Submitted:
30 June 2025
Posted:
30 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Obtaining and Preserving Isolates
2.2. Total DNA Extraction
2.3. Amplification and Sequencing
2.4. Phylogenetic Analyses
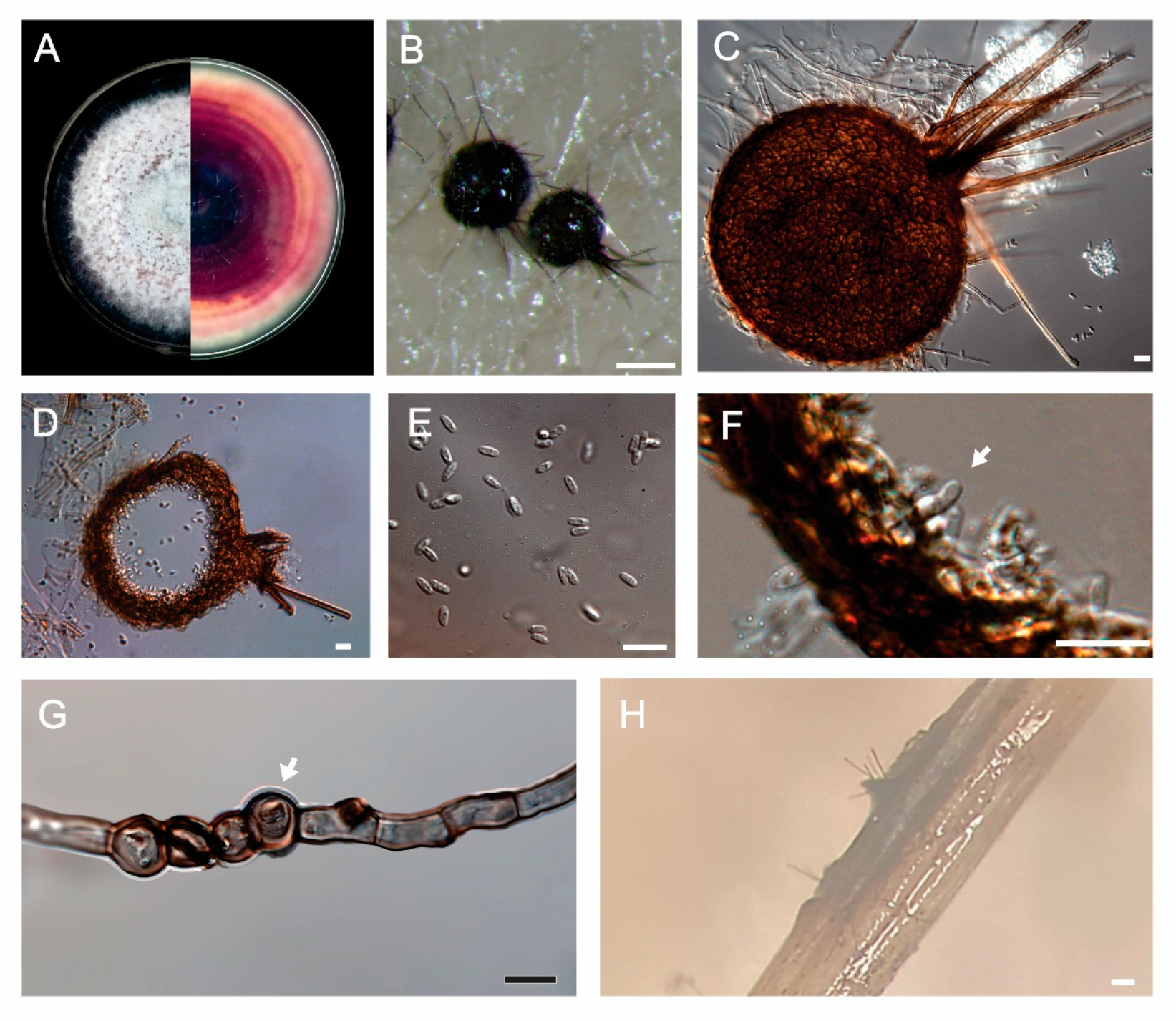
2.5. Morphological Characterization of Setophoma terrestris
2.6. Pathogenicity
3. Results
3.1. Phylogenetic Analysis
3.2. Taxonomy
3.3. Pathogenicity Test
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO - Food and Agriculture Organization. 2024. Faostats – FAO Statistical Databases. Available at: https://www.fao.org/faostat/en/#data [Accessed 11th April 2025].
- Reis, A.; Oliveira, V.R.; Lourenço Junior, V. Identificação e Manejo Da Raiz Rosada Da Cebola. Embrapa: Brasília, DF. 6p. (Comunicado Técnico 114) 2016, 6.
- Sumner, D.R. Pink Root. In Compendium of onion and garlic diseases and pests; Schwartz, H.F., Mohan, K., Eds.; American Phytopathological Society Press: St. Paul, Minnesota, 2008; pp. 18–20. [Google Scholar]
- Taubenhaus, J.J.; Johnson, A.D. Pink Root, a New Root Disease of Onions in Texas. Phytopathology 1917, 7, 59. [Google Scholar]
- Chaves, G.M.; Erickson, H.T. Pyrenochaeta terrestris (Hansen) Gorenz, J. C. Walker y Larson, Um Novo Fungo Da Cebola (Allium cepa L.) Em Minas Gerais. Revista Ceres 1960, 63, 112–114. [Google Scholar]
- Luz, N.L. Raízes Rosadas - Uma Nova Moléstia Da Cebola Para o Rio Grande Do Sul. Revista da Faculdade de Agronomia e Veterinária da Universidade Federal do Rio Grande do Sul 1967, 9, 159–165. [Google Scholar]
- Noda, H. Reação Da Cebola (Allium Cepa L.) a Pyrenochaeta Terrestris (Hansen), Gorenz, Walker e Larson, Escola Superior de Agricultura “Luiz de Queiroz”, Universidade de São Paulo: Piracicaba, 1981.
- Taubenhaus, J.J. PINK ROOT OF ONIONS. Science (1979) 1919, 49, 217–218. [Google Scholar] [CrossRef]
- Taubenhaus, J.J.; Mally, F.W. Pink Root Disease of Onions and Its Control in Texas. Texas. Agricultural Experiment Station. Bulletin 273 1921, 42. [Google Scholar]
- Sideris, C.P. Species of Fusarium Isolated from Onion Roots. Phytopathology 1924, 14, 211–216. [Google Scholar]
- Sideris, C.P. Species of Fusarium Isolated from Onion Roots. Phytopathology 1929, 19, 233–268. [Google Scholar]
- Hansen, H.N. “Pink-Root” Of Onions Caused By Phoma sp. Science (1979) 1926, 64, 525–525. [Google Scholar] [CrossRef]
- Hansen, H.N. Etiology of the Pink Root Disease of Onions. Phytopathology 1929, 19, 691–704. [Google Scholar]
- Du Plessis, S.J. Pink Root and Bulbrot of Onions; Union of South Africa: Pretoria, 1933; Vol. 124. [Google Scholar]
- Tims, E.C. Pink Root of Shallots, Allium ascalonicum. Plant Disease Reports 1953, 37, 533–537. [Google Scholar]
- Watson, R.D. Rapid Identification of the Onion Pink Root Fungus. Plant Disease Reports 1961, 45, 289. [Google Scholar]
- Kodama, F.; Sugawara, Y.; Yokoyama, T. Pink Root Rot of Onion Caused by Pyrenochaeta terrestris. Phytopathological Society of Japan 1976, 42, 320–321. [Google Scholar] [CrossRef]
- Awuah, R.T.; Lorbeer, J.W. A Procedure for Isolating Pyrenochaeta terrestris from Onion Roots. Annals of Applied Biology 1989, 114, 205–208. [Google Scholar] [CrossRef]
- Yoshida, N. Seasonal Dynamics of the Pink Root Fungus (Setophoma terrestris) in Rhizosphere Soil: Effect of Crop Species and Rotation. Plant Pathol 2022, 71, 361–372. [Google Scholar] [CrossRef]
- Gorenz, A.M.; Larson, R.H.; Walker, J.C. Factors Affecting Pathogenicity of Pink Root Fungus of Onions. J Agric Res 1949, 78, 1–18. [Google Scholar]
- Gorenz, A.M.; Walker, J.C.; Lardon, R.H. Morphology and Taxonomy of the Onion Pink-Root Fungus. Phytopathology 1948, 38, 831–840. [Google Scholar]
- de Gruyter, J.; Woudenberg, J.H.C.; Aveskamp, M.M.; Verkley, G.J.M.; Groenewald, J.Z.; Crous, P.W. Systematic Reappraisal of Species in Phoma Section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 2010, 102, 1066–1081. [Google Scholar] [CrossRef]
- Biles, C.L.; Holland, M.; Ulloa-Godinez, M.; Clason, D.; Corgan, J. Pyrenochaeta terrestris Microsclerotia Production and Pigmentation on Onion Roots. HortScience 1992, 27, 1213–1216. [Google Scholar] [CrossRef]
- Nalim, F.A.; Samuels, G.J.; Wijesundera, R.L.; Geiser, D.M. New Species from the Fusarium solani Species Complex Derived from Perithecia and Soil in the Old World Tropics. Mycologia 2011, 103, 1302–1330. [Google Scholar] [CrossRef]
- Ellis, M.L.; Paul, P.A.; Dorrance, A.E.; Broders, K.D. Two New Species of Pythium, P. Schmitthenneri and P. Selbyi Pathogens of Corn and Soybean in Ohio. Mycologia 2012, 104, 477–487. [Google Scholar] [CrossRef]
- Liu, F.; Wang, J.; Li, H.; Wang, W.; Cai, L. Setophoma spp. on Camellia sinensis. Fungal Syst Evol 2019. [Google Scholar] [CrossRef] [PubMed]
- Alfenas, A.C.; Ferreira, F.A.; Mafia, R.G.; Gonçalves, R.C. Métodos Em Fitopatologia. In Métodos em Fitopatologia; Alfenas, A.C., Mafia, R.G., Eds.; UFV: Viçosa, 2016; pp. 55–91. [Google Scholar]
- Pinho, D.B.; Firmino, A.L.; Ferreira-Junior, W.G.; Pereira, O.L. An Efficient Protocol for DNA Extraction from Meliolales and the Description of Meliola Centellae Sp. nov. Mycotaxon 2013, 122, 333–345. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two Divergent Intragenomic RDNA ITS2 Types within a Monophyletic Lineage of the Fungus Fusarium Are Nonorthologous. Mol Phylogenet Evol 1997, 7, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J Bacteriol 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; van den Ende, A.H.G.G. Molecular Diagnostics of Clinical Strains of Filamentous Basidiomycetes. Mycoses 1998, 41, 183–189. [Google Scholar] [CrossRef]
- Jacobs, K.; Bergdahl, D.R.; Wingfield, M.J.; Halik, S.; Seifert, K.A.; Bright, D.E.; Wingfield, B.D. Leptographium wingfieldii Introduced into North America and Found Associated with Exotic Tomicus piniperda and Native Bark Beetles. Mycol Res 2004, 108, 411–418. [Google Scholar] [CrossRef]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus Phylogenetics and the Origin of Known, Highly Virulent Pathogens, Inferred from ITS and Glyceraldehyde-3-Phosphate Dehydrogenase Gene Sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic Relationships among Ascomycetes: Evidence from an RNA Polymerse II Subunit. Mol Biol Evol 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Sung, G.-H.; Sung, J.-M.; Hywel-Jones, N.L.; Spatafora, J.W. A Multi-Gene Phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of Localized Incongruence Using a Combinational Bootstrap Approach. Mol Phylogenet Evol 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.-J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a Node or a Foot-Shaped Basal Cell. Stud Mycol 2021, 98, 100116. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModeltest Version 2. Program Distributed by the Author. Evolutionary Biology Center, Uppsala University, Uppsala, 2004. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); IEEE, November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree. 2018. Available at: http://tree.bio.ed.ac.uk/software/figtree/. [Accessed May 14, 2022].
- Crous, P.W.; Phillips, A.J.L.; Baxter, A.P. Phytopathogenic Fungi from South Africa; University of Stellenbosch, Department of Plant Pathology Press: 358, 2000. [Google Scholar]
- Wordell Filho, J.A.; Rowe, E.; Gonçalves, P.A.S.; Debarba, J.F.; Boff, P.; Thomazelli, L.F. Manejo Fitossanitário Na Cultura Da Cebola. In Doenças de origem parasitária; Wordell Filho, J.A., Boff, P., Eds.; Epagri: Florianópolis, 2006; p. 226. [Google Scholar]
- Babadoost, M. Onion Pink Root: Report on Plant Disease. 1990. University of Illinois Extension RPD No. 932. Available at: <https://ipm.illinois.edu/diseases/rpds/932.pdf> [Accessed: March 2, 2021].
- Ikeda, K.; Kuwabara, K.; Urushibara, T.; Soyai, P.; Miki, S.; Shibata, S. Pink Root Rot of Squash Caused by Setophoma Terrestris in Japan. Journal of General Plant Pathology 2012, 78, 372–375. [Google Scholar] [CrossRef]
- Levic, J.; Petrovic, T.; Stankovic, S.; Ivanovic, D. The Incidence of Pyrenochaeta terrestris in Root of Different Plant Species in Serbia. Zb Matice Srp Prir Nauk 2013, 21–30. [Google Scholar] [CrossRef]
- Punithalingram, E.; Holiday, P. CMI Descriptions of Pathogenic Fungi and Bacteria. No. 397. 1973. Available at: https://www.cabi.org/dfb/restricted?target=%2fdfb%2ffulltextpdf%2f2005%2f20056400397.pdf [Accessed: 01 March 2021].
- Yang, Y.; Zuzak, K.; Harding, M.; Neilson, E.; Feindel, D.; Feng, J. First Report of Pink Root Rot Caused by Setophoma (Pyrenochaeta) terrestris on Canola. Canadian Journal of Plant Pathology 2017, 39, 354–360. [Google Scholar] [CrossRef]
- López-López, M.; Léon-Félix, J.; Allende-Molar, R.; Lima, N.B.; Tovar-Pedraza, J.M.; García-Estrada, R.S. First Report of Setophoma terrestris Causing Corky and Pink Root of Tomato in Sinaloa, Mexico. Plant Dis 2020, 104, 1553–1553. [Google Scholar] [CrossRef]
- USDA Fungal Databases. 2024. Available at: https://fungi.ars.usda.gov/ [Accessed May 25th 2025].
- Nico, A.I.; Sánchez, M.G. Response of Different Intermediate-Day Onion Hybrids to Natural Infestation by Phoma terrestris and Fusarium oxysporum f. sp. cepae in Ciudad Real, Spain with Assessment of Different Soil Disinfestation Methods. Eur J Plant Pathol 2012, 134, 783–793. [Google Scholar] [CrossRef]
- Rivera-Méndez, W.; Brenes-Madriz, J.; Alvarado-Marchena, L. Effect of Setophoma terrestris, Sclerotium cepivorum, and Trichoderma spp. on in Vitro Onion (Allium cepa) Root Tissues and the Final Yield at the Field. Eur J Plant Pathol 2021, 160, 53–65. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of Phytopathogenic Fungi: GOPHY 3. Stud Mycol 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Phookamsak, R.; Liu, J.-K.; Manamgoda, D.S.; Chukeatirote, E.; Mortimer, P.E.; Mckenzie, E.H.C.; Hyde, K.D. The Sexual State of Setophoma. Phytotaxa 2014, 176, 260. [Google Scholar] [CrossRef]
- Gao, C.; Montoya, L.; Xu, L.; Madera, M.; Hollingsworth, J.; Purdom, E.; Singan, V.; Vogel, J.; Hutmacher, R.B.; Dahlberg, J.A.; Coleman-Derr, D.; Lemaux, P.G.; Taylor, J.W. Fungal community assembly in drought-stressed sorghum show stochasticity, selection, and universal ecological dynamics. Nature Communications 2020, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wu, B.; Zhang, S.; Li, M.; Jiang, X. Transcriptome dynamics underlying chlamydospore formation in Trichoderma virens GV29-8. Frontiers in Microbiology 2021, 12, 654855. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.-T.; Zhang, L.; Shen, H.-W.; Bao, D.-F.; Luo, Z.-L. Setophoma aseptata sp. nov. and new record of Minutisphaera aspera from Yuanjiang River Basin, China. Phytotaxa 2025, 702, 287–299. [Google Scholar] [CrossRef]
- Absalan, S.; Armand, A.; Jayawardena, R.S.; McKenzie, E.H.C.; Hyde, K.D.; Lumyong, S. Diversity of Pleosporalean Fungi Isolated from Rice (Oryza sativa L.) in Northern Thailand and Descriptions of Five New Species. J. Fungi 2024, 10, 763. [Google Scholar] [CrossRef]
- Li, M.; Liu, D.; Wang, M.; Cui, K.; Chen, L.; He, L.; Zhou, L. Characterization of Setophoma henanensis sp. nov., causing root rot on peanut. BMC Microbiol 2025, 25, 329. [Google Scholar] [CrossRef]
- Liu, H.F.; Choi, H.J.; Paul, N.C.; Ariyawansa, H.A.; Sang, H.K. 2025. Discovering fungal communities in roots of Zoysia japonica and characterising novel species and their antifungal activities. IMA Fungus 2025, 16, 1–43. [Google Scholar] [CrossRef]
- Ferreira, B.W.; Guterres, D. C.; Macedo, D. M.; Barreto, R. Epityfication of Perisporiopsis struthanthi and Perisporiopsis lantanae, and the taxonomic implications for Perisporiopsidaceae, Perisporiopsis and Setophoma. ResearchSquare 2023. [Google Scholar] [CrossRef]
- Sadeghi, S.; Nasr-Esfahani, M.; Maleki, M.; Molahoseini, H.; Khankahdani, H. H.; Mohammadi, M. Fungicidal control of onion pink root caused by Setophoma terrestris and effects on soil enzyme activity. Journal of Phytopathology 2024, 172, e13349. [Google Scholar] [CrossRef]
- Zhao, P.; Feng, Z.; Cai, L.; Phurbu, D.; Duan, W.; Xie, F.; Li, X.; Liu, F. Development of an RPA-CRISPR/Cas12a Assay for Rapid and Sensitive Diagnosis of Plant Quarantine Fungus Setophoma terrestris. J. Fungi 2024, 10, 716. [Google Scholar] [CrossRef]
- Li, X.; Liu, F.; Duan, W.J. TaqMan MGB-based real-time fluorescent PCR method for the rapid detection of Setophoma terrestris. Mycosystema 2024, 43, 240100. [Google Scholar]
- Sayago, P.; Juncosa, F.; Albarracín Orio, A.G.; Luna, D. F.; Molina, G.; Lafi, J.; Ducasse, D.A. Bacillus subtilis ALBA01 alleviates onion pink root by antagonizing the pathogen Setophoma terrestris and allowing physiological status maintenance. European Journal of Plant Pathology 2020, 157, 509–519. [Google Scholar] [CrossRef]
- Zhang, F.B.; Zheng, H.L.; Cui, W.G.; Zhang, M.Q.; Yin, Y.S.; Cui, M.; Gao, M. First report of Setophoma terrestris causing pink root of garlic in China. Plant Dis. 2019, 103, 584. [Google Scholar] [CrossRef]
- López-López, A. M.; Tovar-Pedraza, J. M.; León-Félix, J.; Allende-Molar, R.; Bernardi Lima, N.; Márquez-Zequera, I.; García-Estrada, R. S. Caracterización morfológica, filogenia y patogenicidad de Setophoma terrestris causante de raíz corchosa y rosada de jitomate (Solanum lycopersicum) en Sinaloa, México. Revista Mexicana de Fitopatología 2024, 42. [Google Scholar]
- Le, D.; Audenaert, K.; Haesaert, G. Fusarium basal rot: profile of an increasingly important disease in Allium spp. Tropical Plant Pathology 2021, 46, 241-253. Tropical Plant Pathology 2021, 46, 241–253. [Google Scholar] [CrossRef]
- Abd-El-Baky, A.A.; Yousef, H.; Shalaby, S.I. Garlic pink rot disease and crop yield as affected by salinity and irrigation water deficit. Egyptian Journal of Phytopathology 2019, 47, 141–155. [Google Scholar] [CrossRef]
- El-Elimat, T.; Figueroa, M.; Raja, H. A.; Graf, T. N.; Swanson, S. M.; Falkinham III, J. O.; Oberlies, N. H. Biosynthetically distinct cytotoxic polyketides from Setophoma terrestris. European Journal of Organic Chemistry 2015, 2015, 109–121. [Google Scholar] [CrossRef]
- Ferreira, J.F.; Bosland, P.W.; Williams, P.H. The variability of Pyrenochaeta terrestris isolates based on isozyme polymorphism, cultural characteristics and virulence on differential onion breeding lines. Journal of Phytopathology 1991, 133, 289–296. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




