Submitted:
01 September 2025
Posted:
02 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
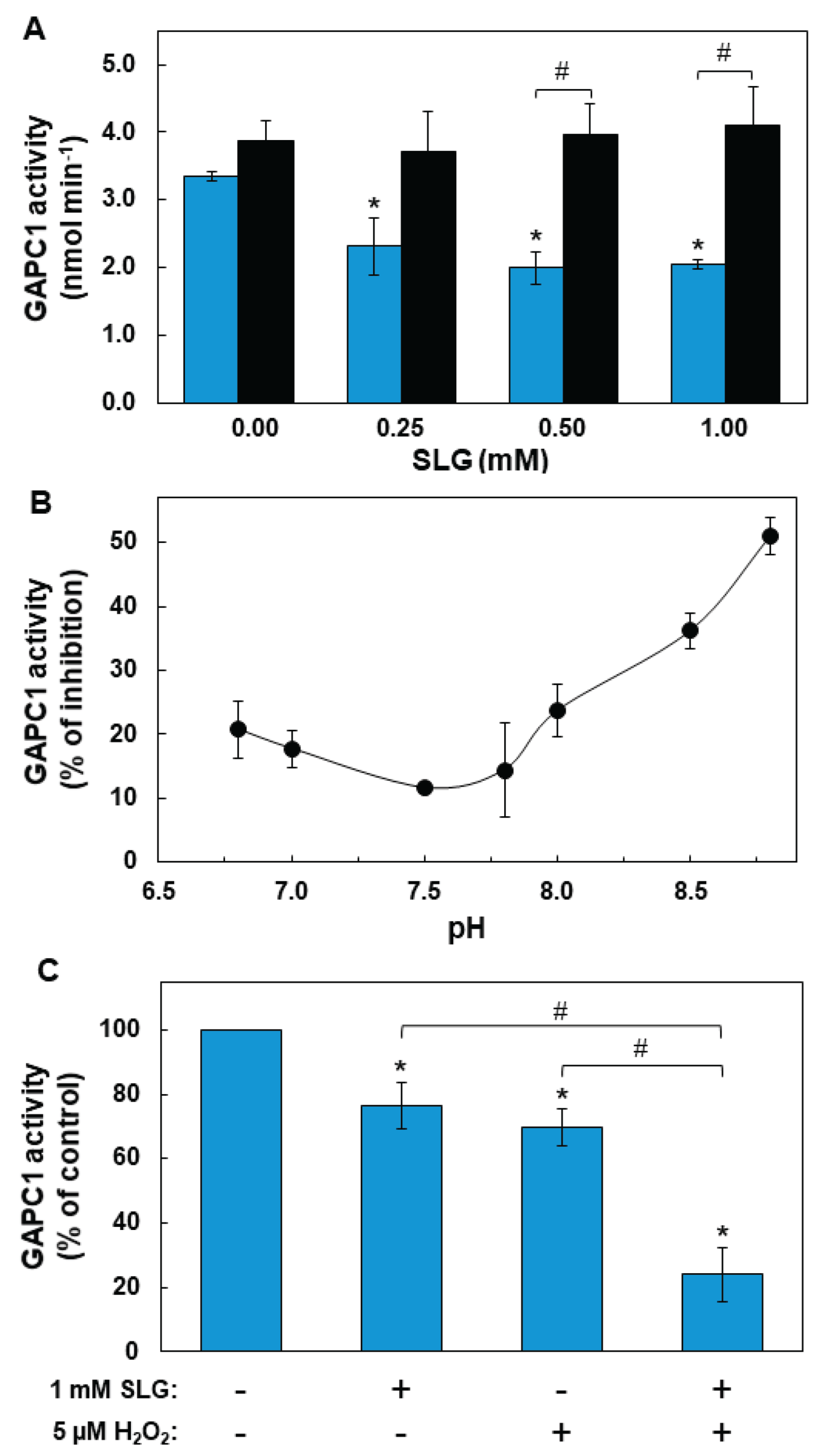
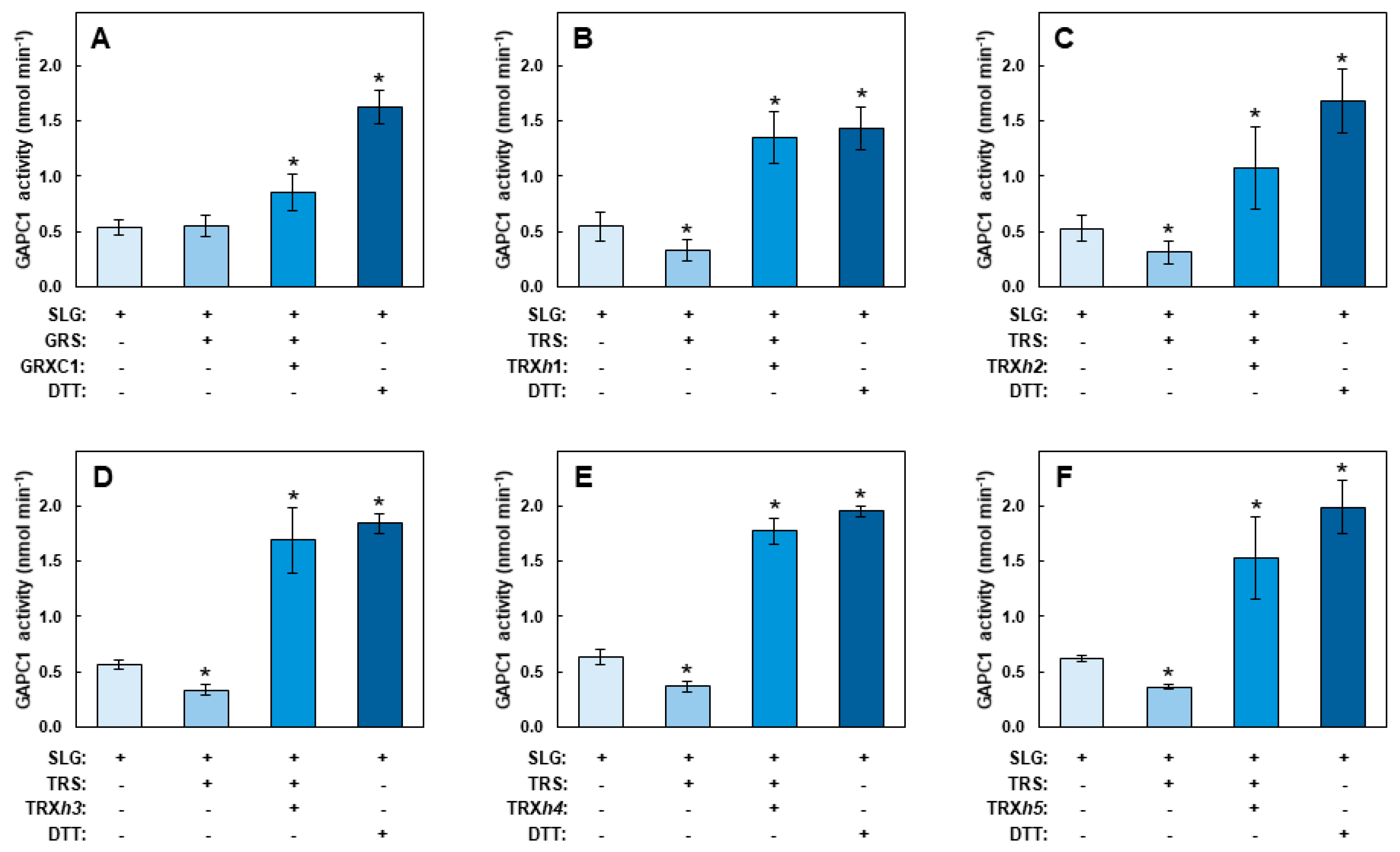
2.1. Inhibition of GAPC1 Activity by SLG and Factors Modulating this Effect
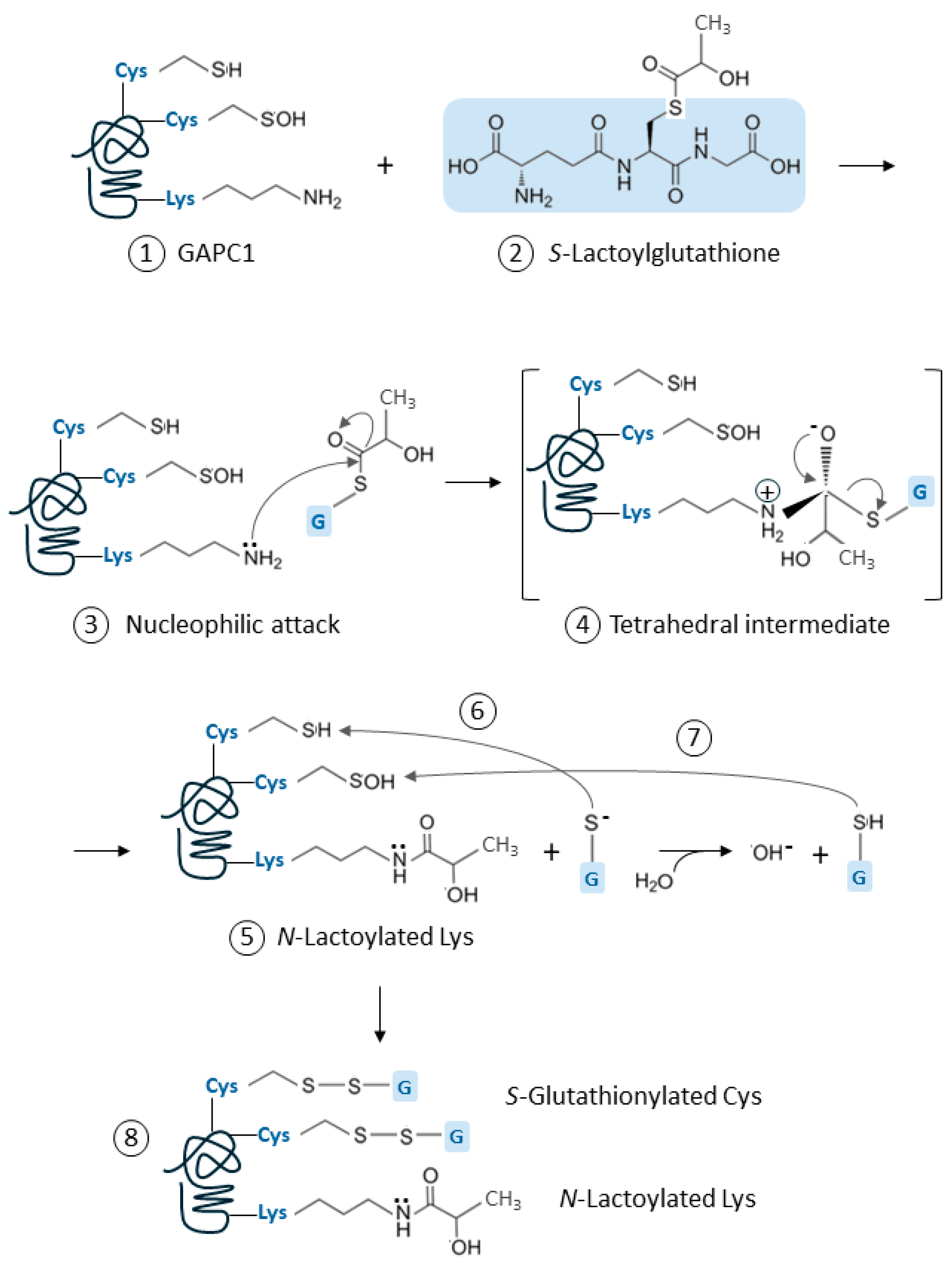
2.2. Inhibition of GAPC1 by SLG is Associated with Covalent Modifications
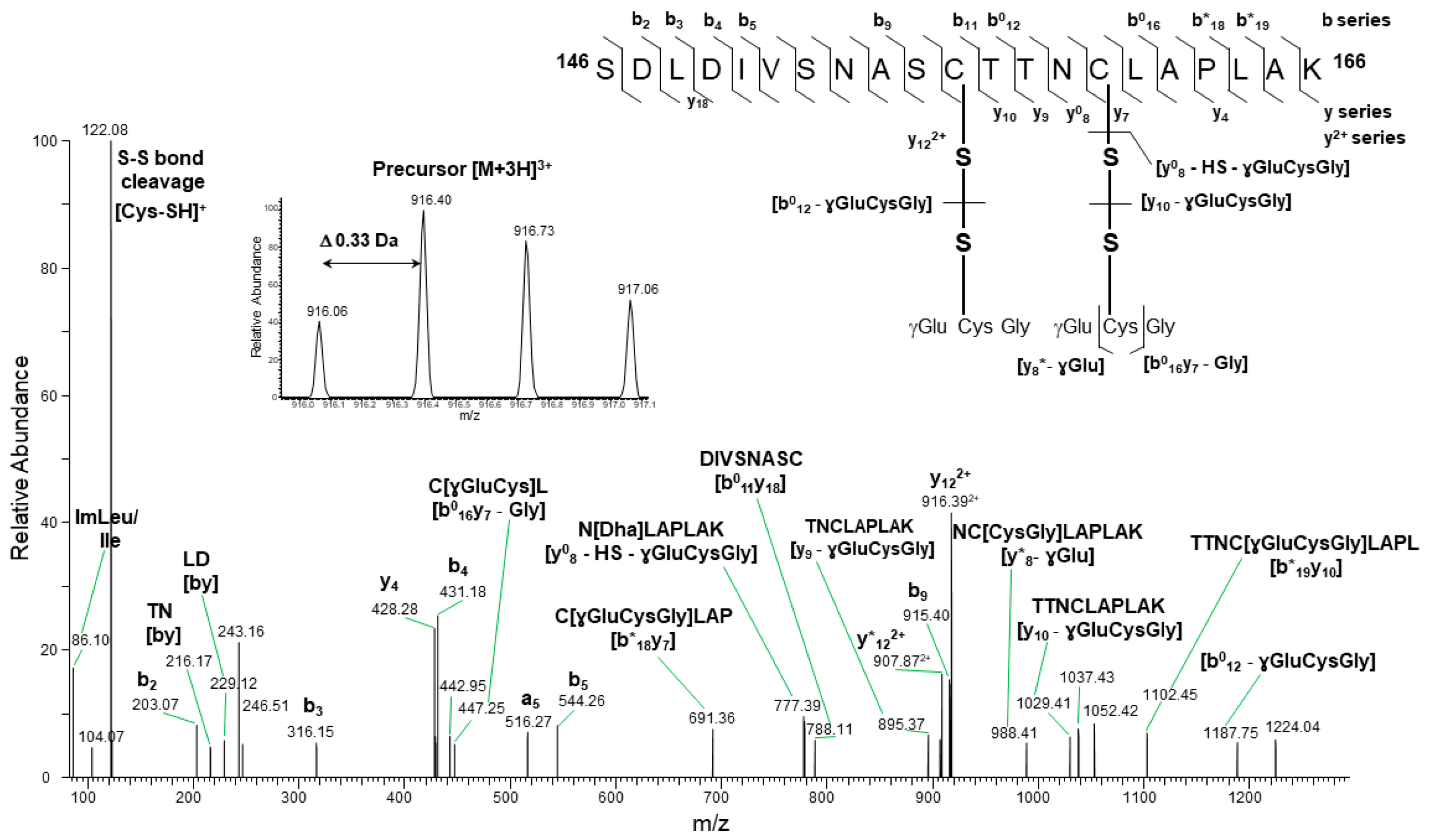
2.2.1. SLG Induces GAPC1 S-Glutathionylation
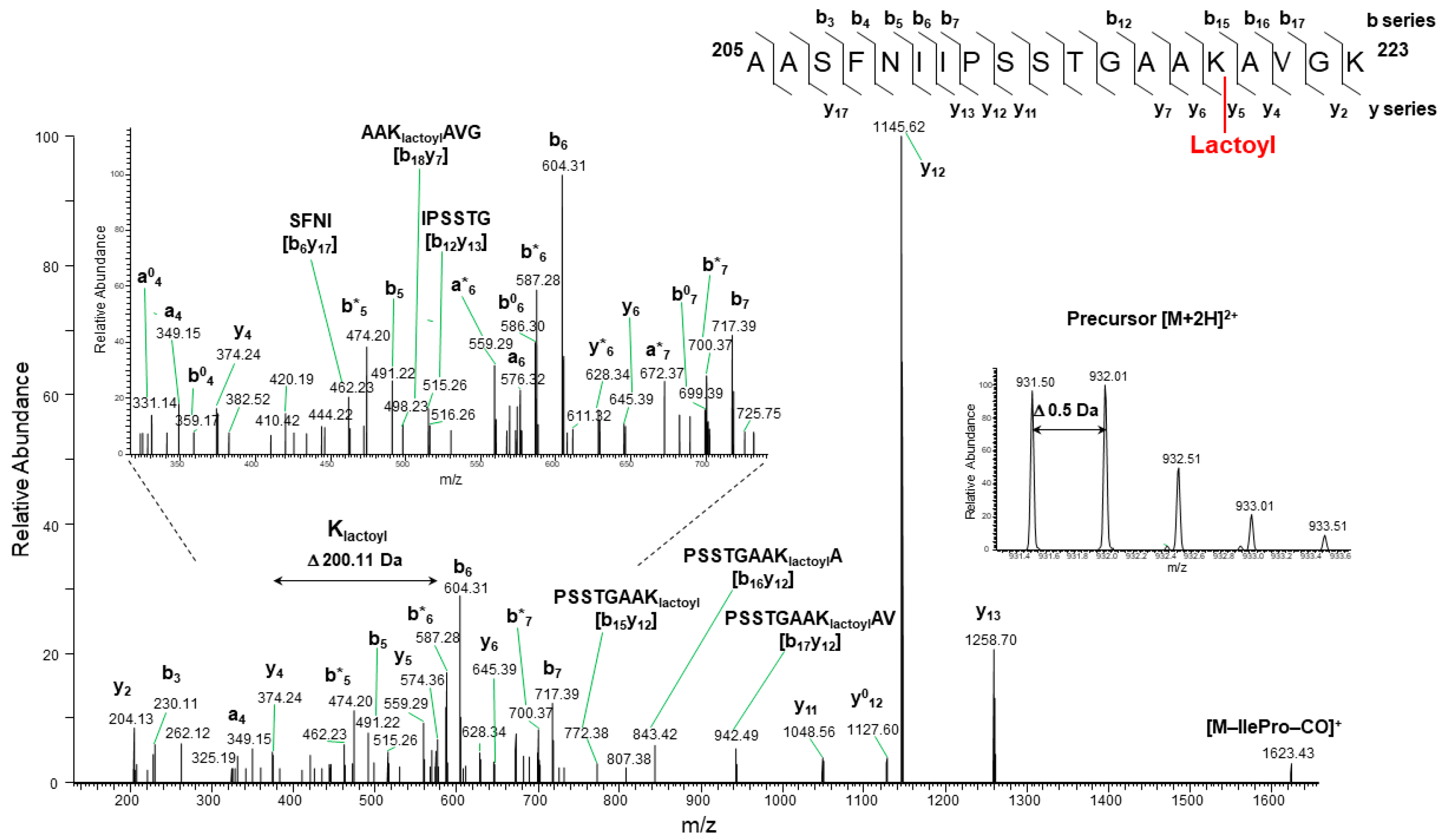
2.2.2. SLG Also Induces Lys N-Lactoylation on GAPC1
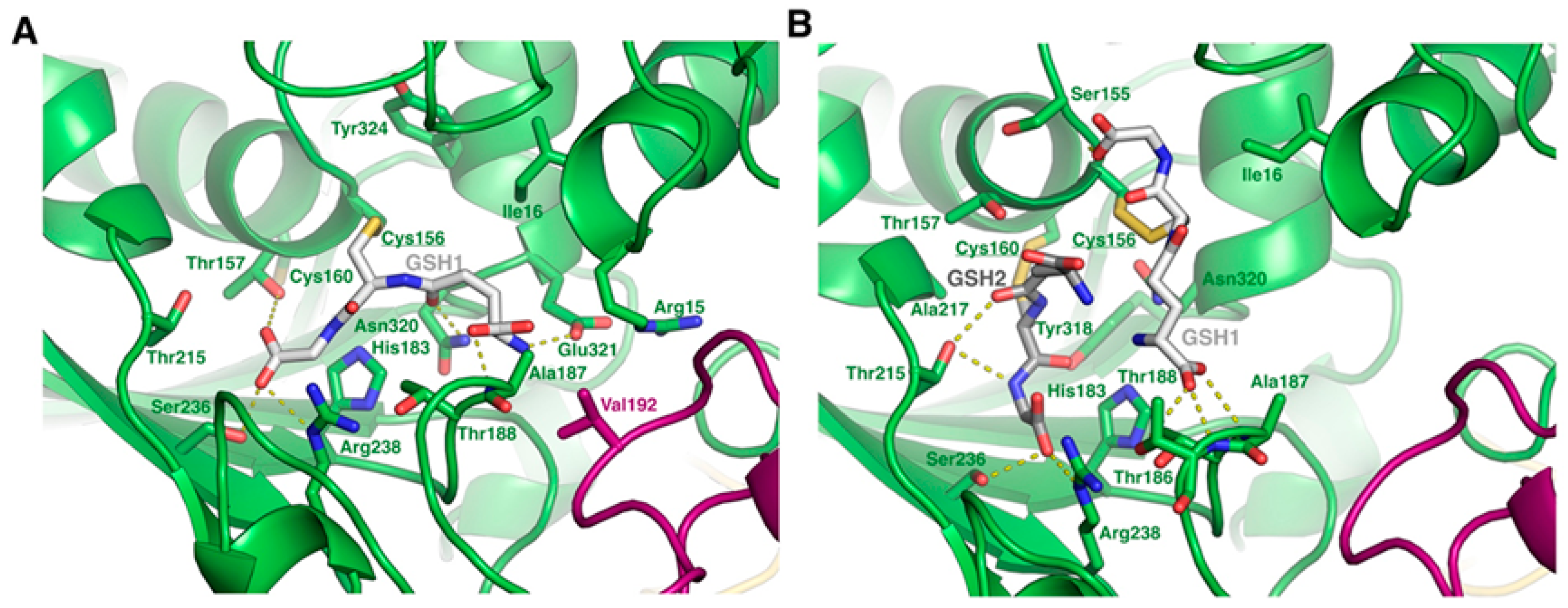
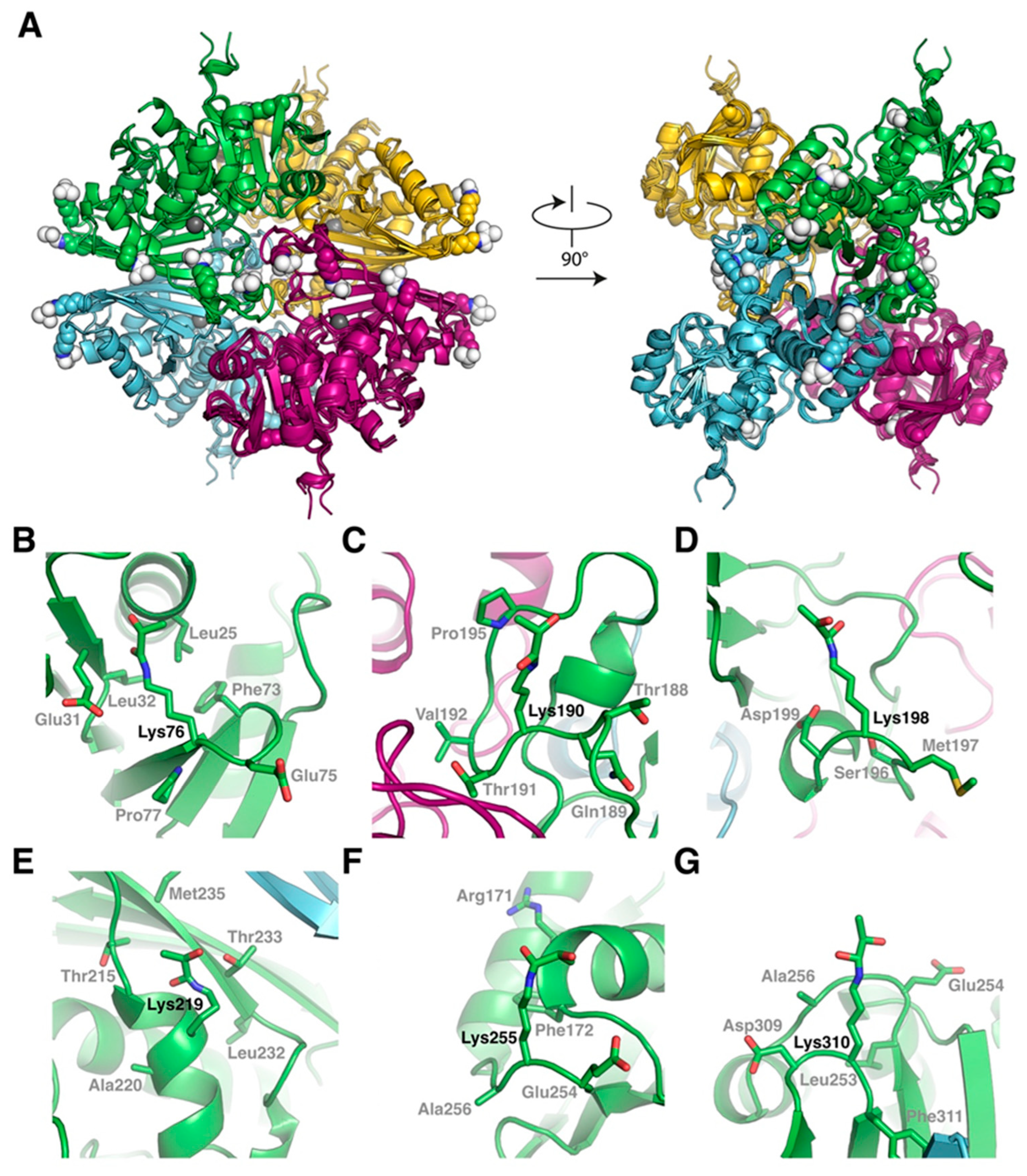
2.3. Modeling of the SLG-Induced S-Glutathionylation Pattern on the GAPC1 Structure
2.4. Modeling of N-Lactoylated GAPC1 and Sequence Features Around N-Lactoylated Lys Residues
2.5. SLG Inhibition of GAPC1 is Reversible In Vitro by Treatments with Redoxins
3. Discussion
3.1. Inhibition of GAPC1 in the Presence of SLG is Reversed by DTT
3.2. SLG Causes Spontaneous Lys N-Lactoylation and Cys S-Glutathionylation of GAPC1
3.3. SLG-Induced Inhibition of GAPC1 Can Be Reversed by GRXC1 and TRXs
3.4. Physiological Relevance of SLG-Dependent PTMs of GAPC1
4. Materials and Methods
4.1. Chemicals
4.2. Plasmid Constructions
4.3. Production, Purification and Quantification of Recombinant Enzymes
4.4. Redox Treatments of Proteins
4.5. Enzyme Activity Measurements
4.6. Sample Preparation and Nano LC-MS/MS Proteomic Analysis
4.7. Proteomics Data Analysis and Interpretation
4.8. Structural Prediction of GAPC1 Structures
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GAPC1 | cytosolic glyceraldehyde 3P dehydrogenase 1 |
| ROS | reactive oxygen species |
| MG | methylglyoxal |
| PTM | post-translational protein modification |
| GSH | glutathione |
| GSSG | oxidized glutathione |
| SLG | S,D-lactoylglutathione |
| GLO1 | glyoxalase I |
| GLO2 | glyoxalase II |
| GAPDH | glyceraldehyde 3P dehydrogenase |
| MDH | malate dehydrogenase |
| DTT | dithiotreitol |
| GRX | Glutaredoxin |
| TRX | thioredoxin |
| GR | NADPH-dependent glutathione reductase |
| NTR | NADPH-dependent thioredoxin reductase |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| FWHM | full width at half-maximum |
| HCD | Higher-energy Collision Dissociation |
| Dha | dehydroalanine |
| GRS | glutaredoxin recycling system |
| TRS | thioredoxin recycling system |
References
- Mareri, L.; Parrotta, L.; Cai, G. Environmental Stress and Plants. Int. J. Mol. Sci. 2022, 23, 5416. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S.; Hashem, A.; Abd_Allah, E.F.; Ansari, M.I. Plant Defense Responses to Biotic Stress and Its Interplay With Fluctuating Dark/Light Conditions. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nature Reviews Genetics 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione metabolism in plants under stress: Beyond reactive oxygen species detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef]
- Foyer, C.H.; Kunert, K. The ascorbate–glutathione cycle coming of age. J. Exp. Bot. 2024, 75, 2682–2699. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Ghosh, A.; Li, Z.G.; Siddiqui, M.N.; Fujita, M.; Tran, L.S.P. Methylglyoxal - a signaling molecule in plant abiotic stress responses. Free Radical Biology and Medicine 2018, 122, 96–109. [Google Scholar] [CrossRef]
- Singh, D. Juggling with reactive oxygen species and antioxidant defense system – A coping mechanism under salt stress. Plant Stress 2022, 5, 100093. [Google Scholar] [CrossRef]
- Boutin, C.; Clément, C.; Rivoal, J. Post-Translational Modifications to Cysteine Residues in Plant Proteins and Their Impact on the Regulation of Metabolism and Signal Transduction. Int. J. Mol. Sci. 2024, 25, 9845. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Rossi, R.; Colombo, G.; Giustarini, D.; Milzani, A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem.Sci. 2009, 34, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ye, Z.-w.; Singh, S.; Townsend, D.M.; Tew, K.D. An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radical Biology and Medicine 2018, 120, 204–216. [Google Scholar] [CrossRef]
- van der Linde, K.; Gutsche, N.; Leffers, H.-M.; Lindermayr, C.; Müller, B.; Holtgrefe, S.; Scheibe, R. Regulation of plant cytosolic aldolase functions by redox modifications. Plant Physiology and Biochemistry 2011, 49, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Bedhomme, M.; Adamo, M.; Marchand, C.H.; Couturier, J.; Rouhier, N.; Lemaire, S.D.; Zaffagnini, M.; Trost, P. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 2012, 445, 337–347. [Google Scholar] [CrossRef]
- Dumont, S.; Bykova, N.V.; Pelletier, G.; Dorion, S.; Rivoal, J. Cytosolic triosephosphate isomerase from Arabidopsis thaliana is reversibly modified by glutathione on cysteines 127 and 218. Front. Plant Sci. 2016, 7, 1942. [Google Scholar] [CrossRef]
- Bender, Kyle W. ; Wang, X.; Cheng, George B.; Kim, Hyoung S.; Zielinski, Raymond E.; Huber, Steven C. Glutaredoxin AtGRXC2 catalyses inhibitory glutathionylation of Arabidopsis BRI1-associated receptor-like kinase 1 (BAK1) in vitro. Biochem. J. 2015, 467, 399–413. [Google Scholar] [CrossRef]
- Chan, K.X.; Mabbitt, P.D.; Phua, S.Y.; Mueller, J.W.; Nisar, N.; Gigolashvili, T.; Stroeher, E.; Grassl, J.; Arlt, W.; Estavillo, G.M.; et al. Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E4567–E4576. [Google Scholar] [CrossRef]
- Gurrieri, L.; Distefano, L.; Pirone, C.; Horrer, D.; Seung, D.; Zaffagnini, M.; Rouhier, N.; Trost, P.; Santelia, D.; Sparla, F. The thioredoxin-regulated α-amylase 3 of Arabidopsis thaliana is a target of S-glutathionylation. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Thornalley, P.J. Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems-role in ageing and disease. Drug Metabol. Drug Interact. 2008, 23, 125–150. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. The glyoxalase system - new developments towards functional-characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990, 269, 1–11. [Google Scholar] [CrossRef]
- Deswal, R.; Chakaravarty, T.N.; Sopory, S.K. The glyoxalase system in higher plants: Regulation in growth and differentiation. Biochem. Soc. Trans. 1993, 21, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Maiti, M.K.; Krishnasamy, S.; Owen, H.A.; Makaroff*, C.A. Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol. Biol. 1997, 35, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Engqvist, M.; Drincovich, M.F.; Flugge, U.I.; Maurino, V.G. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J. Biol. Chem. 2009, 284, 25026–25037. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, D.O.; Jennings, E.Q.; Anderson, C.C.; Marentette, J.O.; Shi, T.; Schou Oxvig, A.-M.; Streeter, M.D.; Johannsen, M.; Spiegel, D.A.; Chapman, E.; et al. Non-enzymatic Lysine Lactoylation of Glycolytic Enzymes. Cell Chemical Biology 2020, 27, 206–213.e206. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M.N.; Jennings, E.Q.; Hoffman, E.A.; Zhang, H.; Phoebe, A.M.; Mastin, G.E.; Kitamura, N.; Reisz, J.A.; Megill, E.; Kantner, D.; et al. Lactoylglutathione promotes inflammatory signaling in macrophages through histone lactoylation. Molecular Metabolism 2023, 81. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Zhao, W.; Xin, J.; Yu, X.; Li, Z.; Li, N. Recent advances of lysine lactylation in prokaryotes and eukaryotes. Frontiers in Molecular Biosciences 2025, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hao, D.; Zhao, S.; Zhang, Z.; Zeng, Z.; Wang, X. Lactate and Lactylation: Clinical Applications of Routine Carbon Source and Novel Modification in Human Diseases. Mol. Cell. Proteomics 2023, 22, 100641. [Google Scholar] [CrossRef]
- Ercolani, L.; Scirè, A.; Galeazzi, R.; Massaccesi, L.; Cianfruglia, L.; Amici, A.; Piva, F.; Urbanelli, L.; Emiliani, C.; Principato, G.; et al. A possible S-glutathionylation of specific proteins by glyoxalase II: An in vitro and in silico study. Cell Biochemistry and Function 2016, 34, 620–627. [Google Scholar] [CrossRef]
- Holtgrefe, S.; Gohlke, J.; Starmann, J.; Druce, S.; Klocke, S.; Altmann, B.; Wojtera, J.; Lindermayr, C.; Scheibe, R. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 2008, 133, 211–228. [Google Scholar] [CrossRef]
- Dumont, S.; Rivoal, J. Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front. Plant Sci. 2019, 10, 166. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Marchand, C.H.; Malferrari, M.; Murail, S.; Bonacchi, S.; Genovese, D.; Montalti, M.; Venturoli, G.; Falini, G.; Baaden, M.; et al. Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 26057–26065. [Google Scholar] [CrossRef] [PubMed]
- Michalski, A.; Neuhauser, N.; Cox, J.; Mann, M. A Systematic Investigation into the Nature of Tryptic HCD Spectra. J. Proteome Res. 2012, 11, 5479–5491. [Google Scholar] [CrossRef] [PubMed]
- Saminathan, I.S.; Wang, X.S.; Guo, Y.; Krakovska, O.; Voisin, S.; Hopkinson, A.C.; Siu, K.W.M. The extent and effects of peptide sequence scrambling via formation of macrocyclic b ions in model proteins. J. Am. Soc. Mass Spectrom. 2010, 21, 2085–2094. [Google Scholar] [CrossRef]
- Su, D.; Gaffrey, M.J.; Guo, J.; Hatchell, K.E.; Chu, R.K.; Clauss, T.R.W.; Aldrich, J.T.; Wu, S.; Purvine, S.; Camp, D.G.; et al. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radical Biology and Medicine 2014, 67, 460–470. [Google Scholar] [CrossRef]
- Li, X.; Day, N.J.; Feng, S.; Gaffrey, M.J.; Lin, T.-D.; Paurus, V.L.; Monroe, M.E.; Moore, R.J.; Yang, B.; Xian, M.; et al. Mass spectrometry-based direct detection of multiple types of protein thiol modifications in pancreatic beta cells under endoplasmic reticulum stress. Redox Biology 2021, 46, 102111. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Huang, J.J.; Willems, P.; Van Breusegem, F.; Messens, J. Pathways crossing mammalian and plant sulfenomic landscapes. Free Radical Biology and Medicine 2018, 122, 193–201. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Manta, B.; Botti, H.; Radi, R.; Trujillo, M.; Denicola, A. Factors Affecting Protein Thiol Reactivity and Specificity in Peroxide Reduction. Chem. Res. Toxicol. 2011, 24, 434–450. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Fermani, S.; Calvaresi, M.; Orrù, R.; Iommarini, L.; Sparla, F.; Falini, G.; Bottoni, A.; Trost, P. Tuning Cysteine Reactivity and Sulfenic Acid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana. Antioxidants & Redox Signaling 2016, 24, 502–517. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Michelet, L.; Marchand, C.; Sparla, F.; Decottignies, P.; Le Marechal, P.; Miginiac-Maslow, M.; Noctor, G.; Trost, P.; Lemaire, S.D. The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 2007, 274, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J. Mechanisms for reactions, nucleophilic substitution at an acyl group. In Structure in protein chemistry, 2nd ed.; eScholarship Repository, University of California, 2024; pp. 39–54. [Google Scholar]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nϵ-acetylation and Nϵ-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef]
- Grek, C.L.; Zhang, J.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504. [Google Scholar] [CrossRef]
- Matsui, R.; Ferran, B.; Oh, A.; Croteau, D.; Shao, D.; Han, J.; Pimentel, D.R.; Bachschmid, M.M. Redox Regulation via Glutaredoxin-1 and Protein S-Glutathionylation. Antioxidants & Redox Signaling 2020, 32, 677–700. [Google Scholar] [CrossRef]
- Caccuri, A.M.; Antonini, G.; Board, P.G.; Parker, M.W.; Nicotra, M.; Lo Bello, M.; Federici, G.; Ricci, G. Proton release on binding of glutathione to alpha, Mu and Delta class glutathione transferases. Biochem. J. 1999, 344 Pt 2, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Orozco, M.; Vega, C.; Parraga, A.; GarciaSaez, I.; Coll, M.; Walsh, S.; Mantle, T.J.; Luque, F.J. On the reaction mechanism of class pi glutathione S-transferase. Proteins-Structure Function and Genetics 1997, 28, 530–542. [Google Scholar] [CrossRef]
- Townsend, D.M.; Manevich, Y.; He, L.; Hutchens, S.; Pazoles, C.J.; Tew, K.D. Novel role for glutathione S-transferase Pi regulator of protein s-glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 2009, 284, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Zhou, D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017, 45, 12241–12255. [Google Scholar] [CrossRef]
- Liu, Z.; Song, J.; Miao, W.; Yang, B.; Zhang, Z.; Chen, W.; Tan, F.; Suo, H.; Dai, X.; Zou, X.; et al. Comprehensive Proteome and Lysine Acetylome Analysis Reveals the Widespread Involvement of Acetylation in Cold Resistance of Pepper (Capsicum annuum L.). Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Finkemeier, I.; Laxa, M.; Miguet, L.; Howden, A.J.M.; Sweetlove, L.J. Proteins of Diverse Function and Subcellular Location Are Lysine Acetylated in Arabidopsis. Plant Physiol. 2011, 155, 1779–1790. [Google Scholar] [CrossRef]
- Yang, S.S.; Zhai, Q.H. Cytosolic GAPDH: a key mediator in redox signal transduction in plants. Biol. Plant. 2017, 61, 417–426. [Google Scholar] [CrossRef]
- Guo, L.; Devaiah, S.P.; Narasimhan, R.; Pan, X.; Zhang, Y.; Zhang, W.; Wang, X. Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with Phospholipase D+¦ to Transduce Hydrogen Peroxide Signals in the Arabidopsis Response to Stress. Plant Cell 2012, 24, 2200–2212. [Google Scholar] [CrossRef]
- Henry, E.; Fung, N.; Liu, J.; Drakakaki, G.; Coaker, G. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015, 11, e1005199. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-C.; Guo, L.; Wang, X. Nuclear moonlighting of cytosolic glyceraldehyde-3-phosphate dehydrogenase regulates Arabidopsis response to heat stress. Nature Communications 2020, 11, 3439. [Google Scholar] [CrossRef] [PubMed]
- Wegener, M.; Persicke, M.; Dietz, K.-J. Reprogramming the translatome during daily light transitions as affected by cytosolic glyceraldehyde-3-phosphate dehydrogenases GAPC1/C2. J. Exp. Bot. 2023, 75, 2494–2509. [Google Scholar] [CrossRef]
- Peralta, D.A.; Araya, A.; Busi, M.V.; Gomez-Casati, D.F. The E3 ubiquitin-ligase SEVEN IN ABSENTIA like 7 mono-ubiquitinates glyceraldehyde-3-phosphate dehydrogenase 1 isoform in vitro and is required for its nuclear localization in Arabidopsis thaliana. Int. J. Biochem. Cell Biol. 2016, 70, 48–56. [Google Scholar] [CrossRef]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Singh, S.P. New insights on thioredoxins (Trxs) and glutaredoxins (Grxs) by in silico amino acid sequence, phylogenetic and comparative structural analyses in organisms of three domains of life. Heliyon 2022, 8, e10776. [Google Scholar] [CrossRef]
- Chibani, K.; Pucker, B.; Dietz, K.-J.; Cavanagh, A. Genome-wide analysis and transcriptional regulation of the typical and atypical thioredoxins in Arabidopsis thaliana. FEBS Lett. 2021, 595, 2715–2730. [Google Scholar] [CrossRef]
- Riondet, C.; Desouris, J.P.; Guilleminot-Montoya, J.; Chartier, Y.; Meyer, Y.; Reichheld, J.-P. A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant, Cell & Environment 2012, 35, 360–373. [Google Scholar] [CrossRef]
- Bodnar, Y.; Gellert, M.; Hossain, F.M.; Lillig, C.H. Breakdown of Arabidopsis thaliana thioredoxins and glutaredoxins based on electrostatic similarity-Leads to common and unique interaction partners and functions. PLoS One 2023, 18. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Michelet, L.; Massot, V.; Trost, P.; Lemaire, S.D. Biochemical Characterization of Glutaredoxins from Chlamydomonas reinhardtii Reveals the Unique Properties of a Chloroplastic CGFS-type Glutaredoxin*. J. Biol. Chem. 2008, 283, 8868–8876. [Google Scholar] [CrossRef]
- Singla-Pareek, S.L.; Kaur, C.; Kumar, B.; Pareek, A.; Sopory, S.K. Reassessing plant glyoxalases: large family and expanding functions. New Phytol. 2020, 227, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochemical and Biophysical Research Communications 2005, 337, 61–67. [Google Scholar] [CrossRef]
- Gupta, B.K.; Sahoo, K.K.; Ghosh, A.; Tripathi, A.K.; Anwar, K.; Das, P.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice. Plant, Cell & Environment 2018, 41, 1186–1200. [Google Scholar] [CrossRef]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Hossain, M.A.; Hossain, M.Z.; Fujita, M. Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust. J. Crop Sci. 2009, 3, 53–64. [Google Scholar]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. Identification of a maize kernel stress-related protein and its effect on aflatoxin accumulation. Phytopathology 2004, 94, 938–945. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular cloning: A Laboratory Manual 4th Edition; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, N.Y, 2012; Volume 448. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 1979, 254, 9627–9632. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Wang, M.H. Isolation and characterization of thioredoxin and NADPH-dependent thioredoxin reductase from tomato (Solanum lycopersicum). BMB Rep 2011, 44, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Claeyssen, E.; Dorion, S.; Clendenning, A.; He, J.Z.; Wally, O.; Chen, J.; Auslender, E.L.; Moisan, M.-C.; Jolicoeur, M.; Rivoal, J. The futile cycling of hexose phosphates could account for the fact that hexokinase exerts a high control on glucose phosphorylation but not on glycolytic rate in transgenic potato (Solanum tuberosum) Roots. PLoS One 2013, 8, e53898. [Google Scholar] [CrossRef] [PubMed]
- Lima, D.B.; de Lima, T.B.; Balbuena, T.S.; Neves-Ferreira, A.G.C.; Barbosa, V.C.; Gozzo, F.C.; Carvalho, P.C. SIM-XL: A powerful and user-friendly tool for peptide cross-linking analysis. J. Proteomics 2015, 129, 51–55. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




