Submitted:
21 June 2025
Posted:
23 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Chemicals and Drug Preparation
2.2. Cell Culture
2.3. Cell Viability Assays
2.4. Synergy
2.5. Flow Cytometry Analyses of Apoptotic Profiles
2.6. ROS Production Analysis
2.7. Liquid Chromatography-Mass Spectrometry, Label Free Quantification Bottom-Up Proteomics Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Antiproliferative Activity of Postbiotic Combinations, Standard Immunotherapy and Standard Chemotherapy
3.2. Synergistic Potential of APB with Dex against the AGS gastric Adenocarcinoma Cells
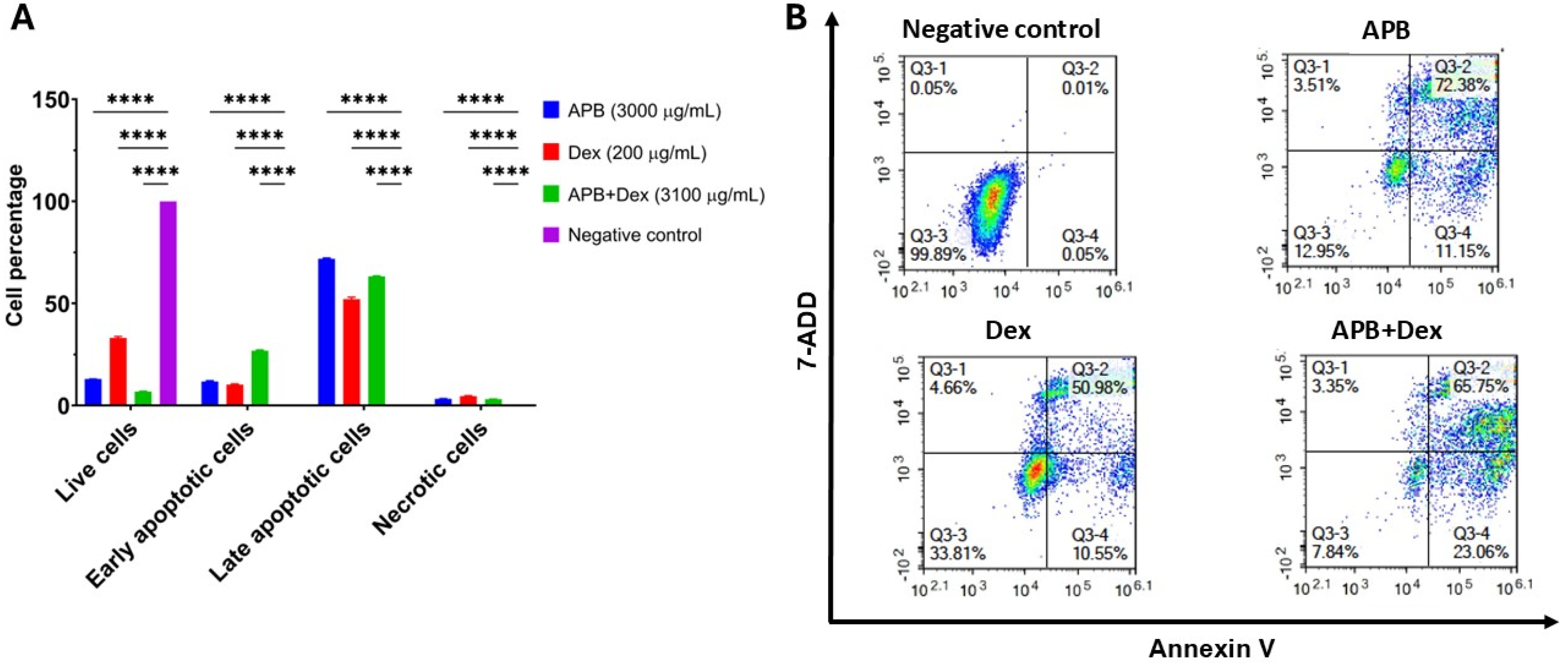
3.3. Flow Cytometric Analyses of Apoptotic Profiles of Mono and Combination Therapies
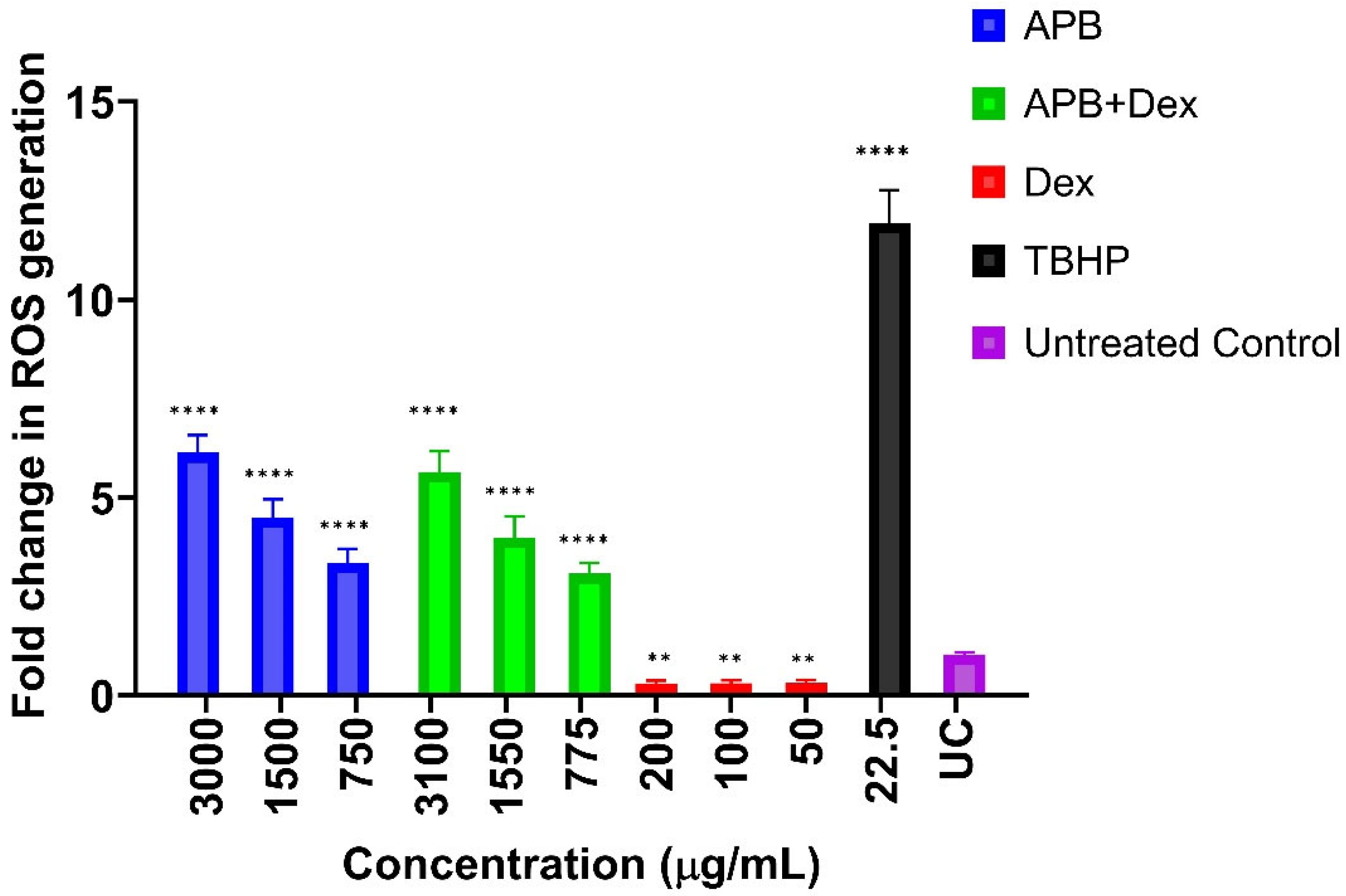
3.4. ROS production in the AGS cells after Treatment with Different Concentrations of APB, Dex and APB+Dex
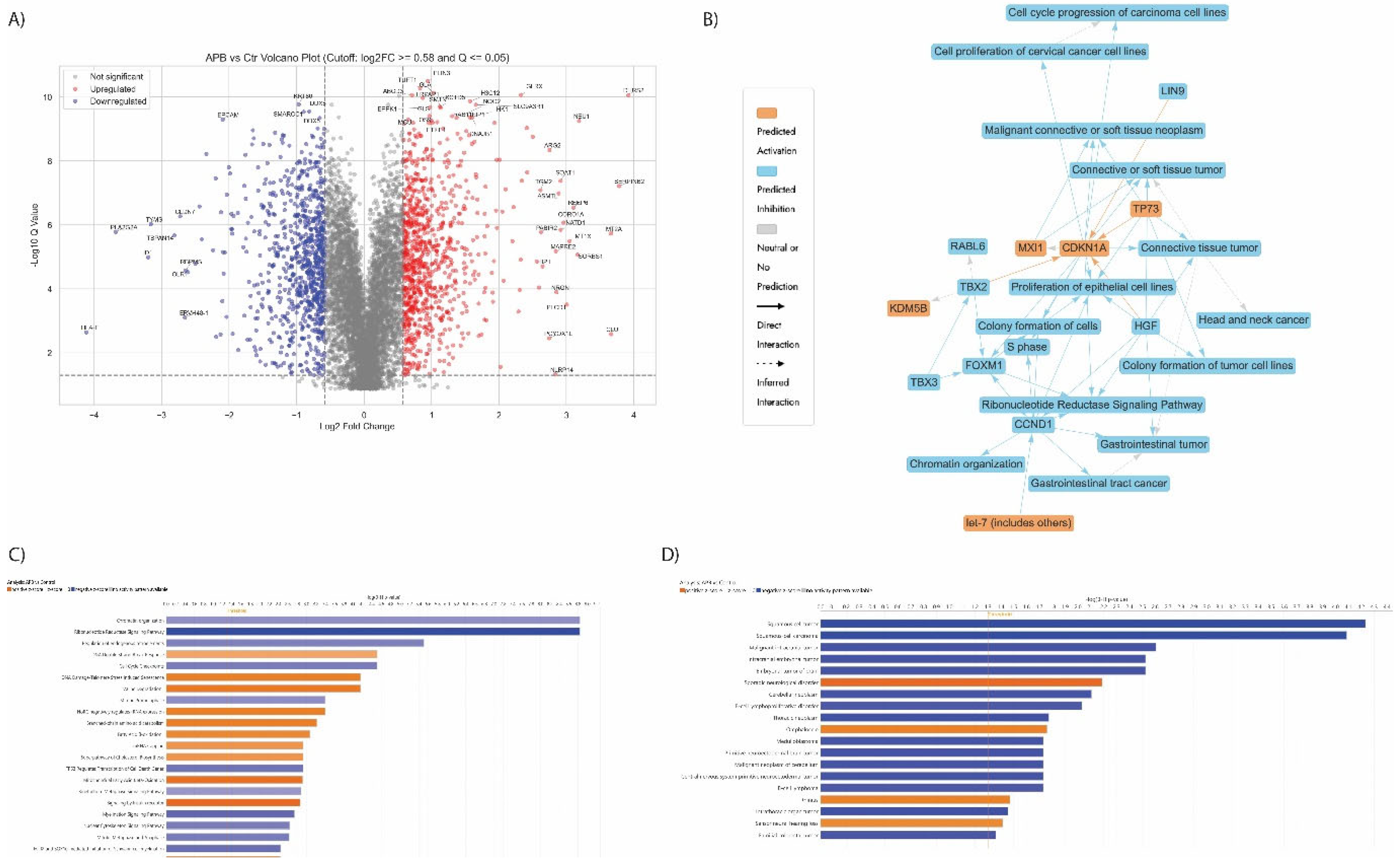
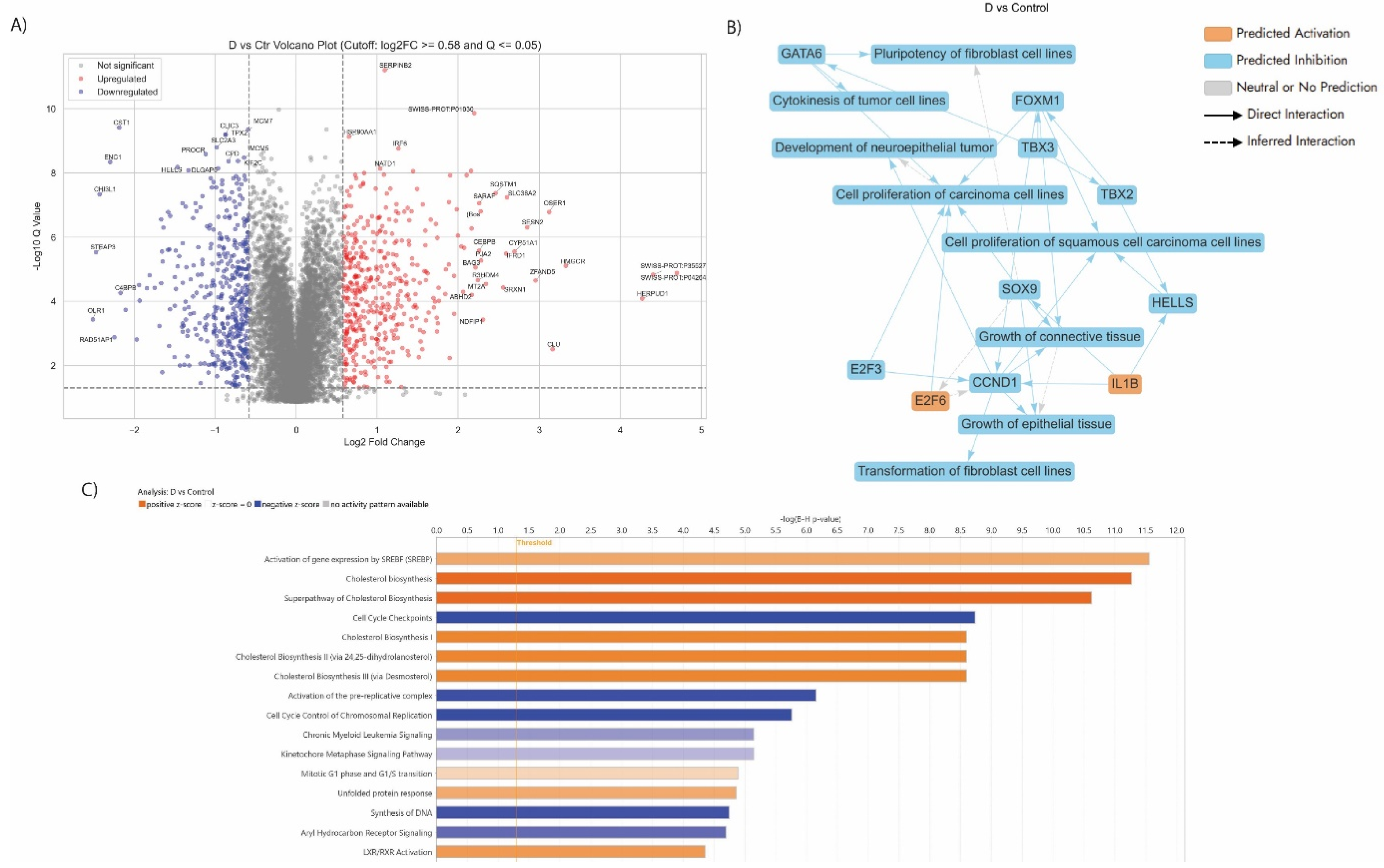
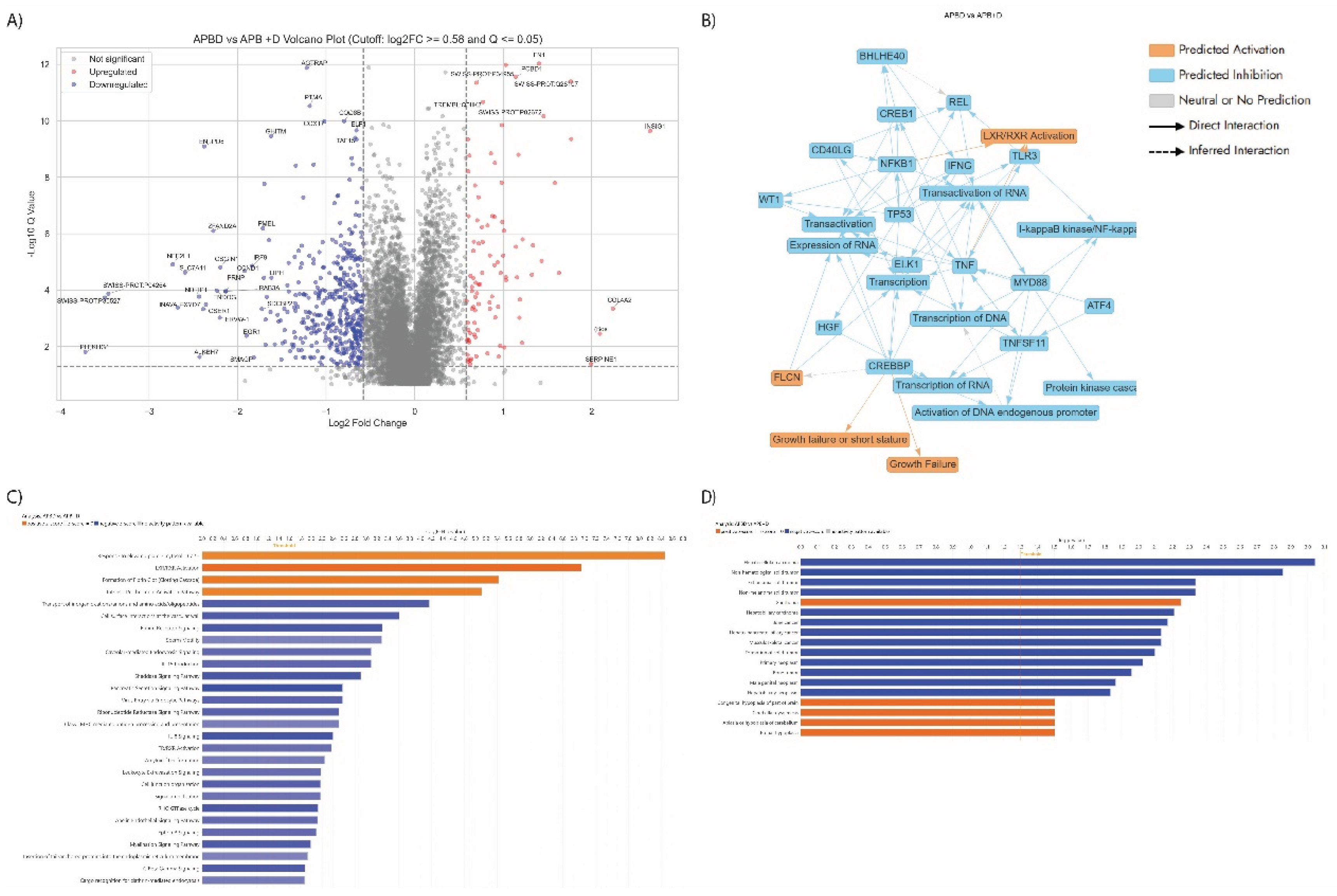
3.5. Proteomics Study of the AGS Cells Treated with the Synergistic Combination vs Mono Treatments
3.5.1. Enrichment Analyses of Differentially Expressed Proteins (DEPS) In Apb-Treated Ags Cells Compared to the Untreated Control Cells
3.5.2. Enriched Pathways of DEPs in Dex Treated AGS Gastric Adenocarcinoma Cells Compared to Control
3.5.3. Enriched Pathways Using DEPs of APB+Dex Combination Treated AGS Cells vs Mono Treatments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Burz, Claudia, Vlad Pop, Ciprian Silaghi, Iulia Lupan, and Gabriel Samasca. "Prognosis and Treatment of Gastric Cancer: A 2024 Update." Cancers 16, no. 9 (2024): 1708. [CrossRef]
- Rawla, Prashanth, and Adam Barsouk. "Epidemiology of Gastric Cancer: Global Trends, Risk Factors and Prevention." Gastroenterology Review/Przegląd Gastroenterologiczny 14, no. 1 (2019): 26-38.
- Joshi, Smita S, and Brian D Badgwell. "Current Treatment and Recent Progress in Gastric Cancer." CA: a cancer journal for clinicians 71, no. 3 (2021): 264-79. [CrossRef]
- Hani, Umme, Riyaz Ali M Osmani, Sabina Yasmin, BH Jaswanth Gowda, Hissana Ather, Mohammad Yousuf Ansari, Ayesha Siddiqua, Mohammed Ghazwani, Adel Al Fatease, and Ali H Alamri. "Novel Drug Delivery Systems as an Emerging Platform for Stomach Cancer Therapy." Pharmaceutics 14, no. 8 (2022): 1576. [CrossRef]
- Jaye, Kayla, Chun Guang Li, Dennis Chang, and Deep Jyoti Bhuyan. "The Role of Key Gut Microbial Metabolites in the Development and Treatment of Cancer." Gut Microbes 14, no. 1 (2022): 2038865.
- Eladwy, Radwa A, Hang Thi Vu, Ravi Shah, Chun Guang Li, Dennis Chang, and Deep Jyoti Bhuyan. "The Fight against the Carcinogenic Epstein-Barr Virus: Gut Microbiota, Natural Medicines, and Beyond." International Journal of Molecular Sciences 24, no. 2 (2023): 1716.
- Chambers, Laura M, Emily L Esakov Rhoades, Rashmi Bharti, Chad Braley, Surabhi Tewari, Lexie Trestan, Zahraa Alali, Defne Bayik, Justin D Lathia, and Naseer Sangwan. "Disruption of the Gut Microbiota Confers Cisplatin Resistance in Epithelial Ovarian Cancer." Cancer research 82, no. 24 (2022): 4654-69.
- Facchin, Sonia, Luisa Bertin, Erica Bonazzi, Greta Lorenzon, Caterina De Barba, Brigida Barberio, Fabiana Zingone, Daria Maniero, Marco Scarpa, and Cesare Ruffolo. "Short-Chain Fatty Acids and Human Health: From Metabolic Pathways to Current Therapeutic Implications." Life 14, no. 5 (2024): 559. [CrossRef]
- Sun, Jinzhe, Shiqian Chen, Dan Zang, Hetian Sun, Yan Sun, and Jun Chen. "Butyrate as a Promising Therapeutic Target in Cancer: From Pathogenesis to Clinic." International Journal of Oncology 64, no. 4 (2024): 44. [CrossRef]
- Son, Mi-Young, and Hyun-Soo Cho. "Anticancer Effects of Gut Microbiota-Derived Short-Chain Fatty Acids in Cancers." Journal of Microbiology and Biotechnology 33, no. 7 (2023): 849. [CrossRef]
- Filippone, Alessia, Giovanna Casili, Sarah Adriana Scuderi, Deborah Mannino, Marika Lanza, Michela Campolo, Irene Paterniti, Anna Paola Capra, Cristina Colarossi, and Annalisa Bonasera. "Sodium Propionate Contributes to Tumor Cell Growth Inhibition through Ppar-Γ Signaling." Cancers 15, no. 1 (2022): 217. [CrossRef]
- Mirzaei, Rasoul, Azam Afaghi, Sajad Babakhani, Masoud Reza Sohrabi, Seyed Reza Hosseini-Fard, Kiandokht Babolhavaeji, Shabnam Khani Ali Akbari, Rasoul Yousefimashouf, and Sajad Karampoor. "Role of Microbiota-Derived Short-Chain Fatty Acids in Cancer Development and Prevention." Biomedicine & Pharmacotherapy 139 (2021): 111619.
- Twycross, Robert. "The Risks and Benefits of Corticosteroids in Advanced Cancer." Drug Safety 11 (1994): 163-78.
- Eladwy, Radwa A, Muhammad A Alsherbiny, Dennis Chang, Mohamed Fares, Chun-Guang Li, and Deep Jyoti Bhuyan. "The Postbiotic Sodium Butyrate Synergizes the Antiproliferative Effects of Dexamethasone against the Ags Gastric Adenocarcinoma Cells." Frontiers in Nutrition 11 (2024): 1372982.
- Dissanayake, Indeewarie Hemamali, Muhammad A Alsherbiny, Dennis Chang, Chun Guang Li, and Deep Jyoti Bhuyan. "Antiproliferative Effects of Australian Native Plums against the Mcf7 Breast Adenocarcinoma Cells and Uplc-Qtof-Im-Ms-Driven Identification of Key Metabolites." Food Bioscience 54 (2023): 102864.
- Alsherbiny, Muhammad A, Deep J Bhuyan, Mitchell N Low, Dennis Chang, and Chun Guang Li. "Synergistic Interactions of Cannabidiol with Chemotherapeutic Drugs in Mcf7 Cells: Mode of Interaction and Proteomics Analysis of Mechanisms." International journal of molecular sciences 22, no. 18 (2021): 10103.
- Jaye, Kayla, Muhammad A Alsherbiny, Dennis Chang, Chun-Guang Li, and Deep Jyoti Bhuyan. "Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells." International journal of molecular sciences 24, no. 20 (2023): 15053.
- Donohoe, Dallas R, Nikhil Garge, Xinxin Zhang, Wei Sun, Thomas M O'Connell, Maureen K Bunger, and Scott J Bultman. "The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon." Cell metabolism 13, no. 5 (2011): 517-26.
- Li, Yangbo, Pengzhan He, Yinghui Liu, Mingming Qi, and Weiguo Dong. "Combining Sodium Butyrate with Cisplatin Increases the Apoptosis of Gastric Cancer in Vivo and in Vitro Via the Mitochondrial Apoptosis Pathway." Frontiers in Pharmacology 12 (2021): 708093.
- Li, Yuanqing, Yaxuan Huang, Haili Liang, Wen Wang, Bo Li, Ting Liu, Yuqi Huang, Zhe Zhang, Yutao Qin, and Xiaoying Zhou. "The Roles and Applications of Short-Chain Fatty Acids Derived from Microbial Fermentation of Dietary Fibers in Human Cancer." Frontiers in Nutrition 10 (2023): 1243390. [CrossRef]
- Den Besten, Gijs, Karen Van Eunen, Albert K Groen, Koen Venema, Dirk-Jan Reijngoud, and Barbara M Bakker. "The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism." Journal of lipid research 54, no. 9 (2013): 2325-40.
- Elmore, Susan. "Apoptosis: A Review of Programmed Cell Death." Toxicologic pathology 35, no. 4 (2007): 495-516. [CrossRef]
- Hayes, John D, Albena T Dinkova-Kostova, and Kenneth D Tew. "Oxidative Stress in Cancer." Cancer cell 38, no. 2 (2020): 167-97.
- Sudaarsan, Aruna Senthil Kumar, and Asit Ranjan Ghosh. "Appraisal of Postbiotics in Cancer Therapy." Frontiers in Pharmacology 15 (2024): 1436021.
- Martín-García, Desirée, Marilina García-Aranda, and Maximino Redondo. "Therapeutic Potential of Clusterin Inhibition in Human Cancer." Cells 13, no. 8 (2024): 665. [CrossRef]
- Vange, Pål, Torunn Bruland, Bjørn Munkvold, Elin Synnøve Røyset, Martin Gleave, and Ingunn Bakke. "Subtle Protective Roles of Clusterin in Gastric Metaplasia after Acute Oxyntic Atrophy." Cellular and Molecular Gastroenterology and Hepatology 7, no. 1 (2019): 246-50. e1. [CrossRef]
- Zhu, Jinliang, Xin Wang, Huiyuan Guan, Qiong Xiao, Zhonghua Wu, Jinxin Shi, Fei Zhang, Peng Gao, Yongxi Song, and Zhenning Wang. "Hip1r Acts as a Tumor Suppressor in Gastric Cancer by Promoting Cancer Cell Apoptosis and Inhibiting Migration and Invasion through Modulating Akt." Journal of Clinical Laboratory Analysis 34, no. 9 (2020): e23425.
- Schroder, Wayne A, Lee D Major, Thuy T Le, Joy Gardner, Matthew J Sweet, Sabina Janciauskiene, and Andreas Suhrbier. "Tumor Cell-Expressed Serpinb2 Is Present on Microparticles and Inhibits Metastasis." Cancer medicine 3, no. 3 (2014): 500-13. [CrossRef]
- Kim, Hye-Youn, and Suntaek Hong. "Multi-Faceted Roles of Dnajb Protein in Cancer Metastasis and Clinical Implications." International journal of molecular sciences 23, no. 23 (2022): 14970.
- Liu, Jun, John C Schmitz, Xiukun Lin, Ningwen Tai, Wu Yan, Michael Farrell, Michelle Bailly, Tian-min Chen, and Edward Chu. "Thymidylate Synthase as a Translational Regulator of Cellular Gene Expression." Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1587, no. 2-3 (2002): 174-82. [CrossRef]
- Voeller, Donna, Lambratu Rahman, and Maria Zajac-Kaye. "Elevated Levels of Thymidylate Synthase Linked to Neoplastic Transformation of Mammalian Cells." Cell Cycle 3, no. 8 (2004): 1003-05. [CrossRef]
- Guijarro, Maria V, Akbar Nawab, Peter Dib, Sandra Burkett, Xiaoping Luo, Michael Feely, Elham Nasri, Robert P Seifert, Frederic J Kaye, and Maria Zajac-Kaye. "Tyms Promotes Genomic Instability and Tumor Progression in Ink4a/Arf Null Background." Oncogene 42, no. 23 (2023): 1926-39. [CrossRef]
- Liu, Yiyang, Yufei Wang, Sheng Sun, Zeyu Chen, Shuai Xiang, Zeyang Ding, Zhao Huang, and Bixiang Zhang. "Understanding the Versatile Roles and Applications of Epcam in Cancers: From Bench to Bedside." Experimental hematology & oncology 11, no. 1 (2022): 97. [CrossRef]
- Sachdeva, Rohit, Megan Wu, Sandra Smiljanic, Oleksandra Kaskun, Kimia Ghannad-Zadeh, Angela Celebre, Keren Isaev, A Sorana Morrissy, Jennifer Guan, and Jiefei Tong. "Id1 Is Critical for Tumorigenesis and Regulates Chemoresistance in Glioblastoma." Cancer research 79, no. 16 (2019): 4057-71.
- Wang, Xin, Shen Li, Chen Liu, Jiawei Zhao, Gangfeng Ren, Feng Zhang, Xuyang Liu, Shuang Cao, Yuming Xu, and Zongping Xia. "High Expression of Pla2g2a in Fibroblasts Plays a Crucial Role in the Early Progression of Carotid Atherosclerosis." Journal of Translational Medicine 22, no. 1 (2024): 967.
- Hashimoto, Itaru, and Takashi Oshima. "Claudins and Gastric Cancer: An Overview." Cancers 14, no. 2 (2022): 290.
- Nie, Min, Yadong Wang, Zenong Yu, Xinyu Li, Yexuan Deng, Ying Wang, Dongjun Yang, Qixiang Li, Xiangwei Zeng, and Junyi Ju. "Aurkb Promotes Gastric Cancer Progression Via Activation of Ccnd1 Expression." Aging (Albany NY) 12, no. 2 (2020): 1304. [CrossRef]
- Broude, Eugenia V, Zoya N Demidenko, Claire Vivo, Mari E Swift, Brian M Davis, Mikhail V Blagosklonny, and Igor B Roninson. "P21 (Cdkn1a) Is a Negative Regulator of P53 Stability." Cell Cycle 6, no. 12 (2007): 1467-70.
- Bao, Chenhui, and Lin Guo. "Tp73-As1 Promotes Gastric Cancer Proliferation and Invasion by Regulation Mir-27b-3p/Tmed5 Axis." Journal of Cancer 13, no. 4 (2022): 1324.
- Koh, Sung Ae, and Kyung Hee Lee. "Function of Hepatocyte Growth Factor in Gastric Cancer Proliferation and Invasion." Yeungnam University Journal of Medicine 37, no. 2 (2020): 73-78.
- Zeng, Jiping, Lixiang Wang, Qiao Li, Wenjuan Li, Magnus Björkholm, Jihui Jia, and Dawei Xu. "Foxm1 Is up-Regulated in Gastric Cancer and Its Inhibition Leads to Cellular Senescence, Partially Dependent on P27kip1." The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland 218, no. 4 (2009): 419-27.
- Zhao, Long-Fei, Feng-Yu Qi, Jin-Ge Zhang, Jing-Ru Pang, Hong-Mei Ren, Dan-Dan Shen, Li-Juan Zhao, Lin Qi, Hong-Min Liu, and Yi-Chao Zheng. "Identification of the Upstream Regulators of Kdm5b in Gastric Cancer." Life Sciences 298 (2022): 120458. [CrossRef]
- Ma, Yuxi, Na Shen, Max S Wicha, and Ming Luo. "The Roles of the Let-7 Family of Micrornas in the Regulation of Cancer Stemness." Cells 10, no. 9 (2021): 2415. [CrossRef]
- Morikawa, Teppei, Rumi Hino, Hiroshi Uozaki, Daichi Maeda, Tetsuo Ushiku, Aya Shinozaki, Takashi Sakatani, and Masashi Fukayama. "Expression of Ribonucleotide Reductase M2 Subunit in Gastric Cancer and Effects of Rrm2 Inhibition in Vitro." Human pathology 41, no. 12 (2010): 1742-48. [CrossRef]
- Mjelle, Robin, Siv Anita Hegre, Per Arne Aas, Geir Slupphaug, Finn Drabløs, Pål Sætrom, and Hans E Krokan. "Cell Cycle Regulation of Human DNA Repair and Chromatin Remodeling Genes." DNA repair 30 (2015): 53-67. [CrossRef]
- Topchu, Iuliia, Rajendra P Pangeni, Igor Bychkov, Sven A Miller, Evgeny Izumchenko, Jindan Yu, Erica Golemis, John Karanicolas, and Yanis Boumber. "The Role of Nsd1, Nsd2, and Nsd3 Histone Methyltransferases in Solid Tumors." Cellular and Molecular Life Sciences 79, no. 6 (2022): 285. [CrossRef]
- Kataoka, Isao, Sayaka Funata, Kiyotaka Nagahama, Kazunobu Isogaya, Hirohisa Takeuchi, Nobutsugu Abe, and Junji Shibahara. "Dnmt3a Overexpression Is Associated with Aggressive Behavior and Enteroblastic Differentiation of Gastric Adenocarcinoma." Annals of Diagnostic Pathology 44 (2020): 151456.
- Meehan, William J, Rajeev S Samant, James E Hopper, Michael J Carrozza, Lalita A Shevde, Jerry L Workman, Kristin A Eckert, Michael F Verderame, and Danny R Welch. "Breast Cancer Metastasis Suppressor 1 (Brms1) Forms Complexes with Retinoblastoma-Binding Protein 1 (Rbp1) and the Msin3 Histone Deacetylase Complex and Represses Transcription." Journal of Biological Chemistry 279, no. 2 (2004): 1562-69. [CrossRef]
- Guo, Xiu-Li, Ya-Jie Wang, Pei-Lin Cui, Yan-Bin Wang, Pi-Xia Liang, Ya-Nan Zhang, and You-Qing Xu. "Effect of Brms1 Expression on Proliferation, Migration and Adhesion of Mouse Forestomach Carcinoma." Asian Pacific Journal of Tropical Medicine 8, no. 9 (2015): 724-30. [CrossRef]
- Bhat, Vipul, Manisha Koneru, Kristen Knapp, Upasana Joneja, Jamin Morrison, and Young K Hong. "Identification and Treatment of Smarca4 Deficient Poorly Differentiated Gastric Carcinoma." The American Surgeon™ 89, no. 11 (2023): 4987-89.
- Kohashi, Kenichi, and Yoshinao Oda. "Oncogenic Roles of Smarcb 1/Ini 1 and Its Deficient Tumors." Cancer science 108, no. 4 (2017): 547-52.
- Zhou, Zhiyi, Dandan Huang, Shudong Yang, Jiabei Liang, Xuan Wang, and Qiu Rao. "Clinicopathological Significance, Related Molecular Changes and Tumor Immune Response Analysis of the Abnormal Swi/Snf Complex Subunit Pbrm1 in Gastric Adenocarcinoma." Pathology and Oncology Research 28 (2022): 1610479. [CrossRef]
- Foskolou, Iosifina P, Christian Jorgensen, Katarzyna B Leszczynska, Monica M Olcina, Hanna Tarhonskaya, Bauke Haisma, Vincenzo D’Angiolella, William K Myers, Carmen Domene, and Emily Flashman. "Ribonucleotide Reductase Requires Subunit Switching in Hypoxia to Maintain DNA Replication." Molecular cell 66, no. 2 (2017): 206-20. e9. [CrossRef]
- O’Reilly, Lorraine A, Tracy L Putoczki, Lisa A Mielke, Jun T Low, Ann Lin, Adele Preaudet, Marco J Herold, Kelvin Yaprianto, Lin Tai, and Andrew Kueh. "Loss of Nf-Κb1 Causes Gastric Cancer with Aberrant Inflammation and Expression of Immune Checkpoint Regulators in a Stat-1-Dependent Manner." Immunity 48, no. 3 (2018): 570-83. e8.
- Riquelme, Ismael, Oscar Tapia, Jaime A Espinoza, Pamela Leal, Kurt Buchegger, Alejandra Sandoval, Carolina Bizama, Juan Carlos Araya, Richard M Peek, and Juan Carlos Roa. "The Gene Expression Status of the Pi3k/Akt/Mtor Pathway in Gastric Cancer Tissues and Cell Lines." Pathology & Oncology Research 22 (2016): 797-805.
- Weng, Xiaoling, Hong Zhang, Junyi Ye, Mengyuan Kan, Fatao Liu, Ting Wang, Jiaying Deng, Yanfang Tan, Lin He, and Yun Liu. "Hypermethylated Epidermal Growth Factor Receptor (Egfr) Promoter Is Associated with Gastric Cancer." Scientific reports 5, no. 1 (2015): 10154. [CrossRef]
- Zhao, Li-Ping, Cong Xue, Jian-Wei Zhang, Zhi-Huang Hu, Yuan-Yuan Zhao, Jing Zhang, Yan Huang, Hong-Yun Zhao, and Li Zhang. "Expression of Rrm1 and Its Association with Resistancy to Gemcitabine-Based Chemotherapy in Advanced Nasopharyngeal Carcinoma." Chinese journal of cancer 31, no. 10 (2012): 476.
- Jiang, Kailong, Minjie Deng, Wenjing Du, Tao Liu, Jia Li, and Yubo Zhou. "Functions and Inhibitors of Chk1 in Cancer Therapy." Medicine in Drug Discovery (2024): 100185.
- Qi, Zhaolai, Ting Zhang, Lei Song, Hongyong Fu, Haifeng Luo, Jie Wu, Shuyun Zhao, Tianhua Zhang, Lianying Guo, and Lingling Jin. "Pras40 Hyperexpression Promotes Hepatocarcinogenesis." EBioMedicine 51 (2020). [CrossRef]
- Ooi, Akishi, Takeru Oyama, Ritsuko Nakamura, Ryousuke Tajiri, Hiroko Ikeda, Sachio Fushida, and Yoh Dobashi. "Gene Amplification of Ccne1, Ccnd1, and Cdk6 in Gastric Cancers Detected by Multiplex Ligation-Dependent Probe Amplification and Fluorescence in Situ Hybridization." Human pathology 61 (2017): 58-67.
- Park, Sun Yi, Ji-Ho Park, Jung Wook Yang, Eun-Jung Jung, Young-Tae Ju, Chi-Young Jeong, Ju-Yeon Kim, Taejin Park, Tae-Han Kim, and Miyeong Park. "Smarcd3 Overexpression Promotes Epithelial–Mesenchymal Transition in Gastric Cancer." Cancers 16, no. 12 (2024): 2282. [CrossRef]
- Kudaravalli, Sriya, Petra den Hollander, and Sendurai A Mani. "Role of P38 Map Kinase in Cancer Stem Cells and Metastasis." Oncogene 41, no. 23 (2022): 3177-85. [CrossRef]
- Chen, Hongjie, Yangchan Hu, Zirui Zhuang, Dingyi Wang, Zu Ye, Ji Jing, and Xiangdong Cheng. "Advancements and Obstacles of Parp Inhibitors in Gastric Cancer." Cancers 15, no. 21 (2023): 5114.
- Fan, Zhiyuan, Wenjing Yan, Jianfang Li, Min Yan, Bingya Liu, Zhongyin Yang, and Beiqin Yu. "Phf10 Inhibits Gastric Epithelium Differentiation and Induces Gastric Cancer Carcinogenesis." Cancer Gene Therapy 31, no. 10 (2024): 1511-24.
- Grundy, Erin E, Noor Diab, and Katherine B Chiappinelli. "Transposable Element Regulation and Expression in Cancer." The FEBS journal 289, no. 5 (2022): 1160-79.
- Kim, Hye-Young, Yunhee Cho, HyeokGu Kang, Ye-Seal Yim, Seok-Jun Kim, Jaewhan Song, and Kyung-Hee Chun. "Targeting the Wee1 Kinase as a Molecular Targeted Therapy for Gastric Cancer." Oncotarget 7, no. 31 (2016): 49902.
- Hino, Kaori, Tomohiro Nishina, Takeshi Kajiwara, Hideaki Bando, Maho Nakamura, Shigenori Kadowaki, Keiko Minashi, Satoshi Yuki, Takashi Ohta, and Hiroki Hara. "Association of Erbb2 Copy Number and Gene Coalterations with Trastuzumab Efficacy and Resistance in Human Epidermal Growth Factor Receptor 2–Positive Esophagogastric and Gastric Cancer." JCO Precision Oncology 6 (2022): e2200135.
- Xing, Kailin, Bingxin Gu, Ping Zhang, and Xianghua Wu. "Dexamethasone Enhances Programmed Cell Death 1 (Pd-1) Expression During T Cell Activation: An Insight into the Optimum Application of Glucocorticoids in Anti-Cancer Therapy." BMC immunology 16 (2015): 1-9. [CrossRef]
- Kakumoto, Kyoko, Jun-ichiro Ikeda, Masato Okada, Eiichi Morii, and Chitose Oneyama. "Mlst8 Promotes Mtor-Mediated Tumor Progression." PLoS One 10, no. 4 (2015): e0119015.
- Gallo, Alberto, Mirko Ronzio, Eugenia Bezzecchi, Roberto Mantovani, and Diletta Dolfini. "Nf-Y Subunits Overexpression in Gastric Adenocarcinomas (Stad)." Scientific reports 11, no. 1 (2021): 23764.
- Liu, Pei-Feng, Chun-Feng Chen, Chih-Wen Shu, Hui-Min Chang, Cheng-Hsin Lee, Huei-Han Liou, Luo-Ping Ger, Chun-Lin Chen, and Bor-Hwang Kang. "Ube2c Is a Potential Biomarker for Tumorigenesis and Prognosis in Tongue Squamous Cell Carcinoma." Diagnostics 10, no. 9 (2020): 674. [CrossRef]
- Wang, Ying, Feifei Huang, Ming Liu, and Quan Zhao. "Ube2c Mrna Expression Controlled by Mir-300 and Hur Determines Its Oncogenic Role in Gastric Cancer." Biochemical and Biophysical Research Communications 534 (2021): 597-603.
- Chen, Peng, Zhiwei He, Jie Wang, Jian Xu, Xueyi Jiang, Yankun Chen, Xinyuan Liu, and Jianxin Jiang. "Hypoxia-Induced Zwint Mediates Pancreatic Cancer Proliferation by Interacting with P53/P21." Frontiers in cell and developmental biology 9 (2021): 682131.
- Marks, Jeffrey R. "Refining the Role of Brca1 in Combating Oxidative Stress." Breast Cancer Research 15 (2013): 1-2. [CrossRef]
- Zhang, Youwei, and Tony Hunter. "Roles of Chk1 in Cell Biology and Cancer Therapy." International journal of cancer 134, no. 5 (2014): 1013-23. [CrossRef]
- Yin, Yuping, Qian Shen, Peng Zhang, Ruikang Tao, Weilong Chang, Ruidong Li, Gengchen Xie, Weizhen Liu, Lihong Zhang, and Prabodh Kapoor. "Chk1 Inhibition Potentiates the Therapeutic Efficacy of Parp Inhibitor Bmn673 in Gastric Cancer." American journal of cancer research 7, no. 3 (2017): 473.
- Matheson, Christopher J, Donald S Backos, and Philip Reigan. "Targeting Wee1 Kinase in Cancer." Trends in pharmacological sciences 37, no. 10 (2016): 872-81.
- Iliaki, Styliani, Rudi Beyaert, and Inna S Afonina. "Polo-Like Kinase 1 (Plk1) Signaling in Cancer and Beyond." Biochemical Pharmacology 193 (2021): 114747.
- Kanaji, Shingo, Hiroaki Saito, Shunichi Tsujitani, Sachiko Matsumoto, Shigeru Tatebe, Akira Kondo, Mitsuhiko Ozaki, Hisao Ito, and Masahide Ikeguchi. "Expression of Polo-Like Kinase 1 (Plk1) Protein Predicts the Survival of Patients with Gastric Carcinoma." Oncology 70, no. 2 (2006): 126-33.
- Li, Qian, Dongdong Tong, Xintao Jing, Peihan Ma, Fang Li, Qiuyu Jiang, Jinyuan Zhang, Hua Wen, Manli Cui, and Chen Huang. "Mad2l1 Is Transcriptionally Regulated by Tead4 and Promotes Cell Proliferation and Migration in Colorectal Cancer." Cancer Gene Therapy 30, no. 5 (2023): 727-37. [CrossRef]
- Seidlitz, Therese, Bon-Kyoung Koo, and Daniel E Stange. "Gastric Organoids—an in Vitro Model System for the Study of Gastric Development and Road to Personalized Medicine." Cell Death & Differentiation 28, no. 1 (2021): 68-83. [CrossRef]
- Liao, Guo-Bin, Xin-Zhe Li, Shuo Zeng, Cheng Liu, Shi-Ming Yang, Li Yang, Chang-Jiang Hu, and Jian-Ying Bai. "Regulation of the Master Regulator Foxm1 in Cancer." Cell Communication and Signaling 16, no. 1 (2018): 57. [CrossRef]
- Zhang, Yan, Xiangming Fang, Fen Shuang, and Guoxun Chen. "Dexamethasone Potentiates the Insulin-Induced Srebp-1c Expression in Primary Rat Hepatocytes." Food Science and Human Wellness 12, no. 5 (2023): 1519-25.
- Nakamura, Tomoko, Akira Iwase, B Bayasula, Yoshinari Nagatomo, Mika Kondo, Tatsuo Nakahara, Sachiko Takikawa, Maki Goto, Tomomi Kotani, and Tohru Kiyono. "Cyp51a1 Induced by Growth Differentiation Factor 9 and Follicle-Stimulating Hormone in Granulosa Cells Is a Possible Predictor for Unfertilization." Reproductive sciences 22, no. 3 (2015): 377-84.
- Ershov, Pavel, Leonid Kaluzhskiy, Yuri Mezentsev, Evgeniy Yablokov, Oksana Gnedenko, and Alexis Ivanov. "Enzymes in the Cholesterol Synthesis Pathway: Interactomics in the Cancer Context." Biomedicines 9, no. 8 (2021): 895.
- Yang, Yongfei, Meiying Luo, Kexin Zhang, Jun Zhang, Tongtong Gao, Douglas O’ Connell, Fengping Yao, Changwen Mu, Bingyu Cai, and Yuxue Shang. "Nedd4 Ubiquitylates Vdac2/3 to Suppress Erastin-Induced Ferroptosis in Melanoma." Nature communications 11, no. 1 (2020): 433.
- Liu, Pulin, Naifei Xing, Zhikai Xiahou, Jingwei Yan, Zhiheng Lin, and Junlong Zhang. "Unraveling the Intricacies of Glioblastoma Progression and Recurrence: Insights into the Role of Nfyb and Oxidative Phosphorylation at the Single-Cell Level." Frontiers in Immunology 15 (2024): 1368685.
- Beishline, Kate, and Jane Azizkhan-Clifford. "Sp1 and the ‘Hallmarks of Cancer’." The FEBS journal 282, no. 2 (2015): 224-58.
- He, Ying, Shasha Qi, Lu Chen, Jinyu Zhu, Linda Liang, Xudong Chen, Hao Zhang, Lvjia Zhuo, Shujuan Zhao, and Shuiping Liu. "The Roles and Mechanisms of Srebp1 in Cancer Development and Drug Response." Genes & Diseases 11, no. 4 (2024): 100987.
- Guarrera, Luca, Mami Kurosaki, Silvio-Ken Garattini, Maurizio Gianni’, Gianpiero Fasola, Luca Rossit, Michele Prisciandaro, Maria Di Bartolomeo, Marco Bolis, and Paola Rizzo. "Anti-Tumor Activity of All-Trans Retinoic Acid in Gastric-Cancer: Gene-Networks and Molecular Mechanisms." Journal of Experimental & Clinical Cancer Research 42, no. 1 (2023): 298. [CrossRef]
- Lauder, I, AM Zaitoun, and WA Aherne. "The Effects of Dexamethasone on the Cell Kinetics of a Murine Malignant Lymphoma." The Journal of Pathology 129, no. 1 (1979): 1-8. [CrossRef]
- Goya, Luis, Anita C Maiyar, Ying Ge, and Gary L Firestone. "Glucocorticoids Induce a G1/G0 Cell Cycle Arrest of Con8 Rat Mammary Tumor Cells That Is Synchronously Reversed by Steroid Withdrawal or Addition of Transforming Growth Factor-Alpha." Molecular endocrinology 7, no. 9 (1993): 1121-32.
- He, Wenning, and Jun Meng. "Cdc20: A Novel Therapeutic Target in Cancer." American Journal of Translational Research 15, no. 2 (2023): 678.
- Jalali, Pooya, Amir Samii, Malihe Rezaee, Arvin Shahmoradi, Fatemeh Pashizeh, and Zahra Salehi. "Ube2c: A Pan-Cancer Diagnostic and Prognostic Biomarker Revealed through Bioinformatics Analysis." Cancer Reports 7, no. 4 (2024): e2032.
- Mylka, Viacheslav, Julie Deckers, Dariusz Ratman, Lode De Cauwer, Jonathan Thommis, Riet De Rycke, Francis Impens, Claude Libert, Jan Tavernier, and Wim Vanden Berghe. "The Autophagy Receptor Sqstm1/P62 Mediates Anti-Inflammatory Actions of the Selective Nr3c1/Glucocorticoid Receptor Modulator Compound a (Cpda) in Macrophages." Autophagy 14, no. 12 (2018): 2049-64. [CrossRef]
- Zou, Juan, Yaokun Chen, Zeqi Ji, Danyi Liu, Xin Chen, Mengjia Chen, Kexun Chen, Haojia Lin, Yexi Chen, and Zhiyang Li. "Identification of C4bpa as Biomarker Associated with Immune Infiltration and Prognosis in Breast Cancer." Translational Cancer Research 13, no. 1 (2024): 25. [CrossRef]
- Quadros, Edward V, Yasumi Nakayama, and Jeffrey M Sequeira. "Saporin Conjugated Monoclonal Antibody to the Transcobalamin Receptor Tcblr/Cd320 Is Effective in Targeting and Destroying Cancer Cells." Journal of cancer therapy 4, no. 6 (2013): 1074.
- Yan, Hong-fa, Ting Zou, Qing-zhang Tuo, Shuo Xu, Hua Li, Abdel Ali Belaidi, and Peng Lei. "Ferroptosis: Mechanisms and Links with Diseases." Signal transduction and targeted therapy 6, no. 1 (2021): 49.
- Tang, Daolin, Xin Chen, Rui Kang, and Guido Kroemer. "Ferroptosis: Molecular Mechanisms and Health Implications." Cell research 31, no. 2 (2021): 107-25. [CrossRef]
- Li, Huifang, Shuxia Jiang, Chun Yang, Shu Yang, Bin He, Wenqiang Ma, and Ruqian Zhao. "Long-Term Dexamethasone Exposure Down-Regulates Hepatic Tfr1 and Reduces Liver Iron Concentration in Rats." Nutrients 9, no. 6 (2017): 617. [CrossRef]
- Dong, Xiao-Ying, and Sheng-Qiu Tang. "Insulin-Induced Gene: A New Regulator in Lipid Metabolism." Peptides 31, no. 11 (2010): 2145-50.
- Ferrè, Silvia, Jeroen HF de Baaij, Patrick Ferreira, Roger Germann, Johannis BC de Klerk, Marla Lavrijsen, Femke van Zeeland, Hanka Venselaar, Leo AJ Kluijtmans, and Joost GJ Hoenderop. "Mutations in Pcbd1 Cause Hypomagnesemia and Renal Magnesium Wasting." Journal of the American Society of Nephrology 25, no. 3 (2014): 574-86.
- Zeng, Xi, Hao-Ying Wang, Yu-Ping Wang, Su-Yang Bai, Ke Pu, Ya Zheng, Qing-Hong Guo, Quan-Lin Guan, Rui Ji, and Yong-Ning Zhou. "Col4a Family: Potential Prognostic Biomarkers and Therapeutic Targets for Gastric Cancer." Translational Cancer Research 9, no. 9 (2020): 5218. [CrossRef]
- Liu, Shanshan, Wei Zhao, Xuemei Li, La Zhang, Yu Gao, Qiling Peng, Chengyou Du, and Ning Jiang. "Agtrap Is a Prognostic Biomarker Correlated with Immune Infiltration in Hepatocellular Carcinoma." Frontiers in oncology 11 (2021): 713017.
- Liu, Yan, Jihai Zhu, Jun Liu, Xueman Ma, Jun Zhao, and Zhanhai Su. "Knockdown of Gastrin Promotes Apoptosis of Gastric Cancer Cells by Decreasing Ros Generation." BioMed Research International 2021, no. 1 (2021): 5590037. [CrossRef]
- Tang, Li, Chengming Guo, Xu Li, Bo Zhang, and Liuye Huang. "Taf15 Promotes Cell Proliferation, Migration and Invasion of Gastric Cancer Via Activation of the Raf1/Mek/Erk Signalling Pathway." Scientific reports 13, no. 1 (2023): 5846. [CrossRef]
- Wang, Yu, Zhicheng Zhao, GuiMei Li, Qin Jin, and Shu Zhang. "Human Gastric Cancer Decellularized Scaffold Promotes Epithelial-Mesenchymal Transition of Gastric Cancer Cells in Vitro with the Involvement of Elf1." Human Gastric Cancer Decellularized Scaffold Promotes Epithelial-Mesenchymal Transition of Gastric Cancer Cells in Vitro with the Involvement of Elf1.
- Liu, Xingzhu, Chang Xu, Wanglong Xiao, and Nianlong Yan. "Unravelling the Role of Nfe2l1 in Stress Responses and Related Diseases." Redox biology 65 (2023): 102819. [CrossRef]
- Wang, Bin, Hanfei Guo, Hongquan Yu, Yong Chen, Haiyang Xu, and Gang Zhao. "The Role of the Transcription Factor Egr1 in Cancer." Frontiers in oncology 11 (2021): 642547.
- Graziano, Francesco, Nicholas W Fischer, Irene Bagaloni, Maria Di Bartolomeo, Sara Lonardi, Bruno Vincenzi, Giuseppe Perrone, Lorenzo Fornaro, Elena Ongaro, and Giuseppe Aprile. "Tp53 Mutation Analysis in Gastric Cancer and Clinical Outcomes of Patients with Metastatic Disease Treated with Ramucirumab/Paclitaxel or Standard Chemotherapy." Cancers 12, no. 8 (2020): 2049.
- Kim, Min Sung, Nam Jin Yoo, and Sug Hyung Lee. "Expressional and Mutational Analysis of Crebbp Gene in Gastric and Colorectal Cancers with Microsatellite Instability." Pathology & Oncology Research 20 (2014): 221-22.
- Chaithongyot, Supattra, Phatcharida Jantaree, Olga Sokolova, and Michael Naumann. "Nf-Κb in Gastric Cancer Development and Therapy." Biomedicines 9, no. 8 (2021): 870.
- Du, Qiupeng, Chenchen Zhu, Qingqing Shang, Haiyan Mao, Xiaoyun Li, Yingchun Huang, and Na Du. "A Study on the Correlation of Myd88 Expression with Gastric Cancer." International Journal of Clinical and Experimental Pathology 11, no. 10 (2018): 4836.
- Talukdar, Priyanka Dey, and Urmi Chatterji. "Transcriptional Co-Activators: Emerging Roles in Signaling Pathways and Potential Therapeutic Targets for Diseases." Signal transduction and targeted therapy 8, no. 1 (2023): 427.
- Guo, Qing, Yizi Jin, Xinyu Chen, Xiaomin Ye, Xin Shen, Mingxi Lin, Cheng Zeng, Teng Zhou, and Jian Zhang. "Nf-Κb in Biology and Targeted Therapy: New Insights and Translational Implications." Signal transduction and targeted therapy 9, no. 1 (2024): 53. [CrossRef]
- Ivashkiv, Lionel B. "Ifnγ: Signalling, Epigenetics and Roles in Immunity, Metabolism, Disease and Cancer Immunotherapy." Nature Reviews Immunology 18, no. 9 (2018): 545-58.
- Ahn, So Yeong, Jin Kim, Min A Kim, Jiwoon Choi, and Woo Ho Kim. "Increased Hgf Expression Induces Resistance to C-Met Tyrosine Kinase Inhibitors in Gastric Cancer." Anticancer Research 37, no. 3 (2017): 1127-38. [CrossRef]
- Liu, Xinmei, Shasha Huang, Yun Guan, and Qing Zhang. "Long Noncoding Rna Oser1-As1 Promotes the Malignant Properties of Non-Small Cell Lung Cancer by Sponging Microrna-433-3p and Thereby Increasing Smad2 Expression." Oncology reports 44, no. 2 (2020): 599-610.
- Chu, Hang Yin, Zihao Chen, Luyao Wang, Zong-Kang Zhang, Xinhuan Tan, Shuangshuang Liu, Bao-Ting Zhang, Aiping Lu, Yuanyuan Yu, and Ge Zhang. "Dickkopf-1: A Promising Target for Cancer Immunotherapy." Frontiers in Immunology 12 (2021): 658097.
- Ebrahimnezhad, Mohammad, Mohammad Natami, Ghazaleh Hafezi Bakhtiari, Peyman Tabnak, Niloufar Ebrahimnezhad, Bahman Yousefi, and Maryam Majidinia. "Foxo1, a Tiny Protein with Intricate Interactions: Promising Therapeutic Candidate in Lung Cancer." Biomedicine & Pharmacotherapy 169 (2023): 115900. [CrossRef]
- Lin, Ching-Yih, Ying-En Lee, Yu-Feng Tian, Ding-Ping Sun, Ming-Jen Sheu, Chen-Yi Lin, Chien-Feng Li, Sung-Wei Lee, Li-Ching Lin, and I-Wei Chang. "High Expression of Epha4 Predicted Lesser Degree of Tumor Regression after Neoadjuvant Chemoradiotherapy in Rectal Cancer." Journal of Cancer 8, no. 6 (2017): 1089.
- Greuber, Emileigh K, Pameeka Smith-Pearson, Jun Wang, and Ann Marie Pendergast. "Role of Abl Family Kinases in Cancer: From Leukaemia to Solid Tumours." Nature Reviews Cancer 13, no. 8 (2013): 559-71.
- Kim, Eric S, and Ravi Salgia. "Met Pathway as a Therapeutic Target." Journal of Thoracic Oncology 4, no. 4 (2009): 444-47.
- Pelaz, Sara G, and Arantxa Tabernero. "Src: Coordinating Metabolism in Cancer." Oncogene 41, no. 45 (2022): 4917-28. [CrossRef]
- Zhang, Ying, and Zhaoyong Liu. "Stat1 in Cancer: Friend or Foe?" Discovery medicine 24, no. 130 (2017): 19-29.
- Jiménez-Martínez, Marta, Konstantinos Stamatakis, and Manuel Fresno. "The Dual-Specificity Phosphatase 10 (Dusp10): Its Role in Cancer, Inflammation, and Immunity." International journal of molecular sciences 20, no. 7 (2019): 1626.
- Zhang, Keyi, Xiao Ni, Xiaoling Ma, Rui Sun, Jiangnan Qiu, and Chengyan Luo. "Linc01012 Upregulation Promotes Cervical Cancer Proliferation and Migration Via Downregulation of Cdkn2d." Oncology Letters 25, no. 3 (2023): 1-9. [CrossRef]
- Silva-Rodríguez, Paula, Daniel Fernández-Díaz, Manuel Bande, María Pardo, Lourdes Loidi, and María José Blanco-Teijeiro. "Gnaq and Gna11 Genes: A Comprehensive Review on Oncogenesis, Prognosis and Therapeutic Opportunities in Uveal Melanoma." Cancers 14, no. 13 (2022): 3066.
- Vera-Puente, Olga, Carlos Rodriguez-Antolin, Ana Salgado-Figueroa, Patrycja Michalska, Olga Pernia, Brett M Reid, RocÍo Rosas, Alvaro Garcia-Guede, Silvia SacristÁn, and Julia Jimenez. "Mafg Is a Potential Therapeutic Target to Restore Chemosensitivity in Cisplatin-Resistant Cancer Cells by Increasing Reactive Oxygen Species." Translational Research 200 (2018): 1-17.
- Marni, Rakshmitha, Anindita Chakraborty, and RamaRao Malla. "Oncogenic Tetraspanins: Implications for Metastasis, Drug Resistance, Cancer Stem Cell Maintenance and Diagnosis of Leading Cancers in Females." Gene Reports 27 (2022): 101548. [CrossRef]
- Mast, Alan E, Alisa S Wolberg, David Gailani, Michael R Garvin, Christiane Alvarez, J Izaak Miller, Bruce Aronow, and Daniel Jacobson. "Sars-Cov-2 Suppresses Anticoagulant and Fibrinolytic Gene Expression in the Lung." Elife 10 (2021): e64330.
- Shan, Qingqing, Chi Zhang, Yangke Li, Qunying Li, Yifan Zhang, Xue Li, Junqing Shi, and Fengying Hu. "Slc7a11, a Potential Immunotherapeutic Target in Lung Adenocarcinoma." Scientific reports 13, no. 1 (2023): 18302. [CrossRef]
- Jiang, Yun, Jingyi Cui, Ming Cui, and Rongrong Jing. "Slc7a11 Promotes the Progression of Gastric Cancer and Regulates Ferroptosis through Pi3k/Akt Pathway." Pathology-Research and Practice 248 (2023): 154646.
- Sun, Fengfeng, Peng Sun, Xiaofeng Yang, Liangliang Hu, Jianguo Gao, and Tao Tian. "Inhibition of Fstl3 Abates the Proliferation and Metastasis of Renal Cell Carcinoma Via the Gsk-3β/Β-Catenin Signaling Pathway." Aging (Albany NY) 13, no. 18 (2021): 22528.
- Liu, Yuan-Jie, Jie-Pin Li, Ying Zhang, Meng-Jun Nie, Yong-Hua Zhang, Shen-Lin Liu, and Xi Zou. "Fstl3 Is a Prognostic Biomarker in Gastric Cancer and Is Correlated with M2 Macrophage Infiltration." OncoTargets and therapy (2021): 4099-117.
- Nakano, Shun, Masashi Nishikawa, Tomoyo Kobayashi, Eka Wahyuni Harlin, Takuya Ito, Katsuya Sato, Tsuyoshi Sugiyama, Hisashi Yamakawa, Takahiro Nagase, and Hiroshi Ueda. "The Rho Guanine Nucleotide Exchange Factor Plekhg1 Is Activated by Interaction with and Phosphorylation by Src Family Kinase Member Fyn." Journal of Biological Chemistry 298, no. 2 (2022).
- Zhou, Xu-Dong, Ya-Wei Qu, Li Wang, Fu-Hua Jia, Peng Chen, Yin-Pu Wang, and Hai-Feng Liu. "Identification of Potential Hub Genes of Gastric Cancer." Medicine 101, no. 41 (2022): e30741. [CrossRef]
- Raposo, Teresa P, Susanti Susanti, and Mohammad Ilyas. "Investigating Tns4 in the Colorectal Tumor Microenvironment Using 3d Spheroid Models of Invasion." Advanced Biosystems 4, no. 6 (2020): 2000031. [CrossRef]
- Kaneda, Atsushi, Michio Kaminishi, Yukihiro Nakanishi, Takashi Sugimura, and Toshikazu Ushijima. "Reduced Expression of the Insulin-Induced Protein 1 and P41 Arp2/3 Complex Genes in Human Gastric Cancers." International journal of cancer 100, no. 1 (2002): 57-62.
- Jakobsen, Sebastian, and Carsten Uhd Nielsen. "Exploring Amino Acid Transporters as Therapeutic Targets for Cancer: An Examination of Inhibitor Structures, Selectivity Issues, and Discovery Approaches." Pharmaceutics 16, no. 2 (2024): 197.
| Conc. μg/mL 1:1 |
Cell Growth Inhibition (%) of AGS cells |
Cell viability (%) of HS738.St/Int |
||||
| PB | AP | AB | PB | AP | AB | |
| 1500+1500 | 99.22 ± 1.42 a w | 59.79 ± 4.32 ax | 99.49 ± 0.85 ay | 58.86 ± 9.52 ax | 82.39 ± 12.30 ax | 61.08 ± 13.45 az |
| 750+750 | 91.14 ± 1.78 a w | 37.67 ± 9.68 bx | 90.08 ± 1.86 ax | 78.04 ± 4.98 aw | 93.48 ± 12.89 ax | 84.38 ± 13.01 ax |
| 375+375 | 75.93 ± 1.92 b,w | 29.31 ± 6.50 bx | 76.62 ± 2.81 by | 81.09 ± 10.51 aw | 107.71 ± 33.81 ay | 102.51 ± 17.25 ay |
| 187.5 +187.5 | 51.36 ± 7.50 cw | 23.87 ± 6.76 bw | 49.95 ± 7.31 cx | 100.62 ± 16.88 aw | 143.38 ± 26.78 ax | 103.77 ± 10.35 ax |
| 93.75 + 93.75 | 25.11 ± 8.21 dw | 19.20 ± 7.46 bx | 24.28 ± 7.26 dy | 127.45 ± 26.75 ax | 149.37 ± 9.05 ay | 122.20 ± 16.51 az |
| 46.875 + 46.875 | 9.79 ± 7.25 ew | 11.98 ± 12.23 bx | 12.01 ± 8.06 ey | 138.92 ± 9.91 ax | 175.62 ± 4.32 ay | 149.29 ± 21.90 az |
| IC50 | 421.23 ± 15.31 | 1141.13 ± 362.00 | 446.53 ± 19.55 | NA | NA | NA |
| Conc. μg/mL |
Cell Growth Inhibition (%) of AGS cells |
Conc. μg/mL |
Cell Growth Inhibition (%) of AGS cells |
Conc. μg/mL APB+Dex |
Cell Growth Inhibition (%) of AGS cells |
Cell viability (%) of HS738.St/Int |
|---|---|---|---|---|---|---|
| APB | Dex | APB+Dex | ||||
| 3000 | 95.65 ± 7.90 ax | 200 | 80.46 ± 8.08 ax | 3000 + 100 | 103.17 ± 3.06 ax | 68.57 ± 11.74 ax |
| 1500 | 86.25 ± 8.42 ax | 100 | 39.38 ± 24.51bx | 1500 + 50 | 87.81 ± 5.57 bx | 85.91 ± 10.59 ax |
| 750 | 65.54 ± 4.91 bx | 50 | - | 750 + 25 | 62.42 ± 5.34 cx | 110.88 ± 14.35 ay |
| 375 | 40.28 ± 8.05 cx | 25 | - | 375+ 12.5 | 43.49 ± 10.93 dx | 115.29 ± 16.81 ay |
| 187.5 | 22.20 ± 8.55 dx | 12.5 | - | 187.5 + 6.25 | 28.70 ± 11.91 ex | 134.46 ± 24.72 ay |
| 93.75 | 19.20 ± 10.38 dx | 6.25 | - | 93.75 + 3.13 | 16.46 ± 9.71 fx | 159.21 ± 14.16 ay |
| IC50 | 568.33 ± 82.56 | IC50 | 86.60 ±11.85 | IC50 | 643.30 ± 58.26 | NA |
| Combination Index (CI) values at: | ||||
|---|---|---|---|---|
| Combinations | IC50 | IC75 | IC90 | IC95 |
| AP 1:1 (1500 μg/mL A + 1500 μg/mL P) | 3.53 | 7.28 | 17.13 | 31.66 |
| AB 1:1 (1500 μg/mL A + 1500 μg/mL B) | 1.14 | 1.37 | 1.80 | 2.19 |
| BP 1:1 (1500 μg/mL B + 1500 μg/mL P) | 1.41 | 1.68 | 2.11 | 2.51 |
| APB + Dex (3000 μg/mL APB + 100 μg/mL Dex) | 0.76 | 0.48 | 0.31 | 0.23 |
| GENE | PROTEIN | LOG2FC | ROLE | REF |
|---|---|---|---|---|
| OSER1 | Oxidative stress-responsive serine-rich protein 1 | -2.39 | Noted for its role in the negative regulation of intracellular signal transduction. |
[118] |
| DKK1 | Dickkopf-related protein 1 | -1.54 | Overexpressed in cancer, affecting Wnt signaling pathways. Overexpression correlates with poor survival in gastric cancer. |
[119] |
| FOXO1 | Forkhead box protein O1 | -1.21 | A tumour suppressor transcription factor linked to cancer. | [120] |
| EPHA4 | Ephrin type-A receptor 4 | -1.07 | A receptor tyrosine kinase promoting cancer progression. |
[121] |
|
ABL1 ABL2 |
Tyrosine kinase ABL1 and ABL2 | -0.79 -0.76 |
Proto-oncogenes involved in cell differentiation, division, and adhesion and is linked various cancers, especially leukemia. Altered signalling associated with gastric cancer. |
[122] |
| MET | Hepatocyte growth factor receptor | -0.78 | A proto-oncogene involved in several cancers, including gastric cancer. Overexpression and mutations linked to poor prognosis in gastric cancer. |
[123] |
| SRC | Proto-oncogene tyrosine-protein kinase Src | -0.68 | Associated with numerous cancers through oncogenic signalling. Promotes gastric cancer progression through activation of oncogenic pathways. |
[124] |
| STAT1 | Signal transducer and activator of transcription 1-alpha/beta | 0.85 | Influences cancer progression and immune responses |
[125] |
| DUSP10 | Dual specificity protein phosphatase 10 | 1.33 | Regulates pathways connected to cancer development. | [126] |
| CDKN2D | Cyclin-dependent kinase 4 inhibitor D | 1.18 | A cyclin-dependent kinase inhibitor linked to multiple cancers. |
[127] |
| EPCAM | Epithelial cell adhesion molecule | -1.04 | Cell adhesion and signaling; upregulated in gastric tumors for proliferation and metastasis. | [33] |
| GNAQ | Guanine nucleotide-binding protein G(q) subunit alpha | -0.64 | Oncogene in G-protein signaling; implicated in tumor progression. |
[128] |
| GNAS | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas | -0.73 | Oncogenic signaling driver; mutated in some gastric cancers. |
[128] |
| MAFG | Transcription factor MafG | -0.65 | Transcription factor in oxidative stress response; linked to oncogenesis. | [129] |
| TSPAN1, TSPAN6, TSPAN8, TSPAN14, TSPAN15, TSPAN31 | Tetraspanin | -0.78 to -1.49 |
Roles in cell signaling, adhesion, and metastasis. |
[130] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





