Submitted:
14 June 2025
Posted:
18 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Isolation of Culturable Endophytic Streptomycetes from Olive Roots
2.2. In Vitro Selection of Isolates Based on Antifungal Activity in Plate Assays
2.3. Analysis of Antifungal Activity in Small-Scale Soil Tests
2.4. In Vitro Confrontation of Endophytic Streptomyces Strains
2.5. Molecular Identification of Strains and Genome Sequencing
2.6. Cultural and Growth Properties
2.7. Phenotypic and Chemotaxonomic Properties
2.8. Phylogeny and Comparative Genomic Analyses
2.9. In Silico Screening for Secondary Metabolites
2.10. Fermentation, Culture Extract Preparation, and HPLC Analysis of Antifungal Activity
2.11. Purification of Rapamycin and Epoxinnamide by Vacuum Flash Chromatography (VFC)
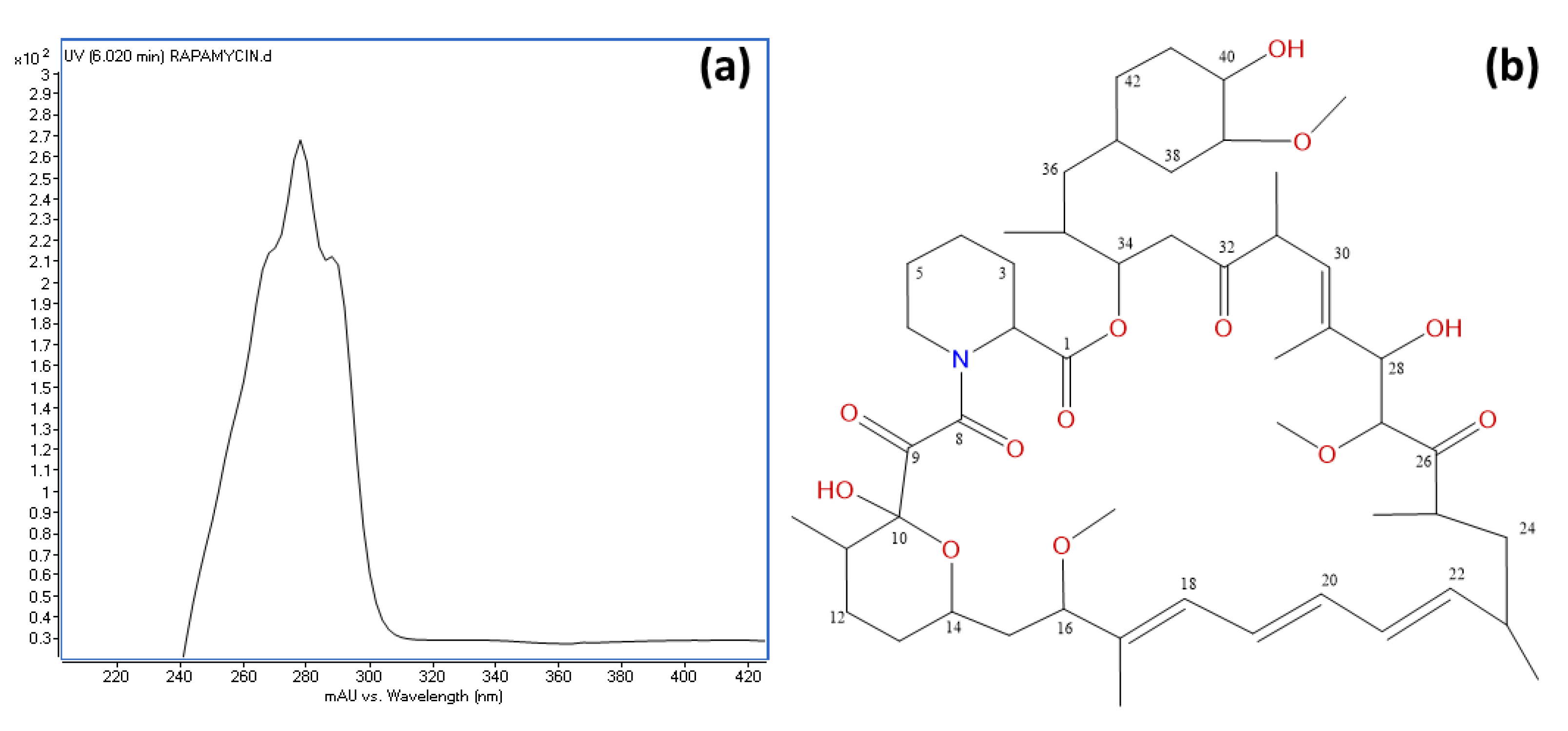
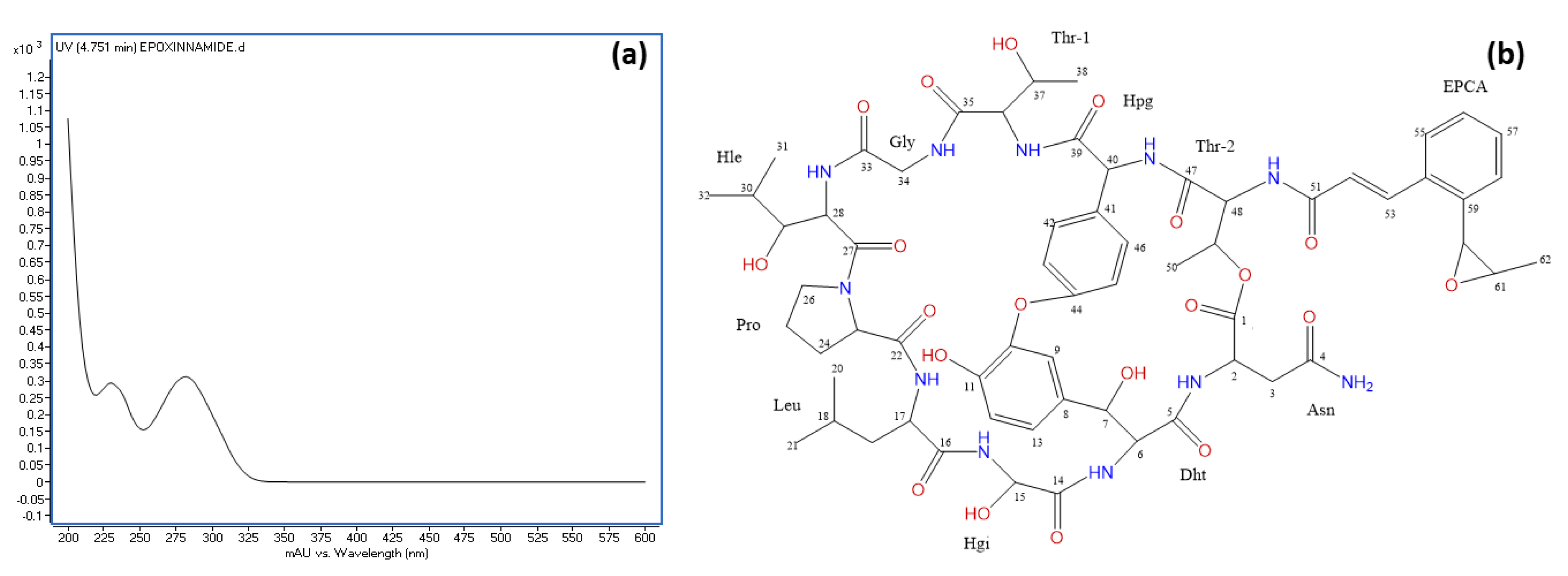
2.12. Structural Elucidation of Rapamycin and Epoxinnamide
2.13. Analysis of Rapamycin’s Inhibitory Effect on Conidiospore Germination and Hyphal Growth
3. Results
3.1. Isolation and Selection of Culturable Endophytic Streptomycetes Associated with Olive Roots and Their Antifungal Activity Against V. dahliae
3.2. Evaluation of Antifungal Activity in a Small-Scale Soil Assay
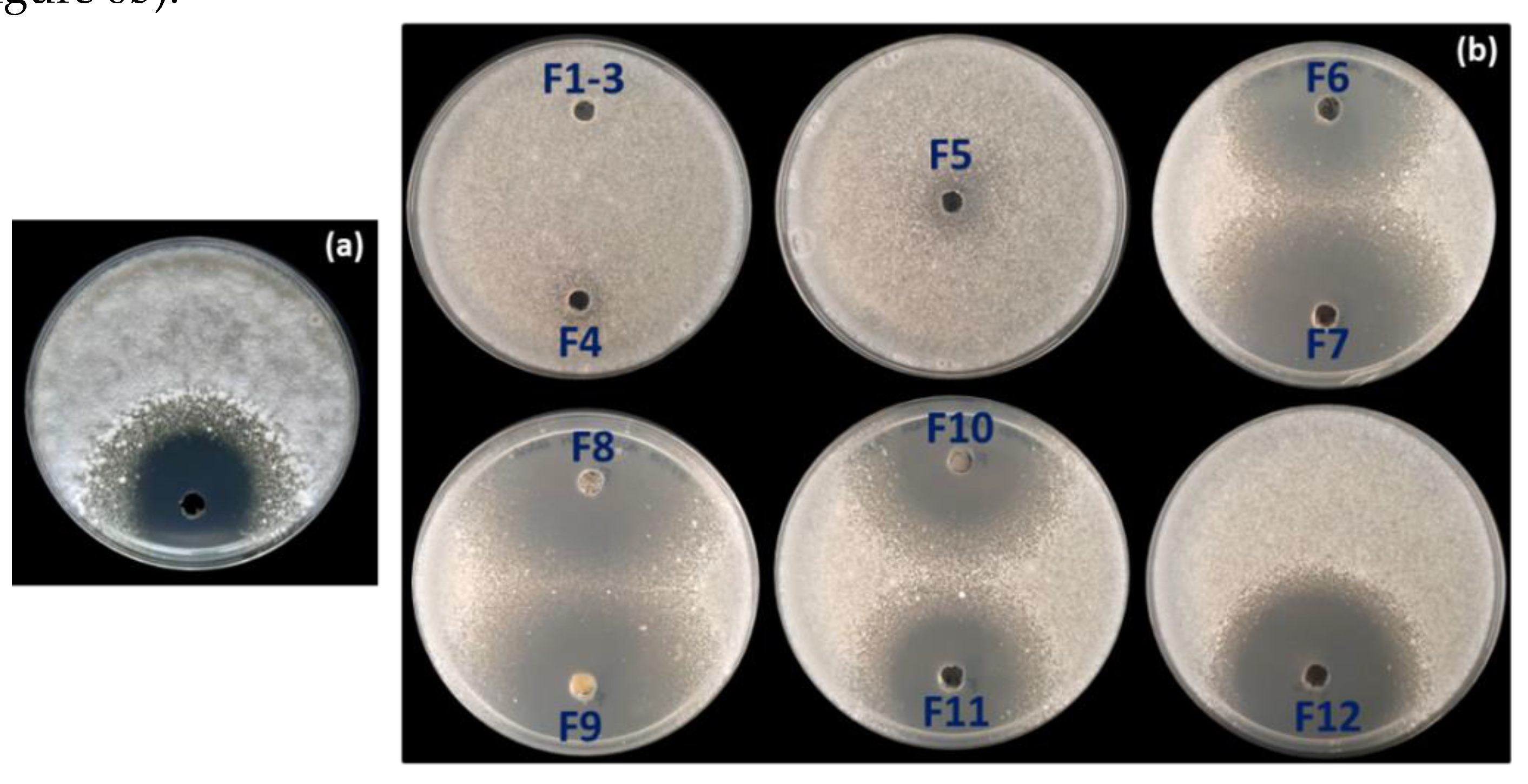
3.3. Strain-Specific Inhibition Patterns in Streptomyces–Streptomyces Interaction Bioassays
3.4. Molecular Identification of Isolates OE54 and OE57T Using 16S rRNA Analysis, Multilocus Sequence Analysis (MLSA), and Comparative Genomics
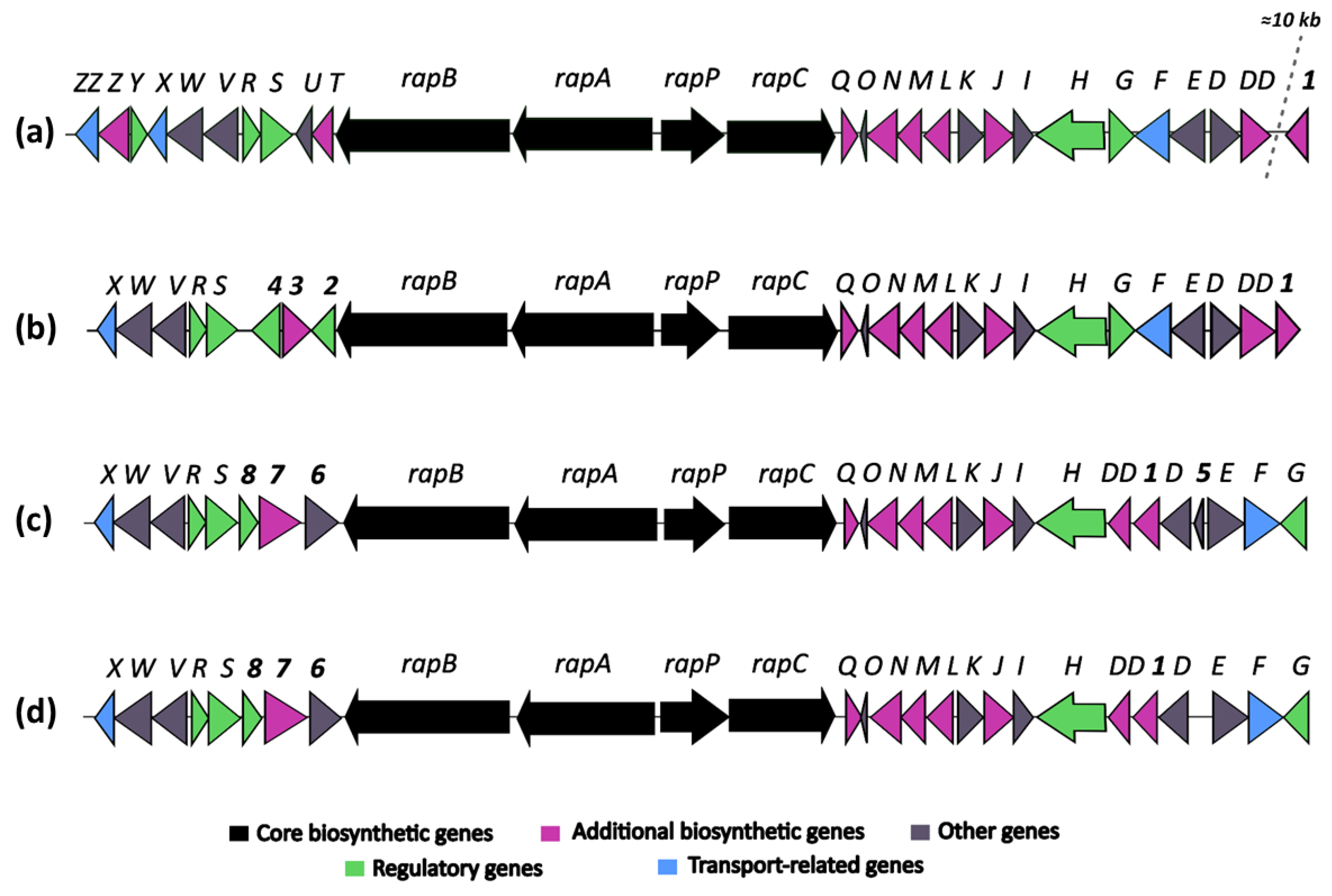
3.5. In Silico Screening for Secondary Metabolite Production in Strains OE54 and OE57T
3.6. Identification of Rapamycin as the Main Antifungal Compound Produced by OE54 and OE57T Strains
3.7. Identification of the Epoxy Cinnamoyl-Containing Nonribosomal Peptide Epoxinnamide Produced by OE54
3.8. Effect of Rapamycin on V. dahliae Conidiospore Germination, Radial Growth Inhibition, and MIC Determination
3.9. Cultural, Morphological, and Phenotypic Properties of OE54 and OE57 Strains
3.10. Chemotaxonomic Features
3.11. Description of Streptomyces lacaronensis sp. nov.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Inderbitzin, P.; Subbarao, K. V. Verticillium Systematics and Evolution: How Confusion Impedes Verticillium Wilt Management and How to Resolve It. Phytopathology 2014, 104, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K. V. Diversity, Pathogenicity, and Management of Verticillium Species. Annu Rev Phytopathol 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Keykhasaber, M.; Thomma, B.P.H.J.; Hiemstra, J.A. Verticillium Wilt Caused by Verticillium dahliae in Woody Plants with Emphasis on Olive and Shade Trees. Eur J Plant Pathol 2018, 150, 21–37. [Google Scholar] [CrossRef]
- Kunej, U.; Jakše, J.; Radišek, S.; Štajner, N. Identification and Characterization of Verticillium nonalfalfae-Responsive MicroRNAs in the Roots of Resistant and Susceptible Hop Cultivars. Plants 2021, 10. [Google Scholar] [CrossRef]
- López-Escudero, F.J.; Mercado-Blanco, J. Verticillium Wilt of Olive: A Case Study to Implement an Integrated Strategy to Control a Soil-Borne Pathogen. Plant Soil 2011, 344, 1–50. [Google Scholar] [CrossRef]
- Coque, J.J.R.; Álvarez-Pérez, J.M.; Cobos, R.; González-García, S.; Ibáñez, A.M.; Diez Galán, A.; Calvo-Peña, C. Advances in the Control of Phytopathogenic Fungi That Infect Crops through Their Root System. In: Advances in Applied Microbiology; Gadd, G.M., Sariaslani, S., Eds.; Academic Press Inc., 2020; Vol. 111, pp. 123–170.
- Mulero-Aparicio, A.; Agustí-Brisach, C.; Varo, Á.; López-Escudero, F.J.; Trapero, A. A Non-Pathogenic Strain of Fusarium oxysporum as a Potential Biocontrol Agent against Verticillium Wilt of Olive. Biological Control 2019, 139, 104045. [Google Scholar] [CrossRef]
- Mulero-Aparicio, A.; Cernava, T.; Turrà, D.; Schaefer, A.; Di Pietro, A.; López-Escudero, F.J.; Trapero, A.; Berg, G. The Role of Volatile Organic Compounds and Rhizosphere Competence in Mode of Action of the Non-Pathogenic Fusarium oxysporum FO12 toward Verticillium Wilt. Front Microbiol 2019, 10, 1808. [Google Scholar] [CrossRef]
- Varo, A.; Raya-Ortega, M.C.; Trapero, A. Selection and Evaluation of Micro-Organisms for Biocontrol of Verticillium dahliae in Olive. J Appl Microbiol 2016, 121, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Carrero-Carrón, I.; Trapero-Casas, J.L.; Olivares-García, C.; Monte, E.; Hermosa, R.; Jiménez-Díaz, R.M. Trichoderma asperellum Is Effective for Biocontrol of Verticillium Wilt in Olive Caused by the Defoliating Pathotype of Verticillium dahliae. Crop Protection 2016, 88, 45–52. [Google Scholar] [CrossRef]
- Lozano-Tovar, M.D.; Garrido-Jurado, I.; Quesada-Moraga, E.; Raya-Ortega, M.C.; Trapero-Casas, A. Metarhizium brunneum and Beauveria bassiana Release Secondary Metabolites with Antagonistic Activity against Verticillium dahliae and Phytophthora megasperma Olive Pathogens. Crop Protection 2017, 100, 186–195. [Google Scholar] [CrossRef]
- Gómez-Lama Cabanás, C.; Sesmero, R.; Valverde-Corredor, A.; López-Escudero, F.J.; Mercado-Blanco, J. A Split-Root System to Assess Biocontrol Effectiveness and Defense-Related Genetic Responses in above-Ground Tissues during the Tripartite Interaction Verticillium dahliae-Olive-Pseudomonas fluorescens PICF7 in Roots. Plant Soil 2017, 417, 433–452. [Google Scholar] [CrossRef]
- Gómez-Lama Cabanás, C.; Legarda, G.; Ruano-Rosa, D.; Pizarro-Tobías, P.; Valverde-Corredor, A.; Niqui, J.L.; Triviño, J.C.; Roca, A.; Mercado-Blanco, J. Indigenous Pseudomonas spp. Strains from the Olive (Olea europaea L.) Rhizosphere as Effective Biocontrol Agents against Verticillium dahliae: From the Host Roots to the Bacterial Genomes. Front Microbiol 2018, 9, 277. [Google Scholar] [CrossRef]
- Markakis, E.A.; Tjamos, S.E.; Antoniou, P.P.; Paplomatas, E.J.; Tjamos, E.C. Biological Control of Verticillium Wilt of Olive by Paenibacillus alvei, Strain K165. BioControl 2016, 61, 293–303. [Google Scholar] [CrossRef]
- Cheffi Azabou, M.; Gharbi, Y.; Medhioub, I.; Ennouri, K.; Barham, H.; Tounsi, S.; Triki, M.A. The Endophytic Strain Bacillus velezensis OEE1: An Efficient Biocontrol Agent against Verticillium Wilt of Olive and a Potential Plant Growth Promoting Bacteria. Biological Control 2020, 142. [Google Scholar] [CrossRef]
- Calvo-Peña, C.; Cobos, R.; Sánchez-López, J.M.; Ibáñez, A.; R. Coque, J.J. Albocycline Is the Main Bioactive Antifungal Compound Produced by Streptomyces sp. OR6 against Verticillium dahliae. Plants 2023, 12, 3612. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Díaz, M.; Antón-Domínguez, B.I.; Raya, M.C.; Bernal-Cabrera, A.; Medina-Marrero, R.; Trapero, A.; Agustí-Brisach, C. Streptomyces spp. Strains as Potential Biological Control Agents against Verticillium Wilt of Olive. Journal of Fungi 2024, 10. [Google Scholar] [CrossRef]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as Symbionts: An Emerging and Widespread Theme? FEMS Microbiol Rev 2012, 36, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S rRNA and 16S rRNA Genes. Appl Environ Microbiol 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Viaene, T.; Langendries, S.; Beirinckx, S.; Maes, M.; Goormachtig, S. Streptomyces as a Plant’s Best Friend? FEMS Microbiol Ecol 2016, 92. [Google Scholar] [CrossRef]
- Li, J.; Guo-zhen, Z.; Huang, H.; Qin, S.; Zhu, W.-Y.; Zhao, L.-X.; Xu, L.-H.; Zhang, S.; Li, W.-J.; Strobel, G. Isolation and Characterization of Culturable Endophytic Actinobacteria Associated with Artemisia annua L. Antonie Van Leeuwenhoek 2012, 101, 515–527. [CrossRef]
- Álvarez-Pérez, J.M.; González-García, S.; Cobos, R.; Olego, M.Á.; Ibañez, A.; Díez-Galán, A.; Garzón-Jimeno, E.; Coque, J.J.R. Use of Endophytic and Rhizosphere Actinobacteria from Grapevine Plants to Reduce Nursery Fungal Graft Infections That Lead to Young Grapevine Decline. Appl Environ Microbiol 2017, 83. [Google Scholar] [CrossRef]
- Ayswaria, R.; Vasu, V.; Krishna, R. Diverse Endophytic Streptomyces Species with Dynamic Metabolites and Their Meritorious Applications: A Critical Review. Crit Rev Microbiol 2020, 46, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Polpass, J.; Maharshi, A.; Jha, B. Actinobacteria in Natural Products Research: Progress and Prospects. Microbiol Res 2021, 246. [Google Scholar] [CrossRef]
- Van Der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; Van Wezel, G.P. Regulation of Antibiotic Production in Actinobacteria: New Perspectives from the Post-Genomic Era. Nat Prod Rep 2018, 35, 575–604. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F.; Biró, S.; Lee, K.J.; Palmer, T.; Schrempf, H. The Complex Extracellular Biology of Streptomyces. FEMS Microbiol Rev 2010, 34, 171–198. [Google Scholar] [CrossRef]
- Cuervo, L.; Álvarez-García, S.; Salas, J.A.; Méndez, C.; Olano, C.; Malmierca, M.G. The Volatile Organic Compounds of Streptomyces Spp.: An In-Depth Analysis of Their Antifungal Properties. Microorganisms 2023, 11. [Google Scholar] [CrossRef]
- Zhao, Q.; Bertolli, S.; Park, Y.J.; Tan, Y.; Cutler, K.J.; Srinivas, P.; Asfahl, K.L.; Fonesca-García, C.; Gallagher, L.A.; Li, Y.; et al. Streptomyces Umbrella Toxin Particles Block Hyphal Growth of Competing Species. Nature 2024, 629, 165–173. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for the Characterization of Streptomyces Species. Int J Syst Bacteriol 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Martin, J.F.; McDaniel, L.E. Biosynthesis of Candicidin by Phosphate-Limited Resting Cells of Streptomyces griseus. European J. Appl. Microbiol 1976, 3, 135–144. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.; Buttner, M.; Chater, K.; Hopwood, D. Practical Streptomyces Genetics; Norwich: John Innes Foundation, 2000; ISBN 0708406238. [Google Scholar]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-García, C.; Valverde-Corredor, A.; Jiménez-Díaz, R.M. Molecular Variability within and among Verticillium dahliae Vegetative Compatibility Groups Determined by Fluorescent Amplified Fragment Length Polymorphism and Polymerase Chain Reaction Markers. Phytopathology 2006, 96, 485–495. [Google Scholar] [CrossRef]
- Schrey, S.D.; Erkenbrack, E.; Früh, E.; Fengler, S.; Hommel, K.; Horlacher, N.; Schulz, D.; Ecke, M.; Kulik, A.; Fiedler, H.P.; et al. Production of Fungal and Bacterial Growth Modulating Secondary Metabolites Is Widespread among Mycorrhiza-Associated Streptomycetes. BMC Microbiol 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A.; Bibb M.J., C.K.F.; Bruton C.J., K.H.M.; Lydiate D., S.C.P.; Ward J.M., S.H. Genetic Manipulation of Streptomyces: A Laboratory Manual; The John Innes Institute: Norwich, England, 1985.
- Lane, D. 16S/23S rRNA Sequencing. In Nucleic acid techniques in bacterial systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, 1991; pp. 115–175. [Google Scholar]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A Prokaryotic 16S RRNA Gene Sequence Database with Phylotypes That Represent Uncultured Species. Int J Syst Evol Microbiol 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J Mol Evol 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Huang, Y. Taxonomic Evaluation of the Streptomyces hygroscopicus Clade Using Multilocus Sequence Analysis and DNA-DNA Hybridization, Validating the MLSA Scheme for Systematics of the Whole Genus. Syst Appl Microbiol 2012, 35, 7–18. [Google Scholar] [CrossRef]
- Guo, Y.P.; Zheng, W.; Rong, X.Y.; Huang, Y. A Multilocus Phylogeny of the Streptomyces griseus 16S rRNA Gene Clade: Use of Multilocus Sequence Analysis for Streptomycete Systematics. Int J Syst Evol Microbiol 2008, 58, 149–159. [Google Scholar] [CrossRef]
- Rong, X.; Guo, Y.; Huang, Y. Proposal to Reclassify the Streptomyces albidoflavus Clade on the Basis of Multilocus Sequence Analysis and DNA-DNA Hybridization, and Taxonomic Elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol 2009, 32, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long- -Read Assembly via Adaptive k- -Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res 2014, 42, 206–214. [Google Scholar] [CrossRef]
- Schleifer, K.H.; Kandler, O. Peptidoglycan Types of Bacterial Cell Walls and Their Taxonomic Implications. Bacteriol Rev 1972, 36, 407–477. [Google Scholar] [CrossRef]
- Lechevalier, M.P.; Lechevalier, H.A. Chemical Composition as a Criterion in the Classification of Aerobic Actinomycetes. Int J Syst Bacteriol 1970, 20, 435–443. [Google Scholar] [CrossRef]
- Minnikin A’, D.E.; O’donnell A’ B, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An Integrated Procedure for the Extraction of Bacterial Isoprenoid Quinones and Polar Lipids. J Microbiol Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Kroppenstedt, R.M.; Goodfellow, M. The Family Thermomonosporaceae: Actinocorallia, Actinomadura, Spirillispora and Thermomonospora. In The Prokaryotes: A Handbook on the Biology of Bacteria. Archaea, Bacteria, Firmicutes, Actinomycetes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, H.H., Stackebrandt, E., Eds.; Springer: NY, USA, 2006; Vol. 3, pp. 682–724. [Google Scholar]
- Sasser, M. MIDI Technical Note 101. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. In MIDI technical note 101; MIDI inc: Newark, DE, USA, 2017; pp. 1–7. [Google Scholar]
- Vieira, S.; Huber, K.J.; Neumann-Schaal, M.; Geppert, A.; Luckner, M.; Wanner, G.; Overmann, J. Usitatibacter rugosus Gen. Nov., Sp. Nov. and Usitatibacter palustris Sp. Nov., Novel Members of Usitatibacteraceae Fam. Nov. within the Order Nitrosomonadales Isolated from Soil. Int J Syst Evol Microbiol 2021, 71, 1–12. [Google Scholar] [CrossRef]
- Moss, C.W.; Lambert-Fair, M.A. Location of Double Bonds in Monounsaturated Fatty Acids of Campylobacter cryaerophila with Dimethyl Disulfide Derivatives and Combined Gas Chromatography-Mass Spectrometry. Journal of clinical microbiology 1989, 27, 1467–1470. [Google Scholar] [CrossRef] [PubMed]
- Schumann, P.; Kalensee, F.; Cao, J.; Criscuolo, A.; Clermont, D.; Köhler, J.M.; Meier-Kolthoff, J.P.; Neumann-Schaal, M.; Tindall, B.J.; Pukall, R. Reclassification of Haloactinobacterium glacieicola as Occultella glacieicola Gen. Nov., Comb. Nov., of Haloactinobacterium album as Ruania alba Comb. Nov, with an Emended Description of the Genus Ruania, Recognition That the Genus Names Haloactinobacterium and Ruania Are Heterotypic Synonyms and Description of Occultella aeris Sp. Nov., a Halotolerant Isolate from Surface Soil Sampled at an Ancient Copper Smelter. Int J Syst Evol Microbiol 2021, 71. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; ¨ Oker, M.G. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinformatics 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int J Syst Evol Microbiol 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat Commun 2019, 10. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.P. When Should a DDH Experiment Be Mandatory in Microbial Taxonomy? Arch Microbiol 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene Cluster. Nat Chem Biol 2015, 11, 625–631. [Google Scholar] [CrossRef]
- van den Belt, M.; Gilchrist, C.; Booth, T.J.; Chooi, Y.H.; Medema, M.H.; Alanjary, M. CAGECAT: The CompArative GEne Cluster Analysis Toolbox for Rapid Search and Visualisation of Homologous Gene Clusters. BMC Bioinformatics 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat Chem Biol 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Das, R.; Romi, W.; Das, R.; Sharma, H.K.; Thakur, D. Antimicrobial Potentiality of Actinobacteria Isolated from Two Microbiologically Unexplored Forest Ecosystems of Northeast India. BMC Microbiol 2018, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Awla, H.K.; Rashid, T.S. HPLC Fractionation: A Comparative Analysis of Anti-Fungal Compounds from Different Streptomyces Isolates Inhibiting Colletotrichum acutatum. Biocatal Agric Biotechnol 2020, 27, 101688. [Google Scholar] [CrossRef]
- López-Moral, A.; Agustí-Brisach, C.; Leiva-Egea, F.M.; Trapero, A. Influence of Cultivar and Biocontrol Treatments on the Effect of Olive Stem Extracts on the Viability of Verticillium dahliae Conidia. Plants 2022, 11. [Google Scholar] [CrossRef]
- EUCAST Definitive Document. Methods for the determination of susceptibility of bacteria to antimicrobial agents. Terminology. Clin Microbiol Infect 1998, 4, 291–296. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S rRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int J Syst Evol Microbiol 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Kämpfer, P. Genus Streptomyces Waksman and Henrici 1943, 339AL Emend. Witt and Stackebrandt 1990, 370, Emend. Wellington, Stackebrandt, Sanders, Wolstrup and Jorgensen, 1992, 159. In: Bergey’s Manual of Systematic Bacteriology; Springer: New York (U.S.A), 2012; Vol. 5, pp. 1455–1767.
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler,’, 0; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt,’, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. International journal of systematic bacteriology 1987, 37, 463–464. [CrossRef]
- Sun, F.; Xu, S.; Jiang, F.; Liu, W. Genomic-Driven Discovery of an Amidinohydrolase Involved in the Biosynthesis of Mediomycin A. Appl Microbiol Biotechnol 2018, 102, 2225–2234. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, S.H.; Palaniyandi, S.A.; Han, J.S.; Yoon, T.M.; Kim, T.J.; Suh, J.W. Azalomycin F Complex Is an Antifungal Substance Produced by Streptomyces malaysiensis MJM1968 Isolated from Agricultural Soil. J Appl Biol Chem 2010, 53, 545–552. [Google Scholar] [CrossRef]
- Bastidas, R.J.; Shertz, C.A.; Lee, S.C.; Heitman, J.; Cardenas, M.E. Rapamycin Exerts Antifungal Activity In Vitro and In Vivo against Mucor circinelloides via FKBP12-Dependent Inhibition of Tor. Eukaryot Cell 2012, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Rao, Y.; Mei, L.; Wang, Y. Antifungal Activity of Rapamycin on Botryosphaeria dothidea and Its Effect against Chinese Hickory Canker. Pest Manag Sci 2021, 77, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, T.; Song, Y.; Luo, X.; Feng, L.; Zhuo, F.; Li, F.; Ren, M. Functional Characterization of Target of Rapamycin Signaling in Verticillium dahliae. Front Microbiol 2019, 10. [Google Scholar] [CrossRef]
- Netzker, T.; Schroeckh, V.; Gregory, M.A.; Flak, M.; Krespach, M.K.C.; Leadlay, P.F.; Brakhage, A.A. An Efficient Method to Generate Gene Deletion Mutants of the Rapamycin-Producing Bacterium Streptomyces iranensis HM 35. Appl Environ Microbiol 2016, 82, 3481–3492. [Google Scholar] [CrossRef]
- Fang, A.; Wong, G.K.; Demain, A.L. Enhancement of the Antifungal Activity of Rapamycin by the Coproduced Elaiophylin and Nigerin. J Antibiot (Tokyo) 2000, 53, 158–162. [Google Scholar] [CrossRef]
- Mandala, S.M.; Thornton, R.A.; Milligan, J.; Rosenbach, M.; Garcia-Calvo, M.; Bull, H.G.; Harris, G.; Abruzzo, G.K.; Flattery, A.M.; Gill, C.J.; et al. Rustmicin, a Potent Antifungal Agent, Inhibits Sphingolipid Synthesis at Inositol Phosphoceramide Synthase. J Biol Chem 1998, 273, 14942–14949. [Google Scholar] [CrossRef]
- Fujita, K.; Sugiyama, R.; Nishimura, S.; Ishikawa, N.; Arai, M.A.; Ishibashi, M.; Kakeya, H. Stereochemical Assignment and Biological Evaluation of BE-14106 Unveils the Importance of One Acetate Unit for the Antifungal Activity of Polyene Macrolactams. J Nat Prod 2016, 79, 1877–1880. [Google Scholar] [CrossRef]
- Goh, F.; Zhang, M.M.; Lim, T.R.; Low, K.N.; Nge, C.E.; Heng, E.; Yeo, W.L.; Sirota, F.L.; Crasta, S.; Tan, Z.; et al. Identification and Engineering of 32 Membered Antifungal Macrolactone Notonesomycins. Microb Cell Fact 2020, 19. [Google Scholar] [CrossRef] [PubMed]
- Shentu, X.P.; Cao, Z.Y.; Xiao, Y.; Tang, G.; Ochi, K.; Yu, X.P. Substantial Improvement of Toyocamycin Production in Streptomyces diastatochromogenes by Cumulative Drug-Resistance Mutations. PLoS One 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.G.; Adidjaja, J.J.; Kim, D.K.; Park, B.S.; Lee, N.; Cho, B.K.; Kim, H.U.; Oh, M.K. Comparative Genomic Analysis of Streptomyces rapamycinicus NRRL 5491 and Its Mutant Overproducing Rapamycin. Sci Rep 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Yoo, Y.J.; Ban, Y.H.; Yoon, Y.J. Biosynthesis of Rapamycin and Its Regulation: Past Achievements and Recent Progress. Journal of Antibiotics 2010, 63, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.J.; Hwang, J. yeon; Shin, H. luyung; Cui, H.; Lee, J.; Yoon, Y.J. Characterization of Negative Regulatory Genes for the Biosynthesis of Rapamycin in Streptomyces rapamycinicus and Its Application for Improved Production. J Ind Microbiol Biotechnol 2015, 42, 125–135. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Nodwell, J.R. The TetR Family of Regulators. Microbiology and Molecular Biology Reviews 2013, 77, 440–475. [Google Scholar] [CrossRef]
- Molnár, I.; Aparicio, J.F.; Haydock, S.F.; Khaw, L.; Schwecke, T.; König, A.; Staunton, J.; Leadlay, P.F. Organisation of the Biosynthetic Gene Cluster for Rapamycin in Streptomyces hygroscopicus: Analysis of Genes Flanking the Polyketide Synthase. Gene 1996, 169, 1–7. [Google Scholar] [CrossRef]
- Mrak, P.; Krastel, P.; Lukančič, P.P.; Tao, J.; Pistorius, D.; Moore, C.M. Discovery of the Actinoplanic Acid Pathway in Streptomyces rapamycinicus Reveals a Genetically Conserved Synergism with Rapamycin. Journal of Biological Chemistry 2018, 293, 19982–19995. [Google Scholar] [CrossRef]
- McAlpine, J.B.; Swanson, S.J.; Jackson, M.; Whittern, D.N. Revised NMR Assignments for Rapamycin. J Antibiot 1991, 44, 688–690. [Google Scholar] [CrossRef]
- Kang, S.; Han, J.; Jang, S.C.; An, J.S.; Kang, I.; Kwon, Y.; Nam, S.J.; Shim, S.H.; Cho, J.C.; Lee, S.K.; et al. Epoxinnamide: An Epoxy Cinnamoyl-Containing Nonribosomal Peptide from an Intertidal Mudflat-Derived Streptomyces sp. Mar Drugs 2022, 20. [CrossRef]
- Hamedi, J.; Mohammadipanah, F.; Klenk, H.P.; Pötter, G.; Schumann, P.; Spröer, C.; Von Jan, M.; Kroppenstedt, R.M. Streptomyces iranensis Sp. Nov., Isolated from Soil. Int J Syst Evol Microbiol 2010, 60, 1504–1509. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Clemente, E.; Moreno-González, V.; Ibáñez, A.; Calvo-Peña, C.; Ghoreshizadeh, S.; Radišek, S.; Cobos, R.; Coque, J.J.R. Changes in the Microbial Composition of the Rhizosphere of Hop Plants Affected by Verticillium Wilt Caused by Verticillium Nonalfalfae. Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Vézina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a New Antifungal Antibiotic. J Antibiot (Tokyo) 1975, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Sakakibara, T.; Aoki, F.; Saito, T.; Ichikawa, K.; Inagaki, T.; Kojima, Y.; Yamauchi, Y.; Huangt, L.H.; Guadliana^, M.A.; et al. Generation of Novel Rapamycin Structures by Microbial Manipulations. J Antibiot (Tokyo) 1995, 48, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Trevillian, P. Immunosuppressants-Clinical Applications. Aust Prescr 2006, 29, 102–108. [Google Scholar] [CrossRef]
- Martel, R.R.; Klicius, J.; Galet, S. Inhibition of the Immune Response by Aapamycin, a New Antifungal Antibiotic. Canadian J Physiol Pharmacol 1977, 55, 48–51. [Google Scholar] [CrossRef]
- Law, B.K. Rapamycin: An Anti-Cancer Immunosuppressant? Crit Rev Oncol Hematol 2005, 56, 47–60. [Google Scholar] [CrossRef]
- Tain, L.S.; Mortiboys, H.; Tao, R.N.; Ziviani, E.; Bandmann, O.; Whitworth, A.J. Rapamycin Activation of 4E-BP Prevents Parkinsonian Dopaminergic Neuron Loss. Nat Neurosci 2009, 12, 1129–1135. [Google Scholar] [CrossRef]
- Malagelada, C.; Jin, Z.H.; Jackson-Lewis, V.; Przedborski, S.; Greene, L.A. Rapamycin Protects against Neuron Death in In Vitro and In Vivo Models of Parkinson’s Disease. Journal of Neuroscience 2010, 30, 1166–1175. [Google Scholar] [CrossRef]
- Sharp, Z.D.; Strong, R. Rapamycin, the Only Drug That Has Been Consistently Demonstrated to Increase Mammalian Longevity. An Update. Exp Gerontol 2023, 176. [Google Scholar] [CrossRef]
- Yoo, Y.J.; Kim, H.; Park, S.R.; Yoon, Y.J. An Overview of Rapamycin: From Discovery to Future Perspectives. J Ind Microbiol Biotechnol 2017, 44, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Kuščer, E.; Coates, N.; Challis, I.; Gregory, M.; Wilkinson, B.; Sheridan, R.; Petković, H. Roles of RapH and RapG in Positive Regulation of Rapamycin Biosynthesis in Streptomyces hygroscopicus. J Bacteriol 2007, 189, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Schwecke, T.; Aparicio, J.F.; Molnar, I.; König, A.; Khaw, L.E.; Haydock, S.F.; Oliynyk, M.; Caffrey, P.; Cortés, J.; Lester, J.B.; et al. The Biosynthetic Gene Cluster for the Polyketide Immunosuppressant Rapamycin. Proc. Natl. Acad. Sci USA 1995, 92, 7839–7843. [Google Scholar] [CrossRef] [PubMed]
- Gatto, G.J.; Boyne, M.T.; Kelleher, N.L.; Walsh, C.T. Biosynthesis of Pipecolic Acid by RapL, a Lysine Cyclodeaminase Encoded in the Rapamycin Gene Cluster. J Am Chem Soc 2006, 128, 3838–3847. [Google Scholar] [CrossRef]
- Kobylarz, M.J.; Grigg, J.C.; Takayama, S.I.J.; Rai, D.K.; Heinrichs, D.E.; Murphy, M.E.P. Synthesis of L-2,3-Diaminopropionic Acid, a Siderophore and Antibiotic Precursor. Chem Biol 2014, 21, 379–388. [Google Scholar] [CrossRef]
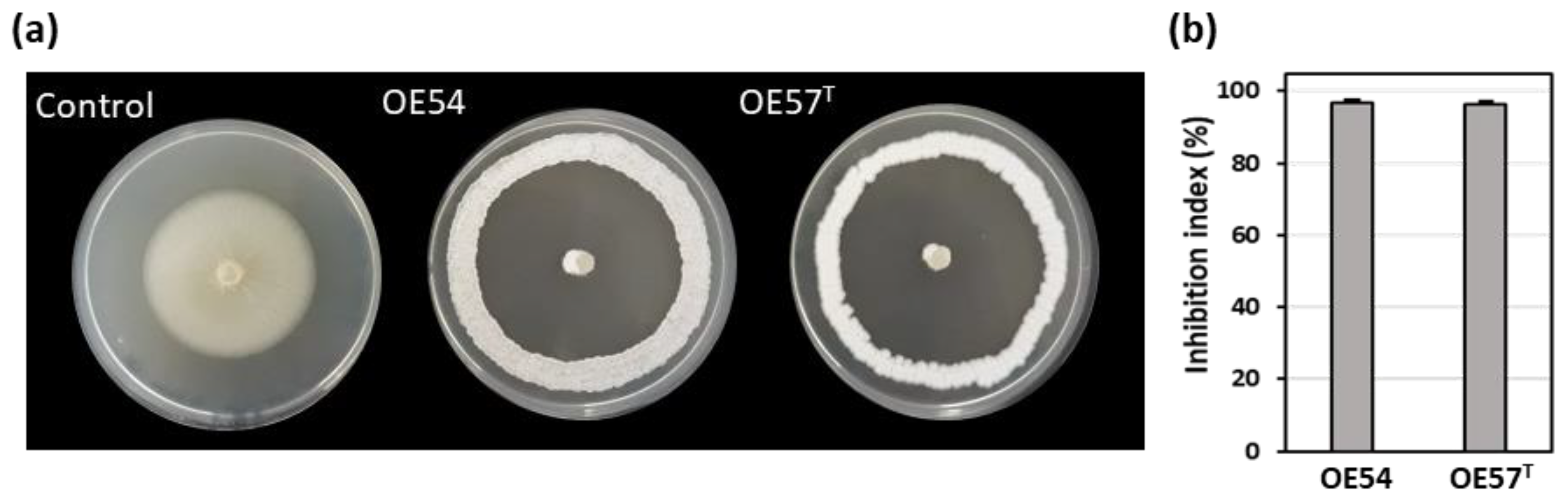
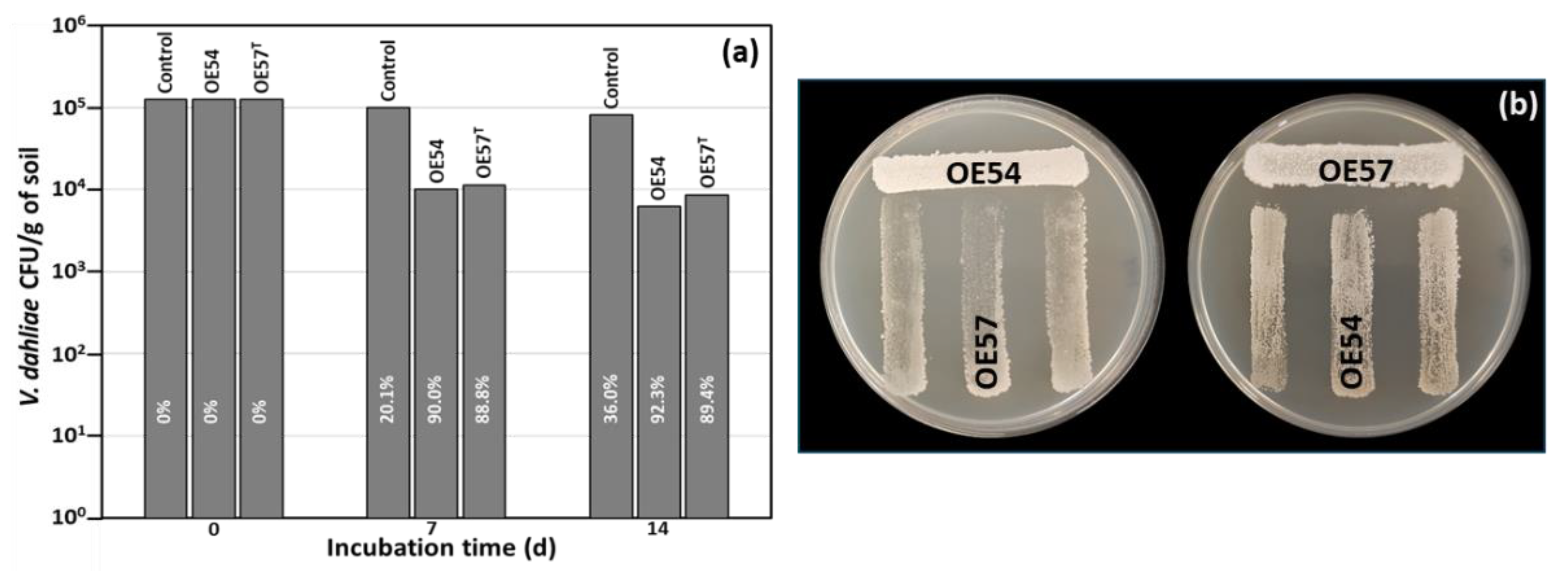
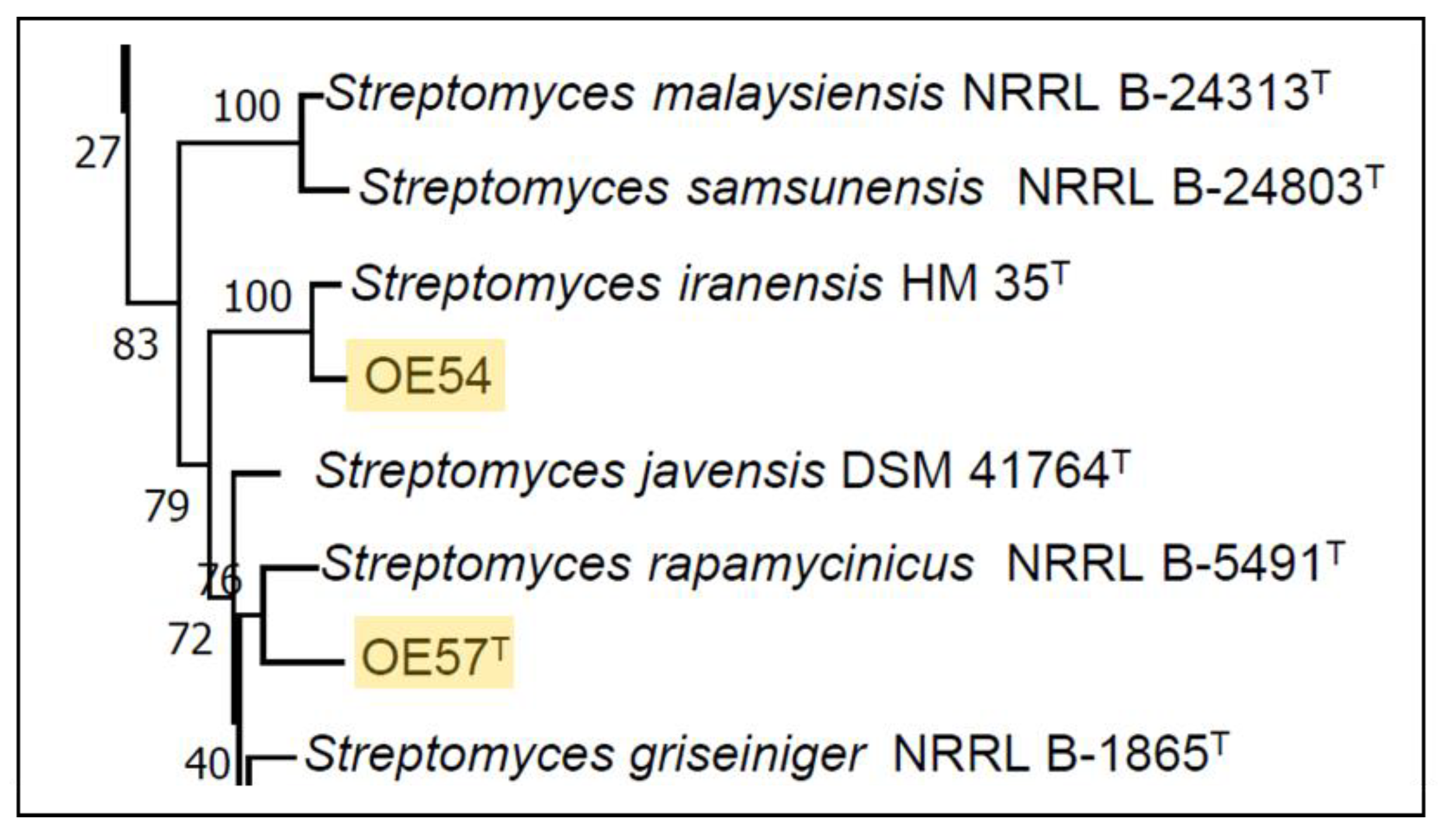
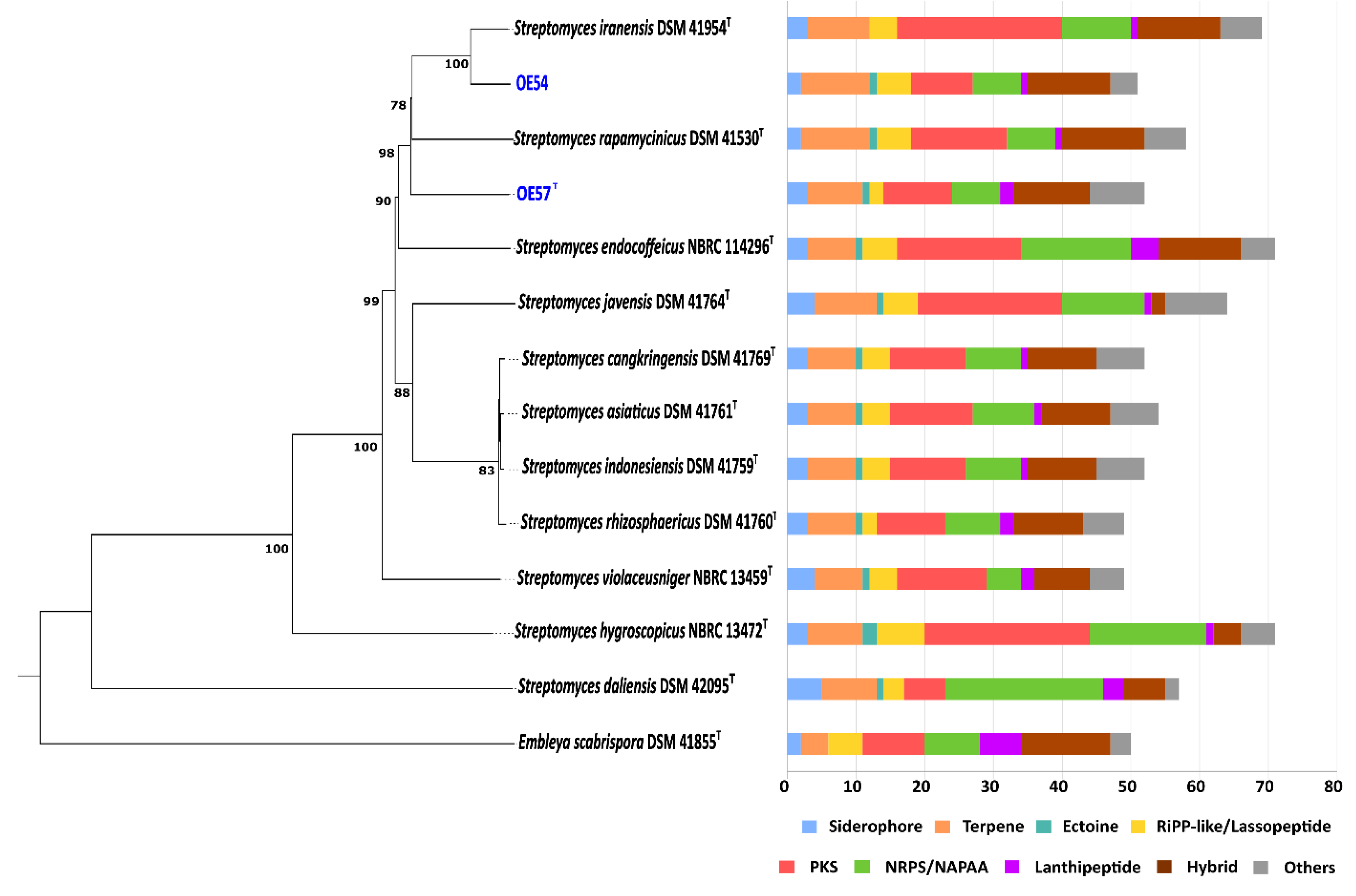
| Strain | GenBank accession numbers | |||||
| 16S rRNA | atpD | gyrB | recA | rpoB | trpB | |
| OE54 | PP980539 | PP979823 | PP979829 | PP979835 | PP979841 | PP979847 |
| OE57T | PP980540 | PP979824 | PP979830 | PP979836 | PP979842 | PP979848 |
| MLSA (Kimura 2-Parameter) Distance* | ||
| Strain | Streptomyces sp. OE54 | Streptomyces sp. OE57T |
| Streptomyces sp. OE57T | 0.021 | |
| Streptomyces iranensis HM 35T | 0.005 | 0.022 |
| Streptomyces rapamycinicus NRRL 5491T | 0.018 | 0.011 |
| Streptomyces javensis DSM 41764T | 0.018 | 0.014 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





