Submitted:
26 September 2025
Posted:
29 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Collection Site
2.2. Media Used in the Study
2.3. Isolation, Purification, and Preservation of Pathogenic Fungi
2.4. Pathogen Identification
2.4.1. Morphological Identification
2.4.2. DNA Extraction, PCR, and Sequencing
2.4.3. Phylogenetic Analysis
2.5. Pathogenicity Assays
2.6. Biological Characteristics of Pathogens
2.6.1. Effects of Different Media
2.6.2. Effects of Temperature
2.6.3. Effects of pH
2.6.4. Effects of Carbon and Nitrogen
2.7. Data Processing
3. Results
3.1. Field Symptoms of Gouqi Root Rot and the Diversity of Pathogenic Fungi
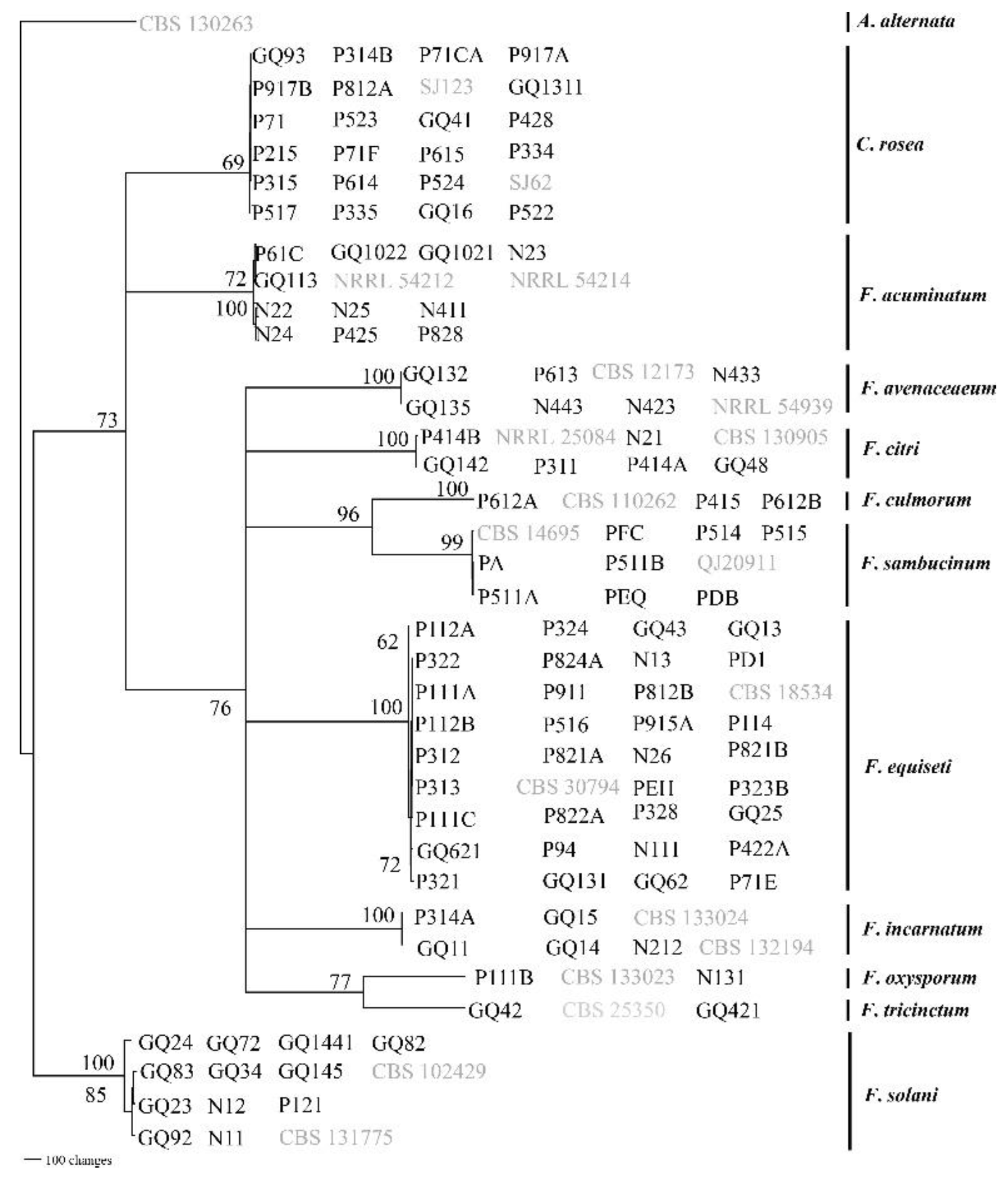
3.2. Morphology and Phylogenetic Analysis of Root Rot Pathogens
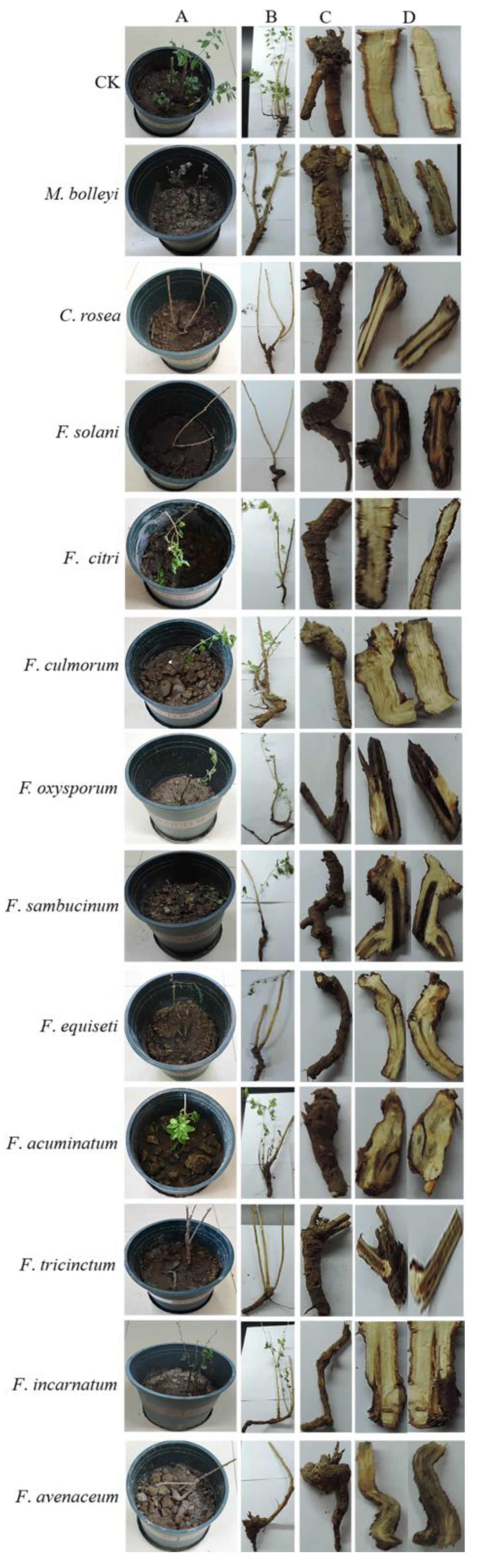
3.3. Pathogenicity Analysis
3.4. Biological Characteristics of Root Rot Pathogens
3.4.1. Effect of Different Media on Mycelial Growth
3.4.2. Effect of Temperature on Mycelial Growth
3.4.3. Effect of pH on Mycelial Growth
3.4.4. Effect of Carbon and Nitrogen Sources on Mycelial Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wetters, S.; Horn, T.; Nick, P. Goji Who? Morphological and DNA Based Authentication of a "Superfood". Front Plant Sci 2018, 9, 1859. [Google Scholar] [PubMed]
- Mocan, A.; Vlase, L.; Vodnar, D.C.; Bischin, C.; Hanganu, D.; Gheldiu, A.-M.; Oprean, R.; Silaghi-Dumitrescu, R.; Crișan, G. Polyphenolic Content, Antioxidant and Antimicrobial Activities of Lycium barbarum L. and Lycium chinense Mill. Leaves. Molecules 2014, 19, 10056–10073. [Google Scholar] [CrossRef]
- Yao, R.; Heinrich, M.; Weckerle, C.S. The genus Lycium as food and medicine: A botanical, ethnobotanical and historical review. J Ethnopharmacol 2018, 212, 50–66. [Google Scholar] [CrossRef]
- Burke, D.S.; Smidt, C.R.; Vuong, L.T. Momordica cochinchinensis, Rosa, roxburghii, wolfberry, and sea buckthorn-highly nutritional fruits supported by tradition and science. Curr Top Nutraceut R 2005, 3(4), 259.
- Han, Y.; Zhou, Y.; Shan, T.; Li, W.; Liu, H. Immunomodulatory Effect of Lycium barbarum Polysaccharides against Liver Fibrosis Based on the Intelligent Medical Internet of Things. J Healthc Eng 2022, 2022, 6280265. [Google Scholar] [CrossRef]
- Luo, Q.; Cai, Y.; Yan, J.; Sun, M.; Corke, H. Hypoglycemic and hypolipidemic effects and antioxidant activity of fruit extracts from Lycium barbarum. Life Sci 2004, 76(2), 137-149.
- Potterat, O. Goji (Lycium barbarum and L. chinense), Phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med 2010, 76(1), 7-19.
- Liang, X.; An, W.; Li, Y.; Qin, X.; Zhao, J.; Su, S. Effects of different nitrogen application rates and picking batches on the nutritional components of Lycium barbarum L. fruits. Front Plant Sci 2024, 15, 355832.
- Zhao, J.; Li, H.; Yin, Y.; An, W.; Qin, X.; Wang, Y.; Fan, Y. Li, Y.; Cao, Y. Fruit ripening in Lycium barbarum and Lycium ruthenicum is associated with distinct gene expression patterns. Febs Open Bi 2020, 10(8), 1550-1567.
- Gnaim, R.; Ledesma-Amaro, R. Synthetic biology of Fusarium for the sustainable production of valuable bioproducts. Blotechnol Adv 2025, 108579.
- Summerell, B.A. Resolving Fusarium, Current status of the genus. Annual review of phytopathol 2019, 57(1), 323-339.
- Doshi, P.; Šerá, B. Role of Non-Thermal Plasma in Fusarium Inactivation and Mycotoxin Decontamination. Plants 2023, 12, 627. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, L.; Hou, W.; Han, Y. Fusarium oxysporum Associated with Fusarium Wilt on Pennisetum sinese in China. Pathogens 2022,11(9), 999-999.
- Khairullina, A.; Micic, N.; Jørgensen, H.J.L.; Bjarnholt, N.; Bülow, L.; Collinge, D.B.; Jensen, B. Biocontrol Effect of Clonostachys rosea on Fusarium graminearum Infection and Mycotoxin Detoxification in Oat (Avena sativa). Plants 2023, 12, 500. [Google Scholar] [CrossRef]
- Thenuwara, G.; Akhtar, P.; Javed, B.; Singh, B.; Byrne, H.J.; Tian, F. Recent Advancements in Lateral Flow Assays for Food Mycotoxin Detection: A Review of Nanoparticle-Based Methods and Innovations. Toxins 2025, 17, 348. [Google Scholar] [CrossRef]
- Ren, X.; Fan, L.; Li, G.; Lyagin, I.V.; Zhang, B.; Ning, M.; Yan, M.; Gao, J.; Wang, F.; Guo, C.; et al. Interaction of Trichoderma Species with Fusarium graminearum Growth and Its Trichothecene Biosynthesis as Further Contribution in Selection of Potential Biocontrol Agents. J. Fungi 2025, 11, 521. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; Foster, G.D. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 2012, 13(4), 414-30.
- Lal, D.; Dev, D.; Kumari, S.; Pandey, S. ; Aparna; Sharma, N.; Nandni, S.; Jha, R.K.; Singh, A. Fusarium wilt pandemic, current understanding and molecular perspectives. Funct Integr Genomic 2024, 24(2), 41.
- Munhoz, T.; Vargas, J.; Teixeira, L.; Staver, C.; Dita, M. Fusarium Tropical Race 4 in Latin America and the Caribbean, status and global research advances towards disease management. Front Plant Sci 2024, 15, 1397617. [Google Scholar] [CrossRef] [PubMed]
- Szabo-Hever, A.; Sharma, J.S.; Faris, J.D.; Zhong, S.; Friesen, T.L.; Fiedler, J.D.; Green, A.J.; Bai, G.; Elias, E.M.; Xu, S.S. Identification and mapping of quantitative trait loci for Fusarium head blight resistance in a synthetic hexaploid × hard red spring wheat population. Plant Genome 2025, 18(3), e70073.
- Chen, S.Y.; Lai, M.H.; Tung, C.W.; Wu, D.H.; Chang, F.Y.; Lin, T.C.; Chung, C.L. Genome-wide association mapping of gene loci affecting disease resistance in the rice-Fusarium fujikuroi pathosystem. Rice (N Y) 2019, 12(1), 85.
- Xie, F.; Sun, Y.; Zhang, H.; Cui, J.; Wang, Q.; Gao, X. ZmBAK1 confers maize resistance to Gibberella stalk rot caused by Fusarium graminearum via activating PAMP-triggered immunity. Plant Signal Behav. 2025, 20(1), 2502739.
- Karlsson, I.; Persson, P.; Friberg, H. Fusarium Head Blight From a Microbiome Perspective. Front Microblol 2021, 12, 628373–628373. [Google Scholar] [CrossRef] [PubMed]
- Pašakinskienė, I.; Stakelienė, V.; Matijošiūtė, S.; Martūnas, J. Diversity of Endophytic Fungi and Bacteria Inhabiting the Roots of the Woodland Grass, Festuca gigantea (Poaceae). Diversity 2024, 16, 453. [Google Scholar] [CrossRef]
- Matušinsky, P.; Florová, V.; Sedláková, B.; Mlčoch, P.; Bleša, D. Colonization dynamic and distribution of the endophytic fungus Microdochium bolleyi in plants measured by qPCR. PLoS One 2024, 19(1), e0297633.
- Ünal, F. Phylogenetic analysis of Microdochium spp. associated with turfgrass and their pathogenicity in cereals. Peer J 2024, 12, e16837–e16837. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.H.; Wan, G.K.; Hyo, W.C.; Lee, S.Y. Identification of Microdochium bolleyi associated with basal rot of creeping bent grass in Korea. Mycobiology 2008, 36(2), 77-80.
- Black, D.L.R.; Brown, A. E. Microdochium bolleyi associated with root rot of flax. Plant Pathol 1986, 35(4), 592-659.
- Liu, J.; Sun, S.; Chen, K.; Gao, S.; Chen, D.; Zhang, Y.; Hu, C. Discerning promotion mechanisms of fungi Clonostachys rosea on growth of freshwater microalga Chlorella sp. by non-contact culture. Algal Research 2025, 86103967-103967.
- Shravani, V.; Iruthayasamy, J.; Nallusamy, S.; Govindasamy, J.; Eswaran, K.; Annaiyan, S. Unveiling Clonostachys rosea bioactives, Investigating antifungal potential against Fusarium oxysporum through in vitro and in silico studies. Research Square 2024.
- Petrucci, A.; Cesarini, M.; Vicente, I.; Merani, L.; Jensen, B.; Collinge, D.B.; Sarrocco, S. Exploring the syner1gistic potential of Trichoderma gamsii T6085 and Clonostachys rosea IK726 for biological control of Fusarium head blight in wheat. Microbiol Res 2025, 296128152.
- Bienapfl, J. C.; Floyd, C. M.; Percich, J. A.; Malvick, D. K. (2012). First report of Clonostachys rosea causing root rot of soybean in the United States. Plant Dis 2012, 96(11), 1700-1700.
- Zhao, J.; Xu, X.; Xu, X.; Guo, L.; Wang, X.; Xiang, W.; Zhao, J. Identification and pathogenicity of Clonostachys spp. and its co-inoculation with Fusarium species on soybean root. Plant Pathol 2024, 73(7), 1801-1811.
- Li, Y.L.; Sun, Z.M.; Li, S.K.; Zhao, X.S.; Abiodun, O. ; Han; Z. M. First report of root rot caused by Clonostachys rosea on Xanthoceras sorbifolium in China. Plant Dis 2024, 108(3), 798.
- Bienapfl, J.C.; Floyd, C.M.; Percich, J.A.; Malvick, D.K. First report of Clonostachys rosea Causing Root Rot of Soybean in the United States. Plant Dis 2012, 96(11), 1700.
- Díaz, R.; Chávez, E.C.; Delgado-Ortiz, J. C.; Velazquez Guerrero, J. J.; Roque, A.; Ochoa, Y.M. First report of Clonostachys rosea causing root rot on garlic in Mexico. Plant Dis 2022, 106(11), 3000.
- Qi, H.; Duan, X.; Xu, W.; Zhou, Y.; Ma, H.; Ma, W.; Ma, G. First report disease of Clonostachys rosea causing root rot on Astragalus membranaceus in China. Plant Dis 2022, 106(6), 1752.
- Li, N.; Chen, W.; Wang, B.; Zhang, C.; Wang, Y.; Li, R.; Yan, Y.; He, J. Arbuscular mycorrhizal fungi improve the disease resistance of Lycium barbarum to root rot by activating phenylpropane metabolism. Front Plant Sci 2024, 15, 1459651. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.Y.; Gao, H.H.; Han, Q.; Jia, C.B.; Swingle, B.; Gao, M.; Su, J.Y. First report of Fusarium redolens causing root rot of Goji berry cv. ‘Ningqi-7’ in China. J Phytopathol 2024, 172(2), e13278.
- Zhang, T.; Zhang, Z.; Li, Y.; He, K. The Effects of Saline Stress on the Growth of Two Shrub Species in the Qaidam Basin of Northwestern China. Sustainability-basel 2019, 11, 828. [Google Scholar] [CrossRef]
- Chen, X. L.; Yang, J.; Peng, Y. L. Large-scale insertional mutagenesis in Magnaporthe oryzae by Agrobacterium tumefaciens-mediated transformation. Methods in molecular biology 2011, 722, 213–224. [Google Scholar]
- Leslie, J.F.; Summerell, B.A. The Fusarium laboratory manual. John Wiley & Sons 2008.
- Xu, J. R.; Hamer, J. E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes development 1996, 10(21), 2696–2706.
- Utama, G.L.; Lestari, W.D.; Kayaputri, I.L.; Balia, R.L. Balia. Indigenous yeast with cellulose-degrading activity in napa cabbage (Brassica pekinensis L.) waste, Characterisation and species identification. Foods and Raw Materials 2019, 321-328.
- White, T. J.; Bruns, T.; Lee, S. J. W. T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols, a guide to methods and applications 1990, 18(1), 315-322.
- Wong, J.Y.; Jeffries, P. Diversity of pathogenic Fusarium populations associated with asparagus roots in decline soils in Spain and the UK. Plant Pathol 2006, 55(3), 331-342.
- Bueno, C.J.; Fischer, I.H.; Rosa D.D.; Firmino, A.C.; Harakava, R.; Oliveira C.M.G.; Furtado, E.L. Fusarium solani F. sp. Passiflorae: a new forma specialis causing collar rot in yellow passion fruit. Plant Pathol 2014, 63(2), 382-389.
- Zhao, J.; Huang, M. Characterization and In Vitro Fungicide Sensitivity of Two Fusarium spp. Associated with Stem Rot of Dragon Fruit in Guizhou; China. J Fungi 2023, 9(12), 1178.
- Swofford, D.L. PAUP* [Phylogenetic Analysis Using Parsimony (*and Other Methods)], Version 4. Sinauer Associates 2003, Sunderland, MA.
- Zhao, X.; Gao, C.; Zhang, X.; Du, M.; Wang, J.; Wang, X.; Lu, B.; Chen, C.; Yang, L.; Zhang, Y.; et al. Identification and biological characteristics of Fusarium tobaccum sp. nov.; a novel species causing tobacco root rot in Jilin Province; China. Microbiol Spectr 2024, 12(12), e00925-24.
- Yang, E.; Fan, L.; Yan, J.; Jiang, Y.; Doucette, C.; Fillmore, S. ; Walker; B. Influence of culture media; pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB express 2018, 8, 1–14. [Google Scholar]
- Bai, L.; Li, X.; Cao, Y.; Song, Z.; Ma, K.; Fan, Y.; Ma, M. Fusarium culmorum and Fusarium equiseti causing root rot disease on Lycium barbarum (goji berry) in China. Plant Dis 2020, 104(11), 3066.
- Uwaremwe, C.; Yue, L.; Liu, Y.; Tian, Y.; Zhao, X.; Wang, Y.; Xie, Z.; Zhang, Y.; Cui, Z.; Wang, R. Molecular identification and pathogenicity of Fusarium and Alternaria species associated with root rot disease of wolfberry in Gansu and Ningxia provinces; China. Plant Pathol 2021, 70(2), 397-406.
- Feng, Zhilin; Xiao, Y. ; Li, N.; Gao, Q.; Wang J.; Chen, S.; Xing, R. Effects of root rot on microbial communities associated with goji berry (Lycium barbarum) in the Qaidam Basin; China European Journal of Plant Pathol 2023, 167, 853–866.
- Zhang, Z.; Zhang, W.; Wang, X.; Kou, Z.; Wang, Y.; Islam, R.; Zhang, J.Q.; Liu, L.; Xie, T.; Tian, Y.Q. Isolation and identification of antagonistic bacteria of Angelica root rot and their mechanism as biological control. Biological Control 2023, 177, 105120. [Google Scholar] [CrossRef]
- Qi, H.; Qi, X.; Xu, W.; An, Z.; Fan, T. First Report of Root Rot Caused by Fusarium avenaceum and Fusarium flocciferum on Astragalus membranaceus var. mongholicus in China. J Phytopathol 2024, 172(5), e13397.
- Sabahi, F.; Banihashemi, Z.; Mirtalebi, M.; Rep, M.; Cacciola, S.O. Molecular Variability of the Fusarium solani Species Complex Associated with Fusarium Wilt of Melon in Iran. J. Fungi 2023, 9, 486. [Google Scholar] [CrossRef]
- Williams, H. Root and stem rot of cucumber caused by Fusarium oxysporum f. sp. radicis-cucumerinum f. sp. nov. Plant Dis 1996, 80(3), 313.
- Ye, Q.; Wang, R.; Ruan, M.; Yao, Z.; Cheng, Y.; Wan, H.; Li, Z.; Yang, Y.; Zhou, G. Genetic diversity and identification of wilt and root rot pathogens of tomato in China. Plant Dis 2020, 104(6), 1715-1724.
- Ding, X.L.; Lin, S.X.; Zhao, R.W.; Ye, J.R. First report of needle blight on Pinus thunbergii Caused by Fusarium proliferatum in China. Plant Dis 2022, 106(11), 2989.
- Tahat, M.M.; Aldakil, H.A.; Alananbeh, K.M. First report of damping off disease caused by Fusarium oxysporum on Pinus pinea in Jordan. Plant Dis 2021, 105(12), 4153.
- Desjardins, A.E.; Proctor, R.H. Molecular biology of Fusarium mycotoxins. International journal of food microbiol 2007, 119(1-2), 47-50.
- Wang, Q.; Xu, L. Beauvericin, a Bioactive Compound Produced by Fungi: A Short Review. Molecules 2012, 17, 2367–2377. [Google Scholar] [CrossRef]
- He, J.; Zhang, X.; Wang, Q.; Li, N.; Ding, D.; Wang, B. Optimization of the fermentation conditions of Metarhizium robertsii and its biological control of wolfberry root rot disease. Microorganisms 2023, 11(10), 2380.
- Lemanceau, P.; Alabouvette, C. Biological control of fusarium diseases by fluorescent Pseudomonas and non-pathogenic Fusarium. Crop protection 1991, 10(4), 279–286. [Google Scholar] [CrossRef]
- Sivan, A; Chet, I. Biological control of Fusarium spp. in cotton; wheat and muskmelon by Trichoderma harzianum. J. Phytopathol 1986, 116(1), 39-47.
- Li, J.; He, K.; Zhang, Q.; Wu, X.; Li, Z.; Pan, X.; Wang, Y.; Li, C.; Zhang, M. Draft Genome and Biological Characteristics of Fusarium solani and Fusarium oxysporum Causing Black Rot in Gastrodia elata. Int. J. Mol. Sci 2023, 24, 4545. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, D.A.; Ivanova, E.A.; Semenov, M.V.; Zhelezova, A.D.; Ksenofontova, N.A.; Tkhakakhova, A.K.; Kholodov, V.A. Diversity, Ecological Characteristics and Identification of Some Problematic Phytopathogenic Fusarium in Soil: A Review. Diversity 2023, 15, 49. [Google Scholar] [CrossRef]
- Li, F.; Jiang, X.; Sun, M.; Xu, H.; Shi, L.; Shan, W. Screening of culture conditions for pathogens of potato dry rot. Acta Agr Scand B-S P 2014, 64(8), 694-699.

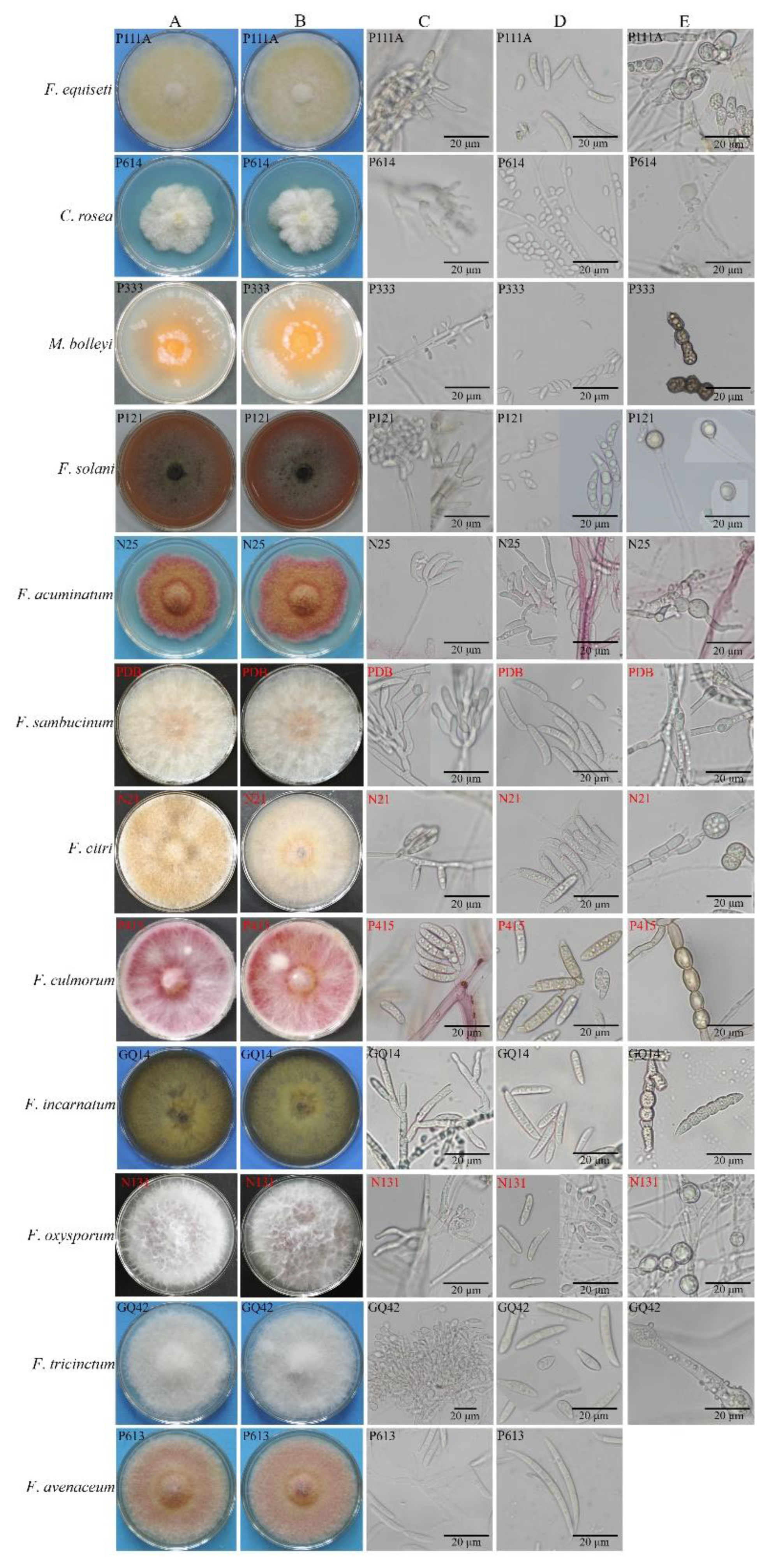

| Types of spores | F. equiseti | C. rosea | M. bolleyi | F. solani | F. acuminatum | F. sambucinum | F. avenaceum | F. citri | F. incarnatum | F. culmorum | F. oxysporum | F. tricinctum | |
| conidia | microconidium | 11.56±1.46×3.85±0.56 | 7.04±0.41×4.36±0.37 | 9.55±2.04×3.39±0.78 | 7.63±0.91×4.7±0.68 | 8.82±1.65×4.32±0.83 | 7.72±1.36×4.22±0.92 | rare | rare | rare | 8.24±1.56×4.43 ±1.14 | 7.22±1.12×3.26±0.70 | 13.20±2.16×4.52±1.44 |
| macroconidium | 26.44±2.14×5.69±1.43 | 39.09±4.63 ×7.07±1.21 | 20.16±3.02×4.79±0.88 | 34.75±5.38 ×7.56±1.20 | 46.45±4.96×5.74 ±1.54 | 30.84±5.41×5.72 ±1.02 | 30.02±1.51×6.17±1.07 | 37.99±1.96×8.97±0.46 | 33.68±2.21×11.45±0.42 | 33.97±4.95×7.17±1.34 | |||
| chlamydospore | 16.34±1.58×14.15±2.08 | 11.38±2.25×12.45±2.02 | 11.28±1.46×8.00±0.59 | 15.74±2.02×14.19±1.90 | 13.07±2.15×12.84±1.23 | 13.12±3.80× 9.45±2.15 | not found | 11.32±2.10×10.57±2.93 | 13.06±3.48 ×16.73±2.54 | 13.77±2.71×11.91±1.79 | 13.91±1.36×12.77±1.68 | 14.76±4.45×13.56±2.25 | |
| Media | F. equiseti | C. rosea | M. bolleyi | F. solani | F. acuminatum | F. sambucinum | F. avenaceum | F. citri | F. incarnatum | F. culmorum | F. oxysporum | F. tricinctum |
| PDA | 0.68±0.06b | 0.44±0.04b | 0.47±0.19bc | 0.78±0.01a | 0.52±0.01c | 0.80±0.00a | 0.80±0.01a | 0.80±0.00ab | 0.80±0.00a | 0.80±0.00a | 0.80±0.00a | 0.72±0.08ab |
| CM | 0.79±0.00a | 0.50±0.01b | 0.34±0.02c | 0.77±0.01b | 0.52±0.06c | 0.80±0.01a | 0.62±0.07b | 0.80±0.01a | 0.80±0.01a | 0.80±0.00a | 0.78±0.01c | 0.64±0.09bc |
| CA | 0.79±0.00a | 0.50±0.07b | 0.28±0.02c | 0.72±0.01d | 0.62±0.07b | 0.80±0.00a | 0.77±0.03a | 0.79±0.01b | 0.80±0.00a | 0.80±0.00a | 0.79±0.01b | 0.68±0.07bc |
| MB | 0.80±0.01a | 0.61±0.04a | 0.60±0.16ab | 0.76±0.01b | 0.53±0.01c | 0.76±0.04a | 0.56±0.06c | 0.80±0.00ab | 0.81±0.01a | 0.80±0.00a | 0.80±0.01a | 0.61±0.05c |
| CLA | 0.79±0.01a | 0.48±0.03b | 0.69±0.03a | 0.73±0.00c | 0.74±0.03a | 0.78±0.03a | 0.79±0.01a | 0.80±0.01a | 0.79±0.02b | 0.80±0.00a | 0.80±0.00a | 0.80±0.01a |
| Temperature | F. equiseti | C. rosea | M. bolleyi | F. solani | F. acuminatum | F. sambucinum | F. avenaceum | F. citri | F. incarnatum | F. culmorum | F. oxysporum | F. tricinctum |
| 35℃ | 0.76±0.03a | 0.51±0.05a | 0.36±0.03a | 0.77±0.01a | 0.61±0.01a | 0.79±0.01a | 0.80±0.01a | 0.79±0.01a | 0.79±0.01a | 0.80±0.01a | 0.79±0a | 0.79±0.01a |
| 40℃ | 0.78±0.02a | 0.51±0.01a | 0.34±0.01a | 0.78±0.01a | 0.61±0.03a | 0.79±0.01a | 0.78±0.01a | 0.80±0.01a | 0.79±0.01a | 0.79±0.01a | 0.80±0.01a | 0.76±0.04a |
| 50℃ | 0.77±0.03a | 0.40±0.03b | 0.25±0.01b | 0.77±0.01a | 0 | 0 | 0.17±0b | 0.58±0.12b | 0.79±0.01a | 0 | 0.80±0.01a | 0.63±0.05b |
| 55℃ | 0 | 0 | 0 | 0.47±0b | 0 | 0 | 0 | 0 | 0 | 0 | 0.79±0.01a | 0 |
| 60℃ | 0 | 0 | 0 | 0.35±0c | 0 | 0 | 0 | 0 | 0 | 0 | 0.80±0.01a | 0 |
| 65℃ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 70℃ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pH value |
F. equiseti |
C. rosea |
M. bolleyi |
F. solani |
F. acuminatum |
F. sambucinum |
F. avenaceum |
F. citri |
F. incarnatum |
F. culmorum |
F. oxysporum |
F. tricinctum |
| 4 | 0.59±0.02f | 0.50±0.03c | 0.43±0.01b | 0.68±0.01e | 0.45±0.03d | 0.79±0.01ab | 0.52±0.01e | 0.80±0.01a | 0.78±0.01e | 0.79±0.01ab | 0.70±0.02c | 0.67±0.04d |
| 5 | 0.70±0.02cd | 0.52±0.02abc | 0.36±0.01cde | 0.71±0.00c | 0.57±0.01c | 0.79±0.01ab | 0.70±0.02cd | 0.79±0.01ab | 0.79±0.01cd | 0.79±0.00b | 0.79±0.00a | 0.77±0.02abc |
| 6 | 0.73±0.01abc | 0.54±0.03ab | 0.35±0.01e | 0.73±0.00ab | 0.63±0.03b | 0.79±0.01ab | 0.76±0.03b | 0.79±0.01ab | 0.79±0.01bc | 0.79±0.00b | 0.79±0.00a | 0.78±0.01ab |
| 7 | 0.75±0.01a | 0.53±0.00ab | 0.35±0.01de | 0.70±0.01d | 0.64±0.03b | 0.79±0.00b | 0.79±0.01ab | 0.79±0.01ab | 0.78±0.01de | 0.79±0.00b | 0.79±0.02a | 0.79±0.01a |
| 8 | 0.73±0.04ab | 0.53±0.03ab | 0.38±0.01c | 0.72±0.01b | 0.73±0.04a | 0.80±0.01a | 0.79±0.01ab | 0.79±0.01ab | 0.80±0.00a | 0.79±0.00b | 0.75±0.01b | 0.78±0.01ab |
| 9 | 0.73±0.02abc | 0.54±0.02a | 0.59±0.02a | 0.73±0.01ab | 0.58±0.02c | 0.80±0.01a | 0.76±0.03b | 0.80±0.01a | 0.80±0.00a | 0.79±0.01ab | 0.79±0.01a | 0.78±0.01ab |
| 10 | 0.66±0.03e | 0.51±0.01bc | 0.36±0.04cd | 0.74±0.01a | 0.59±0.03c | 0.79±0.01ab | 0.72±0.02c | 0.79±0.00b | 0.80±0.00a | 0.80±0.01a | 0.80±0.01a | 0.74±0.05c |
| 11 | 0.70±0.02bcd | 0.52±0.01abc | 0.36±0.02cde | 0.73±0.00ab | 0.58±0.02c | 0.79±0.01ab | 0.68±0.03d | 0.79±0.00b | 0.80±0.01ab | 0.79±0.01ab | 0.79±0.01a | 0.75±0.02bc |
| Carbon source | F. equiseti | C. rosea | M. bolleyi | F. solani | F. acuminatum | F. sambucinum | F. avenaceum | F. citri | F. incarnatum | F. culmorum | F. oxysporum | F. tricinctum |
| CK | 0.80±0.01ab | 0.44±0.03cd | 0.30±0.01c | 0.78±0.01a | 0.64±0.01b | 0.75±0.02b | 0.62±0.03cd | 0.73±0.03bc | 0.79±0.01ab | 0.79±0.00a | 0.79±0.01ab | 0.65±0.02d |
| glucose | 0.76±0.05b | 0.43±0.02d | 0.35±0.02b | 0.79±0.01a | 0.48±0.02d | 0.72±0.06bc | 0.59±0.03de | 0.73±0.04bc | 0.77±0.03b | 0.80±0.00a | 0.80±0.01a | 0.74±0.03b |
| sucrose | 0.80±0.00ab | 0.43±0.01cd | 0.63±0.02a | 0.79±0.00a | 0.46±0.02d | 0.79±0.01a | 0.64±0.05cd | 0.80±0.00a | 0.80±0.00a | 0.80±0.00a | 0.80±0.01a | 0.79±0.01a |
| mannitol | 0.80±0.01ab | 0.45±0.03cd | 0.37±0.20c | 0.80±0.01a | 0.39±0.02e | 0.75±0.01b | 0.46±0.03g | 0.79±0.00a | 0.79±0.01ab | 0.79±0.01a | 0.80±0.01a | 0.71±0.06c |
| dextrin | 0.79±0.01ab | 0.48±0.07bc | 0.30±0.03c | 0.74±0.01c | 0.52±0.01c | 0.80±0.01a | 0.75±0.04a | 0.79±0.01a | 0.80±0.01a | 0.80±0.01a | 0.80±0.01a | 0.80±0.00a |
| maltose | 0.70±0.05c | 0.50±0.06b | 0.29±0.02c | 0.73±0.02c | 0.66±0.05a | 0.61±0.04d | 0.69±0.02b | 0.75±0.02bc | 0.80±0.01a | 0.79±0.01a | 0.80±0.01a | 0.72±0.02bc |
| soluble starch | 0.79±0.01ab | 0.43±0.03d | 0.30±0.03c | 0.76±0.02b | 0.69±0.03a | 0.80±0.01a | 0.76±0.04a | 0.79±0.01a | 0.80±0.01a | 0.80±0.01a | 0.80±0.01a | 0.79±0.01a |
| lactose | 0.65±0.03d | 0.44±0.08bcd | 0.24±0.03de | 0.73±0.01c | 0.39±0.03e | 0.71±0.01c | 0.56±0.03ef | 0.63±0.04d | 0.64±0.02d | 0.69±0.17b | 0.78±0.01b | 0.47±0.02f |
| cellulose | 0.79±0.01ab | 0.51±0.04b | 0.37±0.02b | 0.79±0.01a | 0.72±0.03a | 0.79±0.01a | 0.70±0.03b | 0.79±0.00a | 0.17±0.02e | 0.79±0.00a | 0.79±0.01ab | 0.78±0.01a |
| fructose | 0.79±0.01ab | 0.57±0.03a | 0.25±0.02de | 0.79±0.00a | 0.53±0.02c | 0.79±0.00a | 0.58±0.02def | 0.79±0.00a | 0.78±0.01ab | 0.79±0.00a | 0.80±0.01a | 0.70±0.02c |
| xylose | 0.72±0.04c | 0.44±0.02cd | 0.21±0.03e | 0.76±0.01b | 0.46±0.04d | 0.74±0.02bc | 0.54±0.02f | 0.72±0.01c | 0.75±0.02c | 0.79±0.01a | 0.79±0.01ab | 0.59±0.02e |
| Nitrogen source | F. equiseti | C. rosea | M. bolleyi | F. solani | F. acuminatum | F. sambucinum | F. avenaceum | F. citri | F. incarnatum | F. culmorum | F. oxysporum | F. tricinctum |
| CK | 0.80±0.00a | 0.44±0.04bc | 0.63±0.02c | 0.71±0.02b | 0.71±0.05a | 0.69±0.03d | 0.54±0.02c | 0.62±0.08c | 0.61±0.04e | 0.80±0.01ab | 0.74±0.02c | 0.73±0.01b |
| beef extract | 0.80±0.00a | 0.43±0.01bc | 0.80±0.00a | 0.76±0.02a | 0.57±0.10cd | 0.80±0.00a | 0.75±0.03a | 0.71±0.00b | 0.80±0.00a | 0.80±0.00a | 0.80±0.00a | 0.80±0.01a |
| urea | 0.80±0.01a | 0.38±0.01cd | 0.69±0.01b | 0.64±0.01c | 0.37±0.01f | 0.71±0.02cd | 0.49±0.08d | 0.68±0.03b | 0.72±0.04c | 0.80±0.00a | 0.79±0.01ab | 0.49±0.02d |
| peptone | 0.80±0.00a | 0.51±0.10a | 0.80±0.00a | 0.72±0.02b | 0.64±0.05bc | 0.74±0.04bc | 0.76±0.02a | 0.78±0.04a | 0.80±0.00a | 0.80±0.00a | 0.80±0.01ab | 0.80±0.01a |
| yeast powder | 0.80±0.01a | 0.45±0.01ab | 0.80±0.00a | 0.71±0.01b | 0.59±0.06bc | 0.80±0.01a | 0.78±0.02a | 0.80±0.00a | 0.80±0.00a | 0.80±0.01ab | 0.80±0.00a | 0.80±0.00a |
| sodium nitrate | 0.79±0.01a | 0.42±0.02bc | 0.25±0.01f | 0.79±0.01a | 0.51±0.02d | 0.77±0.02ab | 0.69±0.04b | 0.79±0.01a | 0.78±0.01ab | 0.80±0.00a | 0.79±0.01ab | 0.73±0.04b |
| ammonium chloride | 0.64±0.05b | 0.28±0.04e | 0.59±0.05d | 0.49±0.05e | 0.35±0.05f | 0.53±0.07e | 0.34±0.04e | 0.42±0.03d | 0.50±0.04f | 0.78±0.03c | 0.67±0.03d | 0.45±0.05e |
| ammonium nitrate | 0.79±0.00a | 0.35±0.03d | 0.71±0.03b | 0.60±0.02d | 0.44±0.01e | 0.78±0.01ab | 0.48±0.04d | 0.63±0.06c | 0.65±0.02d | 0.79±0.01abc | 0.78±0.01b | 0.65±0.03c |
| saltpeter | 0.80±0.01a | 0.51±0.06a | 0.34±0.01e | 0.78±0.02a | 0.52±0.01d | 0.78±0.01ab | 0.66±0.06b | 0.73±0.03b | 0.76±0.03b | 0.79±0.00abc | 0.78±0.01b | 0.79±0.01a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





