Submitted:
28 February 2024
Posted:
29 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Isolation of Actinobacteria
2.2. Screening of Antagonistic Actinobacteria against Foc TR4
2.3. Measurement of a Broad-Spectrum Antifungal Activity
2.4. Cultural and Morphological Characteristics of Strain SCA4-21T
2.5. Physiological and Biochemical Characteristics of Strain SCA4-21T
2.6. Chemotaxonomic Characteristics of SCA4-21T
2.7. Phylogenetic Analysis of Strain SCA4-21T
2.8. Genome sequencing and Feature Analysis of Strain SCA4-21T
2.9. The overall Genome Related Indexes
2.10. Determination of Antifungal Activity of Strain SCA4-21T Fermentation Broth
2.11. CAZymes Analysis of Fermentation Broth of Strain SCA4-21T
3. Results
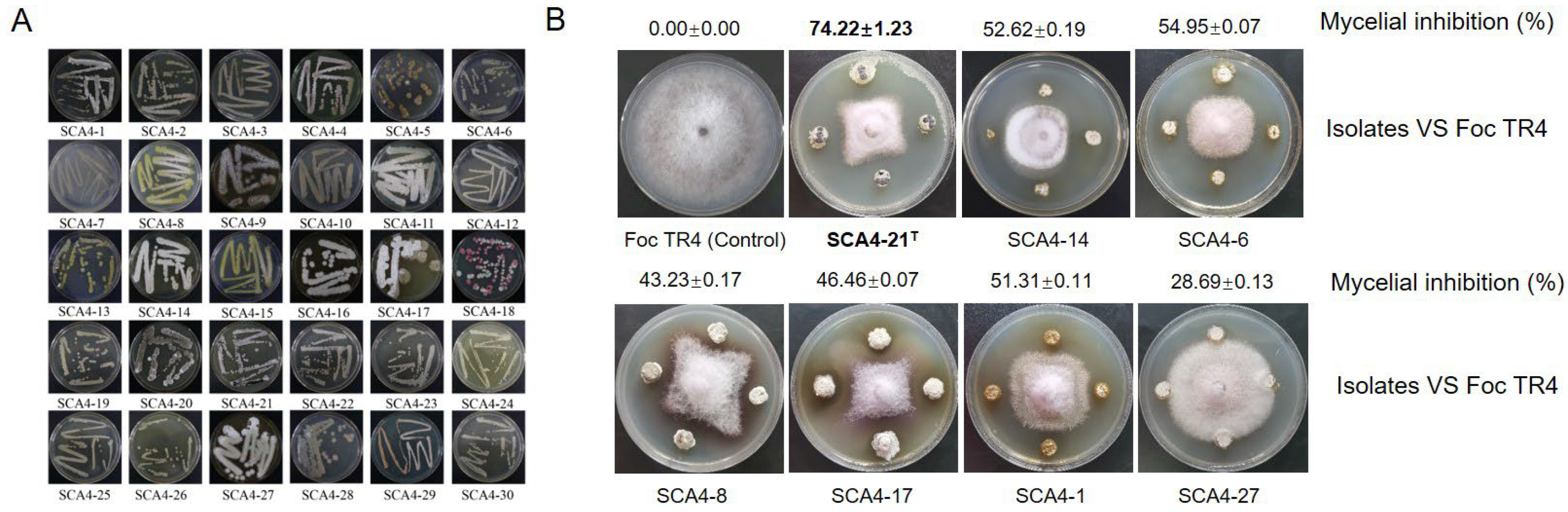
3.1. Isolation and Antifungal Activity Screening of Actinobacteria against Foc TR4
3.2. Strain SCA4-21T Exhibited a Broad-Spectrum Antifungal Activity
3.3. Cultural and Morphological Characteristics of Strain SCA4-21T
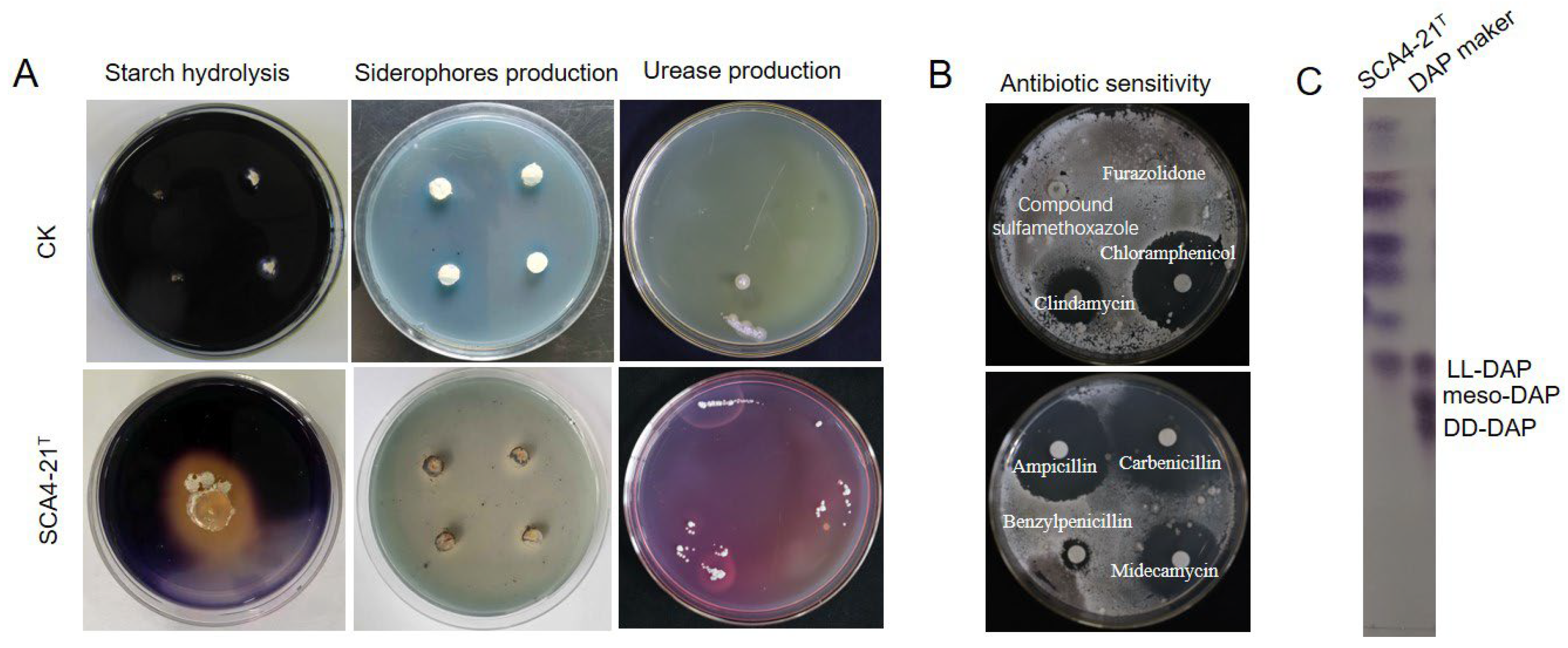
3.4. Physiological and Biochemical Characteristics of Strain SCA4-21T
3.5. Chemotaxonomic Characteristics of Strain SCA4-21T
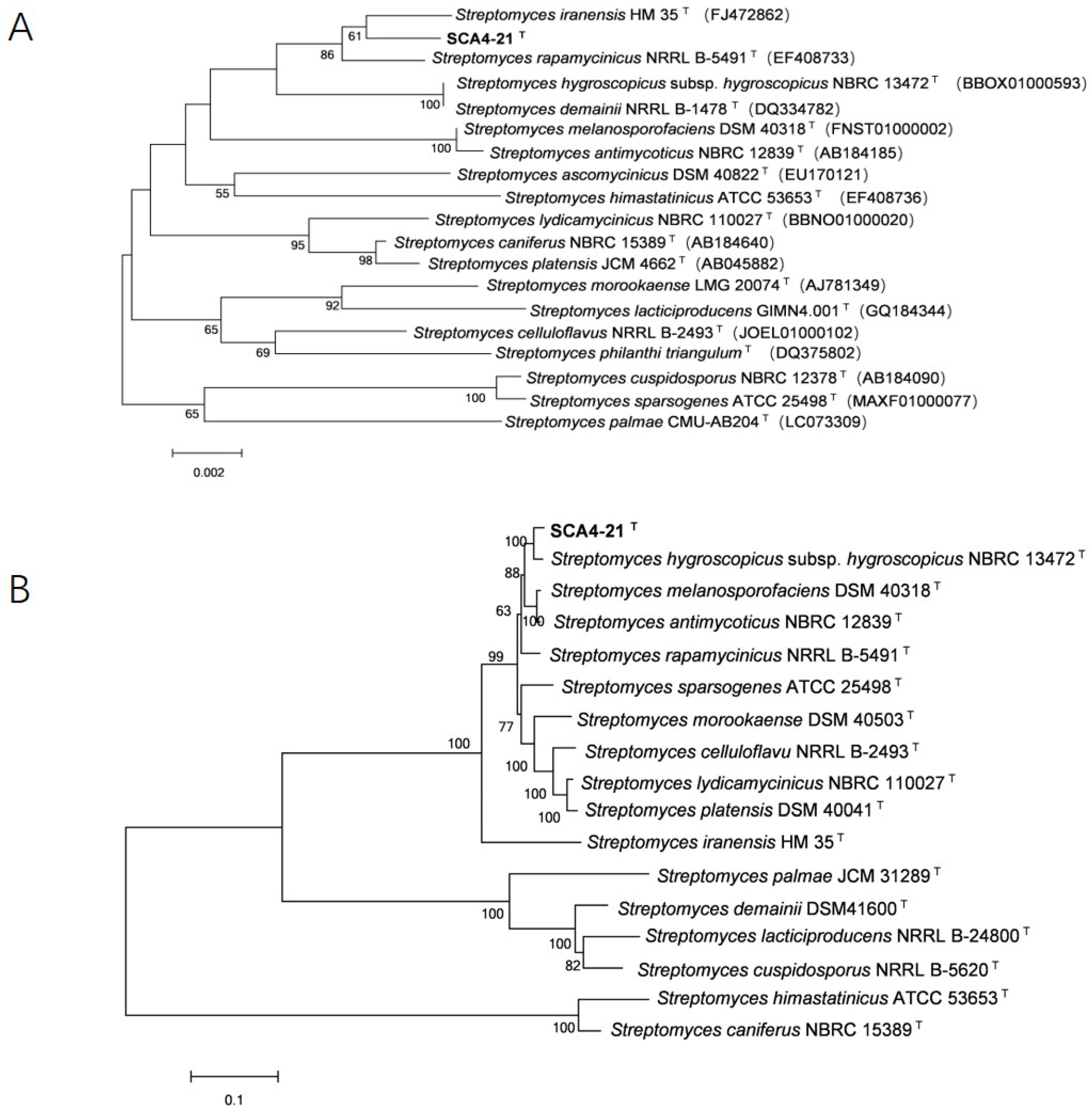
3.6. Phylogenetic Analysis of 16S rRNA and House-Keeping Genes
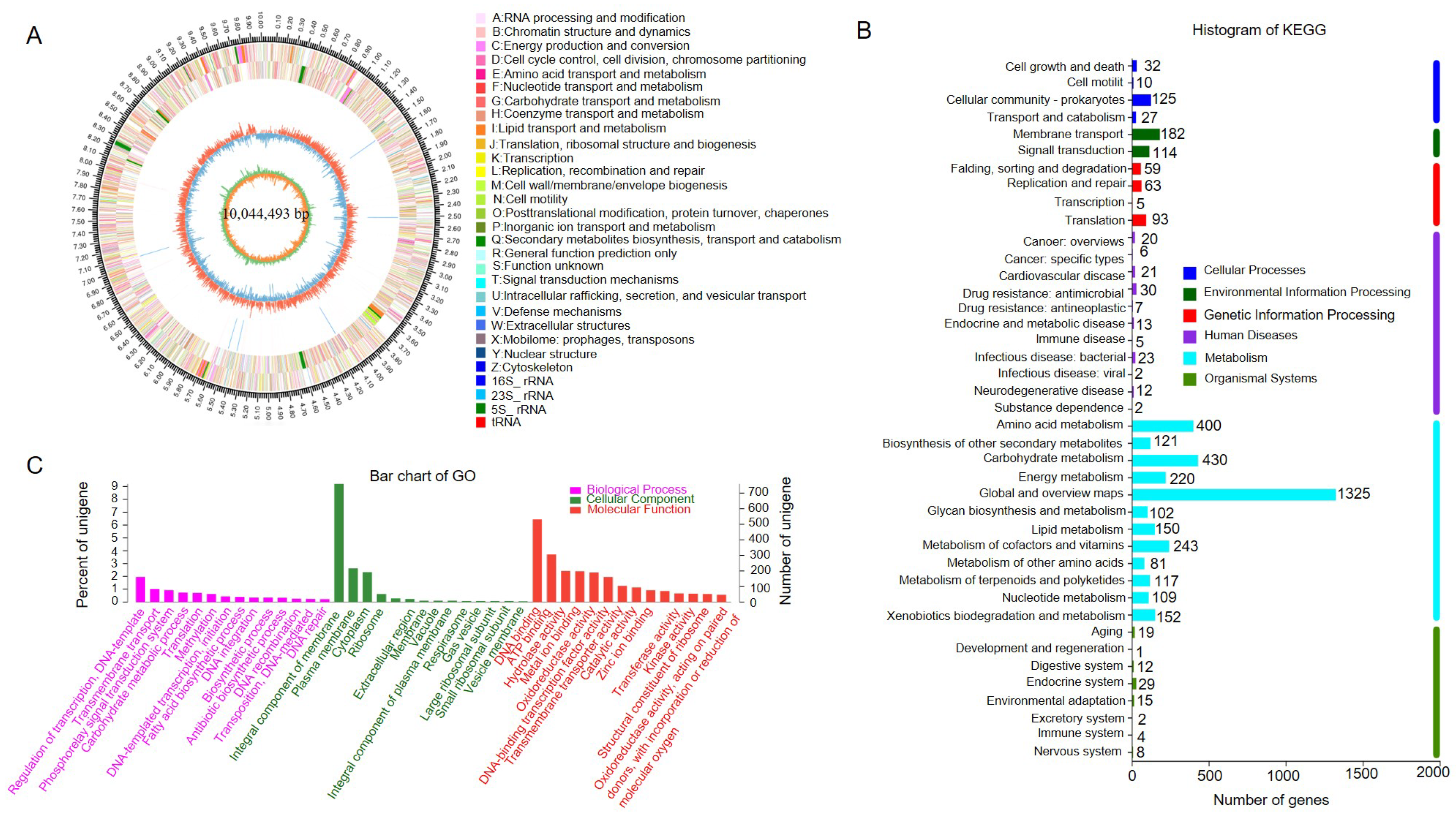
3.7. Genome Sequencing and Feature Analysis of Strain SCA4-21T
3.8. The Overall Genome Related Indexes
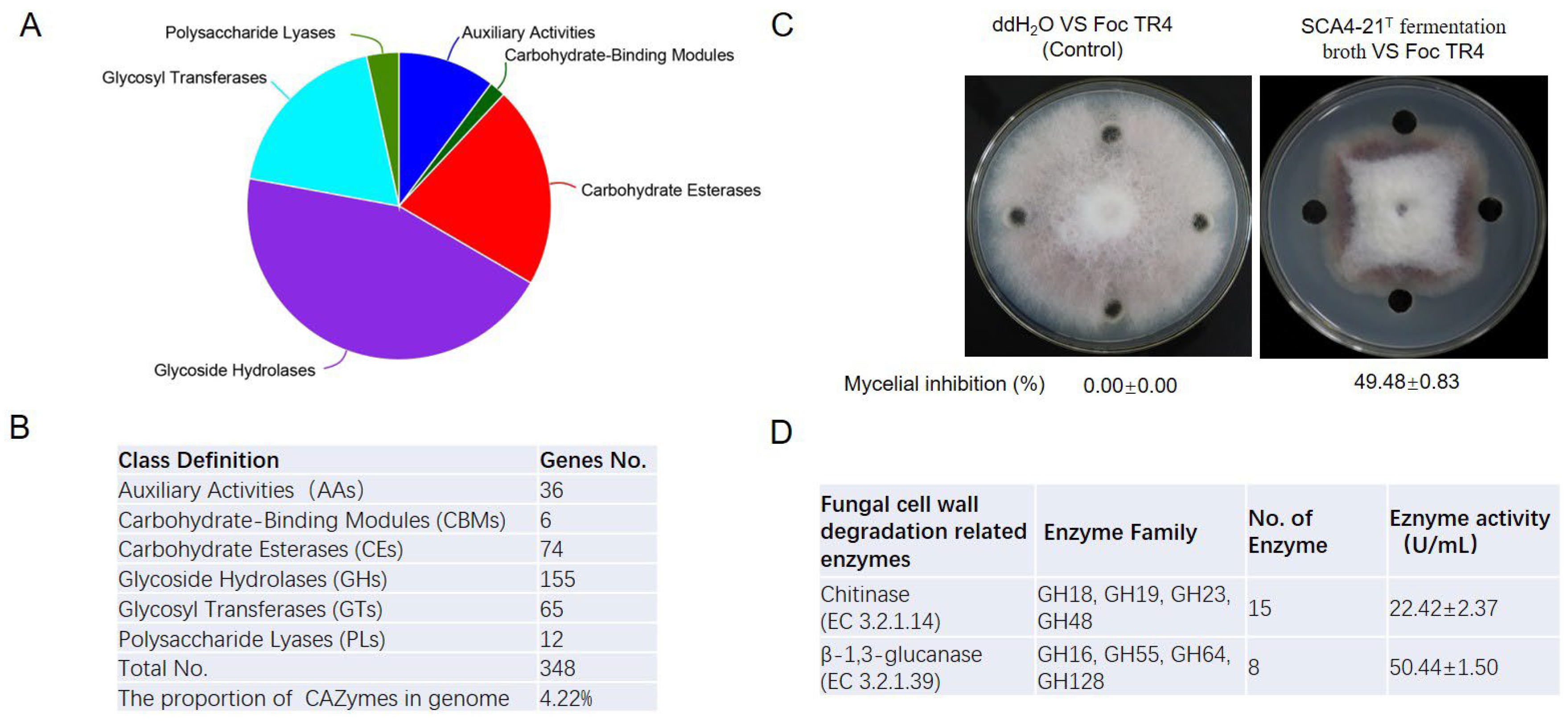
3.9. CAZymes Prediction of the Genome of Strain SCA4-21T
3.10. Antifungal Activity and CAZymes Analysis of Fermentation Broth of Strain SCA4-21T
3.11. Description of Streptomyces luomodiensis sp. nov.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Declaration of competing interest
References
- Mohandas, S., Ravishankar, K.V.) Banana: genomics and transgenic approaches for genetic improvement. Springer, Singapore, 2016, pp 211–226. https:// doi. org/ 10. 1007/ 978- 981- 10- 1585-4.
- Pérez -Vicente, L., Dita, M., Martinez, D.L.P.E. (2014) Technical manual prevention and diagnostic of Fusarium wilt (Panama disease) of banana caused by Fusarium oxysporum f. sp. cubense tropical race 4 (TR4). FAO, Rome, pp 4–13.
- Ploetz, R.C. Management of Fusarium wilt of banana: a review with special reference to tropical race 4. Crop Prot. 2015, 73, 7-15. [CrossRef]
- Fu, L., Penton, C. R., Ruan, Y., Shen, Z., Xue, C., Li, R., Shen, Q. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol Biochem. 2017, 104, 39-48. [CrossRef]
- Bhatti, A. A., Haq, S., Bhat, R. A. Actinomycetes benefaction role in soil and plant health. Microb Pathogenesis. 2017, 111, 458-467. [CrossRef]
- Gonzalez-Franco, A.C., Hernandez, L.R. Actinomycetes as biological control agents of phytopathogenic fungi. Tecnociencia Chihuahua. 2009, 3(2), 62-73.
- Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., Klenk, H. P., Clément, C., Ouhdouch, Y., van Wezel, G. P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol R. 2015, 80 (1), 1-43. [CrossRef]
- Law, J.W., Ser, H.L., Khan, T.M., Chuah, L. H., Pusparajah, P., Chan, K. G., Goh, B. H., Lee, L.H. The Potential of Streptomyces as biocontrol agents against the rice blast Fungus, Magnaporthe oryzae (Pyricularia oryzae). Front Microbiol. 2017, 17, 8, 3. [CrossRef]
- Waksman, S. A., Henrici, A.T. The nomenclature and classification of the actinomycetes. J Bacteriol. 1943, 46, 337-341. [CrossRef]
- Gevers, D., Cohan, F. M., Lawrence, J. G., Spratt, B. G., Coenye, T., Feil, E. J., Stackebrandt, E., Van de Peer, Y., Vandamme, P., Thompson, F. L., Swings, J. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005, 3(9),733-739. [CrossRef]
- Jose, P.A., Maharshi, A., Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiological Research, 2021, 246,126708. [CrossRef]
- Berdy, J. Bioactive microbial metabolites. J Antibiot. 2005, 58, 1-26.
- Mann, A. J., Hahnke, R. L., Huang, S., Werner, J., Xing, P., Barbeyron, T., Huettel, B., Stüber, K., Reinhardt, R., Harder, J., Glöckner, F. O., Amann, R. I., Teeling, H. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol. 2013, 79(21), 6813-6822. [CrossRef]
- Aktuganov, G., Melentjev, A., Galimzianova, N., Khalikova, E., Korpela, T., Susi, P. Wide-range antifungal antagonism of paenibacillus ehimensis IB-x-b and its dependence on chitinase and β-1,3-glucanase production. Can J Microbiol. 2008, 54(7), 577-587.
- Zacky, F. A., Ting, A. S. Y. Investigating the bioactivity of cells and cell-free extracts of streptomyces griseus towards fusarium oxysporum f. sp. cubense race 4. Biol Control. 2013, 66(3), 204-208.
- Yun, T., Jing, T., Zhou, D., Zhang, M., Zhao, Y., Li, K., Zang, X., Zhang, L., Xie, J., Wang, W. (2022) Potential biological control of endophytic Streptomyces sp. 5-4 against Fusarium wilt of banana caused by Fusarium oxysporum f. sp. cubense Tropical Race 4. Phytopathology. 2022 ,112(9):1877-1885. [CrossRef]
- Williams, S.T., Goodfellow, M., Alderson, G., Wellington, E. M. H., Sneath, P. H. A., Sackin, M. J. Numerical classification of streptomyces and related genera. J Gen Microbiol. 1983, 129, 1743-1813. [CrossRef]
- Jing, T., Zhou, D., Zhang, M., Yun, T., Qi, D., Wei, Y., Chen, Y., Zang, X., Wang, W., Xie, J. Newly isolated Streptomyces sp. JBS5-6 as a potential biocontrol agent to control banana fusarium wilt: genome sequencing and secondary metabolite cluster profiles. Front Microbiol. 2020, 11:3036. [CrossRef]
- Qi, D., Zou, L., Zhou, D., Zhang, M., Wei, Y., Zhang, L., Xie, J., Wang, W. Identification and antifungal mechanism of a novel actinobacterium Streptomyces huiliensis sp. nov. against Fusarium oxysporum f. sp. cubense tropical race 4 of banana. Front Microbiol. 2021, 12, 722661. [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, MIDI Technical Note 101. Newark, DE: MIDI Inc. 1990.
- Hasegawa, T., Takizawa, M., Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983, 29, 319-322. [CrossRef]
- Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., Parlett, J. H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Meth. 1984, 2, 233-241.
- Wang, L., Xing, M., Di, R., Luo, Y. Isolation, identification and antifungal activities of Streptomyces aureoverticillatus HN6. J Plant Pathol Microb. 2015, 6, 281. [CrossRef]
- Wang, J., Cai, B., Li, K., Zhao, Y., Li, C., Liu, S., Xiang, D., Zhang, L., Xie, J., Wang, W. Biological control of Fusarium oxysporum f. sp. cubense tropical race 4 in banana plantlets using newly isolated Streptomyces sp. WHL7 from marine soft coral. Plant Dis. 2022, 106 (1), 254-259. [CrossRef]
- Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999, 41, 95-98. [CrossRef]
- Thompson, J. D., Higgins, D. G., Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673-4680. [CrossRef]
- Kumar, S., Stecher, G., Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016, 33, 1870-1874. [CrossRef]
- Rong, X. Y., Huang, Y. Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA-DNA hybridization, with proposalto combine 29 species and three subspecies as 11 genomic species. Int J Syst Evol Microbiol. 2010, 60, 696-703. [CrossRef]
- Bolger, A. M., Lohse, M., Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014, 30 (15), 2114-2120. [CrossRef]
- Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017, 13(6): e1005595. [CrossRef]
- Delcher, A. L., Bratke, K. A., Powers, E. C., Salzberg, S. L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007, 23:673-679.
- Hyatt D, Chen, GL, LoCascio PF, Land ML, Larimer FW, Hauser L J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010, 11, 119. [CrossRef]
- Besemer, J., Borodovsky, M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33(Web Server): W451-W454. [CrossRef]
- Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., Lomsadze, A., Pruitt, K. D., Borodovsky, M., Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614-6624. [CrossRef]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37(Database issue), D233-8. [CrossRef]
- Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017, 110, 1281-1286. [CrossRef]
- Meier-Kolthoff, J. P., Sardà Carbasse, J., Peinado-Olarte, R. L., Göker, M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022, 50, D801-D807. [CrossRef]
- Magaldi, S., Mata-Essayag, S., Hartung de Capriles, C., Perez, C., Colella, M. T., Olaizola, C., Ontiveros, Y. Well diffusion for antifungal susceptibility testing. Int J Infect Dis. 2004, 8, 39-45. [CrossRef]
- Shirling, E. B., Gottlieb, D. Comparative description of type strains of Streptomyces: V. Additional Descriptions. Int J Syst Bacteriol. 1972, 22, 265-394. [CrossRef]
- Hamedi, J., Mohammadipanah, F., Klenk, H. P., Pötter, G., Schumann, P., Spröer, C., von Jan, M., Kroppenstedt, R. M. Streptomyces iranensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2010, 60, 1504-1509. [CrossRef]
- Kieser, T., Bibb, M.J., Buttner, M. J., Chater, K. F., Hopwood, D. A., Charter, K., Hopwood, D.A. Practical streptomyces genetics. John Innes Foundation, 2000, pp2-33.
- Rong, X., Huang, Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol. 2012, 35, 718. [CrossRef]
- Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., Da Costa, M. S., Rooney, A. P., Yi, H., Xu, X.W., De Meyer, S., Trujillo, M. E. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018, 68 (1), 461-466. [CrossRef]
- Qing, F., Shiping, T., Amp, L. H. Production of p-1,3-glucanase and chitinase of two biocon-trol agents and their possible modes of action. Chinese Sci Bull. 2002, 47(4), 292-296.
- Arora, N. K., Kim, M. J., Kang, S.C., Maheshwari, D. K. Role of chitinase and beta-1,3-glucanase activities produced by a fluorescent pseudomonad and in vitro inhibition of Phytophthora capsici and Rhizoctonia solani. Can J Microbiol. 2007, 53(2), 207-12. [CrossRef]
- Heydan, A., Pessarakh, M. A review on biological control of fungal plant pathogens using microbial antagonists. Journal of Biological Sciences. 2010, 10 (4), 273-290.
- Bull, A. T. “Actinobacteria of the extremobiosphere,” in Extremophiles Handbook, eds. Horikoshi, K., Antranikian, G., Bull, A. T., Robb, F., Stelter, K., (Berlin: Springer-Verlag GmbH), 2010, 1203-1240.
- Ruangwong, OU., Kunasakdakul, K., Chankaew, S., Pitija, K., Sunpapao, A. A rhizobacterium, Streptomyces albulus Z1-04-02, displays antifungal activity against sclerotium rot in mungbean. Plants (Basel), 2022, 11(19), 2607. [CrossRef]
- Li, W., Long, Y., Mo, F., Shu, R., Yin, X., Wu, X., Zhang, R., Zhang, Z., He, L., Chen, T., Chen, J. Antifungal activity and biocontrol mechanism of Fusicolla violacea J-1 against Soft Rot in Kiwifruit Caused by Alternaria alternata. J Fungi (Basel). 2021, 7(11), 937. [CrossRef]
- Anitha, A., Rebeeth, M. In vitro antifungal activity of streptomyces griseus against phytopathogenic fungi of tomato field. Acad J Plant Sci. 2009, 2 (2), 119-123.
- Lee, S.Y., Tindwa, H., Lee, Y. S., Naing, K.W., Hong, S. H., Nam, Y., Kim, K.Y. Biocontrol of anthracnose in pepper using chitinase, beta-1,3 glucanase, and 2-furancarboxaldehyde produced by Streptomyces cavourensis SY224. J Microbiol Biotechnol. 2012, 22(10), 1359-66. [CrossRef]
- Guo, Y., Zheng, W., Rong, X., Huang, Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol. 2008, 58, 149-59. [CrossRef]
- Richter, M., Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009, 106, 19126-19131. [CrossRef]
- Rong, X., Guo, Y., Huang, Y. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol. 2009, 32(5), 314-322. [CrossRef]
- Huang, X. L., Kong, F. D., Zhou, S. Q., Huang, D. Y., Zheng, J. P., and Zhu, W. M. Streptomyces tirandamycinicus sp. nov. a novel marine sponge-derived actinobacterium with antibacterial potential against Streptococcus agalactiae. Front Microbiol. 2019, 10, 482. [CrossRef]
- Qi, D., Zou, L., Zhou, D., Zhang, M., Wei, Y., Li, K., Zhao, Y., Zhang, L., Xie, J. Biocontrol potential and antifungal mechanism of a novel Streptomyces sichuanensis against Fusarium oxysporum f. sp. cubense tropical race 4 in vitro and in vivo. Appl Microbiol Biotechnol. 2022, 106(4), 1633-1649. [CrossRef]
- Zhang, L., Song, X., Li, W., Zhao, S., Yang, L., Xia, M., Niu, Q. Phytobacter nematintestini sp. nov. isolated from the intestine of Caenorhabditis elegans and conferring resistance to Bacillus nematocida infection. Ann Microbiol. 2019, 69 (8), 871. [CrossRef]

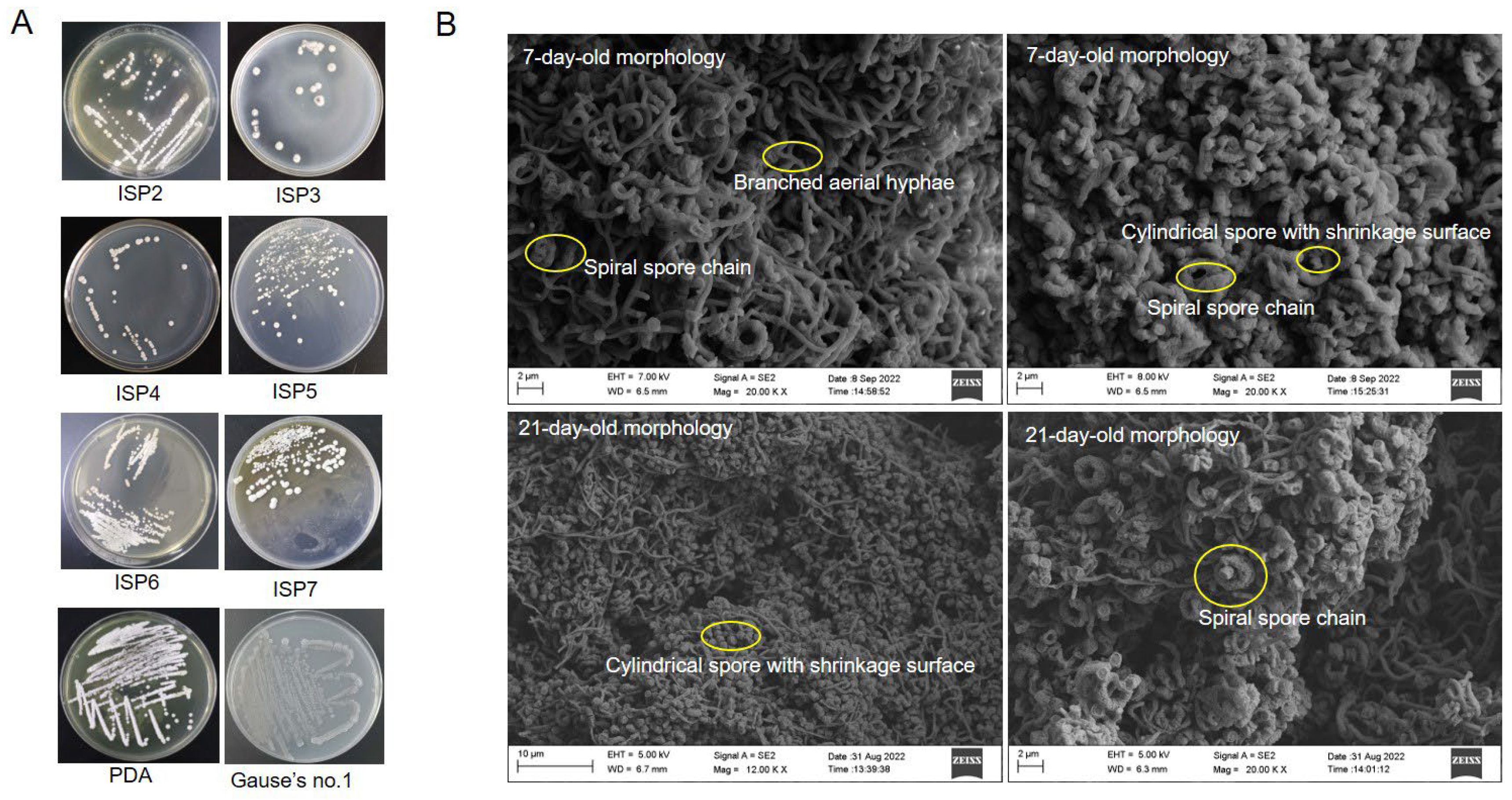
| Cuture Medium | Aerial Hyphae | Vegetative Mycelium | Soluble Pigment | Colony Characteristics | Growth Conditions |
| ISP2 | White | Sand yellow | None | Wrinkled, hard | +++ |
| ISP3 | Cream | Light ivory | None | Wrinkled, hard | + |
| ISP4 | Pure white | Cream | None | compact, wrinkle-free | + |
| ISP5 | Pure white | Cream | None | compact, wrinkle-free | ++ |
| ISP6 | Pure white | Cream | None | Loose, wrinkle-free | ++ |
| ISP7 | Grey white | Sand yellow | Light brown | Loose, wrinkle-free | ++ |
| PDA | Pure white | Lemon yellow | Light yellow | Loose, wrinkle-free | +++ |
| Gause’s no.1 | Silver grey | Silver grey | None | Loose, powery | +++ |
| Characteristic | SCA4-21 |
|---|---|
| Major menaquinones(%) | |
| MK9(H8) | 65.50 |
| MK10(H2) | 34.50 |
| Major fatty acids (>0.5%) | |
| iso-C13:0 | 0.83 |
| anteiso-C13:0 | 0.59 |
| iso-C14:0 | 8.21 |
| C14:0 | 3.34 |
| iso-C15:0 | 9.67 |
| anteiso-C15:0 | 34.78 |
| C16:0 | 19.89 |
| iso-C16:1 H | tr |
| iso-C16:0 | 8.16 |
| anteiso-C16:0 | ND |
| iso-C17:0 | 1.4 |
| anteiso-C17:0 ω9c | 0.53 |
| anteiso-C17:0 | 5.93 |
| C17:0cyclo | 1.78 |
| C17:0 | 0.75 |
| iso-C18:0 | tr |
| Summed feature 3* | 0.54 |
| Summed feature 9* | tr |
| Main amino acid of the cell wall | LL-diaminopimelic acid |
| MLSA distance (Kimura two-parameter) | |||||||||||||||||
| Strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 1 | |||||||||||||||||
| 2 | 0.187 | ||||||||||||||||
| 3 | 0.050 | 0.153 | |||||||||||||||
| 4 | 0.021 | 0.188 | 0.048 | ||||||||||||||
| 5 | 0.679 | 0.714 | 0.705 | 0.660 | |||||||||||||
| 6 | 0.040 | 0.180 | 0.041 | 0.040 | 0.676 | ||||||||||||
| 7 | 0.039 | 0.178 | 0.041 | 0.040 | 0.677 | 0.008 | |||||||||||
| 8 | 1.092 | 1.136 | 1.100 | 1.077 | 1.102 | 1.104 | 1.094 | ||||||||||
| 9 | 0.092 | 0.226 | 0.093 | 0.092 | 0.710 | 0.089 | 0.088 | 1.099 | |||||||||
| 10 | 1.038 | 1.091 | 1.039 | 1.025 | 1.080 | 1.029 | 1.027 | 0.105 | 1.040 | ||||||||
| 11 | 0.098 | 0.232 | 0.098 | 0.096 | 0.715 | 0.093 | 0.093 | 1.102 | 0.018 | 1.050 | |||||||
| 12 | 0.094 | 0.231 | 0.094 | 0.094 | 0.728 | 0.086 | 0.089 | 1.120 | 0.047 | 1.057 | 0.053 | ||||||
| 13 | 0.093 | 0.221 | 0.089 | 0.091 | 0.711 | 0.088 | 0.087 | 1.120 | 0.087 | 1.041 | 0.093 | 0.089 | |||||
| 14 | 0.754 | 0.735 | 0.729 | 0.738 | 0.119 | 0.717 | 0.718 | 1.186 | 0.734 | 1.130 | 0.736 | 0.739 | 0.713 | ||||
| 15 | 0.731 | 0.744 | 0.734 | 0.735 | 0.276 | 0.725 | 0.727 | 1.234 | 0.745 | 1.178 | 0.747 | 0.761 | 0.735 | 0.297 | |||
| 16 | 0.714 | 0.726 | 0.708 | 0.707 | 0.084 | 0.696 | 0.694 | 1.133 | 0.723 | 1.091 | 0.732 | 0.747 | 0.724 | 0.109 | 0.298 | ||
| 17 | 0.065 | 0.204 | 0.068 | 0.063 | 0.685 | 0.067 | 0.068 | 1.085 | 0.094 | 1.031 | 0.100 | 0.103 | 0.092 | 0.732 | 0.729 | 0.672 | |
| Feature | Chromosome characteristics |
|---|---|
| Genome size (bp) | 10,044,493 |
| Chromosome No. | 1 |
| Plasmid No. | 0 |
| GC Content (%) | 71.48 |
| Depth | 454.86 |
| Protein-coding genes No. | 8246 |
| Gene total length (bp) | 8,847,075 |
| Gene average length (bp) | 1072.89 |
| Gene density | 0.82 |
| tRNA genes No. | 64 |
| Type of tRNAs No. | 20 |
| rRNA genes No. | 18 |
| 16S rRNA No. | 6 |
| 23S rRNA No. | 6 |
| 5S rRNA No. | 6 |
| sRNA No. | 134 |
| Repeat No. | 1436 |
| CRISPR-Cas No. | 80 |
| Is No. | 6 |
| Genes assigned to COG (bp) | 7548 |
| Genes assigned to KEGG (bp) | 4930 |
| Genes assigned to GO (bp) | 3460 |
| Strains | ANI (%) | DDH (%) |
| Streptomyces iranensis DSM 41954 T | 89.42 | 37.8 |
| Streptomyces rapamycinicus NRRL B-5491 T | 89.54 | 38.1 |
| Streptomyces hygroscopicus subsp.hygroscopicus NBRC 13472 T | 91.26 | 44.3 |
| Streptomyces melanosporofaciens DSM 40318 T | 89.95 | 39.2 |
| Streptomyces antimycoticus NBRC 12839 T | 89.8 | 39 |
| Streptomyces himastatinicus ATCC 53653 T | 84.73 | 28.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





