome Sequence of the Plant Growth–Promoting Bacterium Bacillus Amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [CrossRef]
- Liu, Y.; Qin, W.; Liu, Q.; Zhang, J.; Li, H.; Xu, S.; Ren, P.; Tian, L.; Li, W. Genome-wide Identification and Characterization of Macrolide Glycosyltransferases from a Marine-derived Bacillus Strain and Their Phylogenetic Distribution. Environ. Microbiol. 2016, 18, 4770–4781. [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [CrossRef]
- Foster, T.J. Antibiotic Resistance in Staphylococcus Aureus. Current Status and Future Prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [CrossRef]
- Matsumoto, K.; Kawabata, Y.; Okada, S.; Takahashi, J.; Hashimoto, K.; Nagai, Y.; Tatsuta, J.; Hatanaka, M. Enantioselective Microbial Oxidation of Allyl Alcohols. Chem. Lett. 2007, 36, 1428–1429. [CrossRef]
- Wilson, D.N. Ribosome-Targeting Antibiotics and Mechanisms of Bacterial Resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [CrossRef]
- ADH-Catalyzed Biooxidation of (Hetero)Aromatic sec-Alcohols to Ketones Employing Vinyl Acetate as Acetaldehyde Surrogate - Rudzka - 2024 - ChemCatChem - Wiley Online Library Available online: https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cctc.202400803 (accessed on 12 April 2025).
- Gandomkar, S.; Jost, E.; Loidolt, D.; Swoboda, A.; Pickl, M.; Elaily, W.; Daniel, B.; Fraaije, M.W.; Macheroux, P.; Kroutil, W. Biocatalytic Enantioselective Oxidation of Sec -Allylic Alcohols with Flavin-Dependent Oxidases. Adv. Synth. Catal. 2019, 361, 5264–5271. [CrossRef]
- Babraham Bioinformatics - FastQC A Quality Control Tool for High Throughput Sequence Data Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 May 2025).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [CrossRef]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing Polished Prokaryotic Pangenomes with the Panaroo Pipeline. Genome Biol. 2020, 21, 180. [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-Sites: Rapid Efficient Extraction of SNPs from Multi-FASTA Alignments. Microb. Genomics 2016, 2. [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [CrossRef]
- Ggplot2: Elegant Graphics for Data Analysis | SpringerLink Available online: https://link.springer.com/book/10.1007/978-0-387-98141-3 (accessed on 16 May 2025).
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 2004, 20, 289–290. [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Markossian, S., Grossman, A., Arkin, M., Auld, D., Austin, C., Baell, J., Brimacombe, K., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., Devanarayan, V., Foley, T.L., Glicksman, M., Gorshkov, K., Haas, J.V., Hall, M.D., Hoare, S., Inglese, J., Iversen, P.W., Lal-Nag, M., Li, Z., Manro, J.R., McGee, J., McManus, O., Pearson, M., Riss, T., Saradjian, P., Sittampalam, G.S., Tarselli, M., Trask, O.J., Weidner, J.R., Wildey, M.J., Wilson, K., Xia, M., Xu, X., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD), 2004.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




