1. Introduction
Puccinia triticina Eriks, the causal agent of wheat leaf rust, is responsible for significant global yield and economic losses in various regions including North America, South America, Africa, Europe, Asia, and Australia [
1,
2]. In China, the occurrence of wheat leaf rust is primarily common in the Yangtze River valley, as well as certain parts of wheat fields in the northeast, north, and southwest regions. In typical years, wheat yield losses caused by leaf rust range from 50%, amounting to an annual reduction of approximately 3 million tonnes [
3]. Currently, although wheat leaf rust is not a prominent disease in China, it often coexists with wheat stripe rust within the same wheat fields, thus hindering the collection of accurate data regarding yield losses. Regardless, severe epidemics of wheat leaf rust were recorded in China during 1969, 1973, 1975, and 1979 [
4], and localized outbreaks can still occur. For instance, in 2013, leaf rust was observed in Shandong, Henan, and certain regions of Xinjiang. Moreover, in 2015, Henan suffered from the worst wheat leaf rust outbreak, affecting 1.832 million hectares and resulting in an annual yield loss of 191,000 tonnes [
5].
Resistance to wheat leaf rust can be divided into race-specific and race-non-specific resistance. Race-specific resistance, or all-stage resistance (ASR), is effective against pathogens but is easily overcome by new virulent pathotypes due to its genetic simplicity [
6,
7]. The high level of mutational and virulence diversity in
P. triticina populations allows the fungus to rapidly overcome race-specific resistance [
8]. In contrast, race-non-specific resistance, or adult plant resistance (APR), is quantitatively inherited and genetically more complex, generally conferred by the additive effects of multiple minor genes [
9]. Wheat cultivars with APR often show a susceptible response at the seedling stage, but ultimately exhibit low disease severity at the adult plant stage by delaying infection, growth and reproduction of the pathogen [
10].
Currently, 83 genes have been officially named, and more than 100 leaf rust resistance genes have been documented in wheat [
11]. Most of these genes are race-specific resistance genes and have a relatively short duration of action. Studies have identified only eight slow-rusting APR genes that don’t target specific races, and these include
Lr34 [
12],
Lr46 [
13],
Lr67 [
14],
Lr68 [
15],
Lr74 [
16,
17,
18],
Lr75 [
19],
Lr77 [
20] and
Lr78 [
18].
Lr34 and
Lr67 are the only two successfully cloned and characterized APR genes [
21,
22,
23]. Significant attention towards the APR genes has facilitated the exploration of novel
Lr resistance QTLs, with over 240 being discovered in wheat and dispersed throughout all 21 chromosomes [
24]. It is uncommon for an individual QTL to grant sufficient resistance, especially in the presence of elevated disease pressure. Multiple QTLs or genes, comprising of 4 or 5, are necessary to develop a significant level of rust resistance [
25].
Simple sequence repeat (SSR) markers are a preferable option for QTL mapping of wheat leaf rust owing to their genome-wide coverage, codominant inheritance, considerable information content, feasible detection and reproducibility. Currently, single nucleotide polymorphism (SNP) arrays are increasingly utilized to map genes and conduct genome-wide association studies (GWAS). SNP arrays offer numerous markers closely associated with agronomic traits, making a significant contribution, but they are not cost-efficient for genotyping individuals in a large, temporary segregating population, like an F2. Competitive allele-specific PCR has become a widely accepted technology for performing SNP genotyping using bioscience KASP assays and serves as a global benchmark [
26]. The Chinese Academy of Agricultural Sciences and Affymetrix developed the wheat 55K SNP array specifically for this purpose (
http://bioservices.capitalbio.com/index.shtml).
The wheat variety Xinong1163-4 can be traced back to 84/79 and Xinong 1376. The specific gene(s) responsible for leaf-rust resistance in this Chinese wheat cultivar from Shanxi province which is highly resistant to leaf rust, stripe rust and powdery mildew of wheat, and has good agronomic traits, are not currently understood. So far, there is no report on the genetic patterns of resistance to leaf rust in Xinong1163-4. In the present study, we used the same Xinong1163-4×Thatcher RIL population, SSR markers, and the 55K SNP array genotyping platform to identify the genetic basis of Lr resistance in this cultivar. The study’s findings offer a theoretical and practical framework for examining and using QTLs in cultivating crop varieties resistant to diseases.
2. Materials and Methods
2.1. Plant Materials and Pathogens
The plant materials used for mapping comprised of 195 RILs from the cross between Xinong1163-4 and Thatcher were developed by single-seed descent. Xinong1163-4 is a Chinese cultivar that is sensitive to Pt races at the seedling stage but highly resistant at the adult-plant stage. It has excellent agronomic properties and is resistant to leaf rust, stripe rust, powdery mildew and other diseases. In turn, Thatcher is an adapted high-yield cultivar that is highly susceptible to Pt races at the seedling and adult-plant stages. The sensitive cultivar, Zhengzhou 5389 was used as a susceptible control. Four Pt races (THTT, THTQ, THTS and PHPS) were used to test the RILs in the fields. The Pt races were from the Biological Control Center for Plant Diseases and Plant Pests of Hebei, Hebei Agricultural University.
2.2. Field Experiments
The RIL population and parents were grown at Baoding in Hebei Province in the 2017/2018, 2018/2019 and 2019/2020 cropping seasons for evaluation of leaf rust response (environments hereinafter referred to as 18BD, 19BD, and 20BD). The field trials were conducted in randomized complete blocks with two replicates. Each plot consisted of a single 1 m row with 50 cm between rows. Approximately 20 seeds were sown in each plot. Every tenth row was followed by rows of the highly susceptible line Zhengzhou 5389 as a control. Leaf rust epidemics were initiated by inoculating the spreader rows with a water suspension of urediniospores of equal amounts of
Pt races (THTT, THTQ, THTS and PHPS) with a concentration 2 to 3 mg/mL added with a few drops of Tween 20 (0.03%) at the early jointing stage. Disease severity was recorded two or three times at weekly intervals with the first scoring at 4 weeks after inoculation. Disease severity data (0 to 100%) was recorded as percentage of leaf area covered with uredinia or necrotic stripes according to the modified Cobb scale [
27] where 0% = immune, and 100% = fully susceptible. Final disease severity (MDS%) was used for QTL analysis. Statistical analysis: Significant differences in phenotype and maximum disease severity (MDS) among trials (2018BD, 2019BD, 2022BD) were identified using analysis of variance (ANOVA) in Microsoft Excel.
2.3. Molecular Genotyping
The CTAB method was used to extract genomic DNA from the leaves of each of the 195 RILs and the two parental lines [
28]. DNA concentrations were determined using a Thermo Scientific NanoDrop 2000. DNA samples of the 10 RILs and parents were genotyped with the Affymetrix 55K SNP Array (53,064 markers) by CapitalBio Technology Company (Beijing, China). SSR markers were also used to genotype the entire population. Linkage maps were constructed using the MAP function in IciMapping 4.1 and algorithm of groups ordered using the Kosambi map function to obtain map distances from recombination frequencies, respectively. Linkage maps were graphically visualized with MapChart 2.3 [
29].
2.4. Construction of Linkage Maps and QTL Analysis
Using the genotyping data from the hybrid mapping population, QTLs associated with MDS were identified using QTL IciMapping 3.2. to make a chain diagram. A logarithm of the odds (LOD) threshold of 2.5 was used to identify significant QTL with 1000 permutations at
p < 0.01. Stepwise regressions were used to calculate the phenotypic variance explained (PVE,R2) by each QTL. The effect orientation of each QTL was determined [
30], while the gene effect was determined by calculating the DR value (the ratio of the absolute value of the dominant effect and the additive effect). The identified flanking sequences of all SNP and SSR probes were searched against the Chinese Spring wheat reference sequence (IWGSC RefSeq v1.0,
https://urgi.versailles.inra.fr/blast_iwgsc/blast.php) using BLAST to identify homologous sequences.
2.5. Wheat Primers and PCR Programs
SSR primer pairs were designed from the Chinese Spring wheat genome sequence (IWGSC,
http://wheat-urgi.versailles.inra.fr/) through a BLAST search. The SSR primers were designed by Batch Primer5 software. The reaction program of SSR primers used for PCR amplification was 94 °C for 5 min, 94 °C for 1 min, and 55 °C (depending on the primers, the temperature was adjusted in the range of 50–60 °C, and 55 °C was mostly used for primary screening) annealing for 45 s., 72 °C for 1 min and 72 °C for 10 min. The denaturation to extension process was 35 cycles, and the amplified samples were stored at 4 °C after the reaction was terminated. csLV46G22 PCR programs: The sequence of CAPs labelled csLV46G22 was 5′-TCGACTTTGGAATGGGAGTTGC-3′ upstream and 5′-GGCGAAGATGCCATCATCCACCAG-3′ downstream. Same as SSR PCR programs. The amplified products were digested by
BsPEⅠ under the reaction conditions of 37 °C for 2 h and 80 °C for 30 min, and the digested products were used for electrophoretic detection.
3. Results
3.1. Field Testing
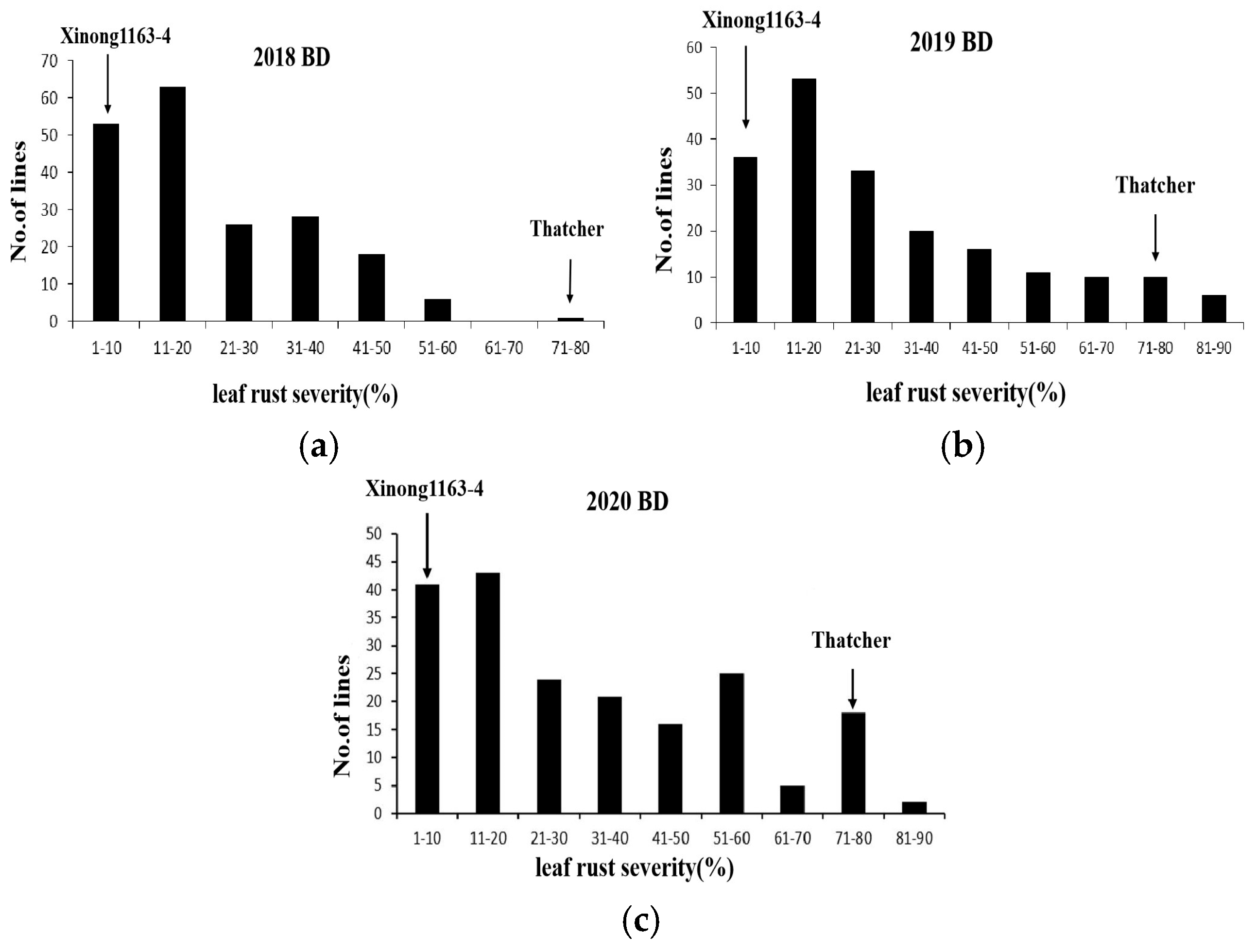
The mean leaf rust MDS scores of Xinong1163-4 and Thatcher were 1 and 80%, respectively, and mean leaf rust severity scores on the RILs ranged from 24.23 to 35.19% across all environments (
Figure 1 and
Table 1). The distribution of mean leaf rust MDS frequencies for the population was continuous, indicating quantitative inheritance. Maximum disease severity scores for leaf rust across the three environments were significantly correlated with coefficients ranging from 0.50 to 0.63 (
p < 0.001) (
Table 2). ANOVA confirmed significant variation among the genotypes, environments, and genotype × environment for leaf rust.
3.2. Genetic Analysis of Markers and Genetic Linkage Mapping
A total of 382 SSR markers showed polymorphism between parents among 1062 SSR markers and were used to test the resistant and susceptible bulks. We found 68 primers with a recombination frequency of less than 30% in small populations, allowing genotyping of the entire population. Finally, the genotyping results of 14 markers were used to construct the linkage map for QTL detection using the IciMapping 4.1 software. Six linkage groups were constructed, and were distributed on the chromosomes 1BL, 3A and 4BL according to wheat consensus maps.
3.3. QTL Mapping
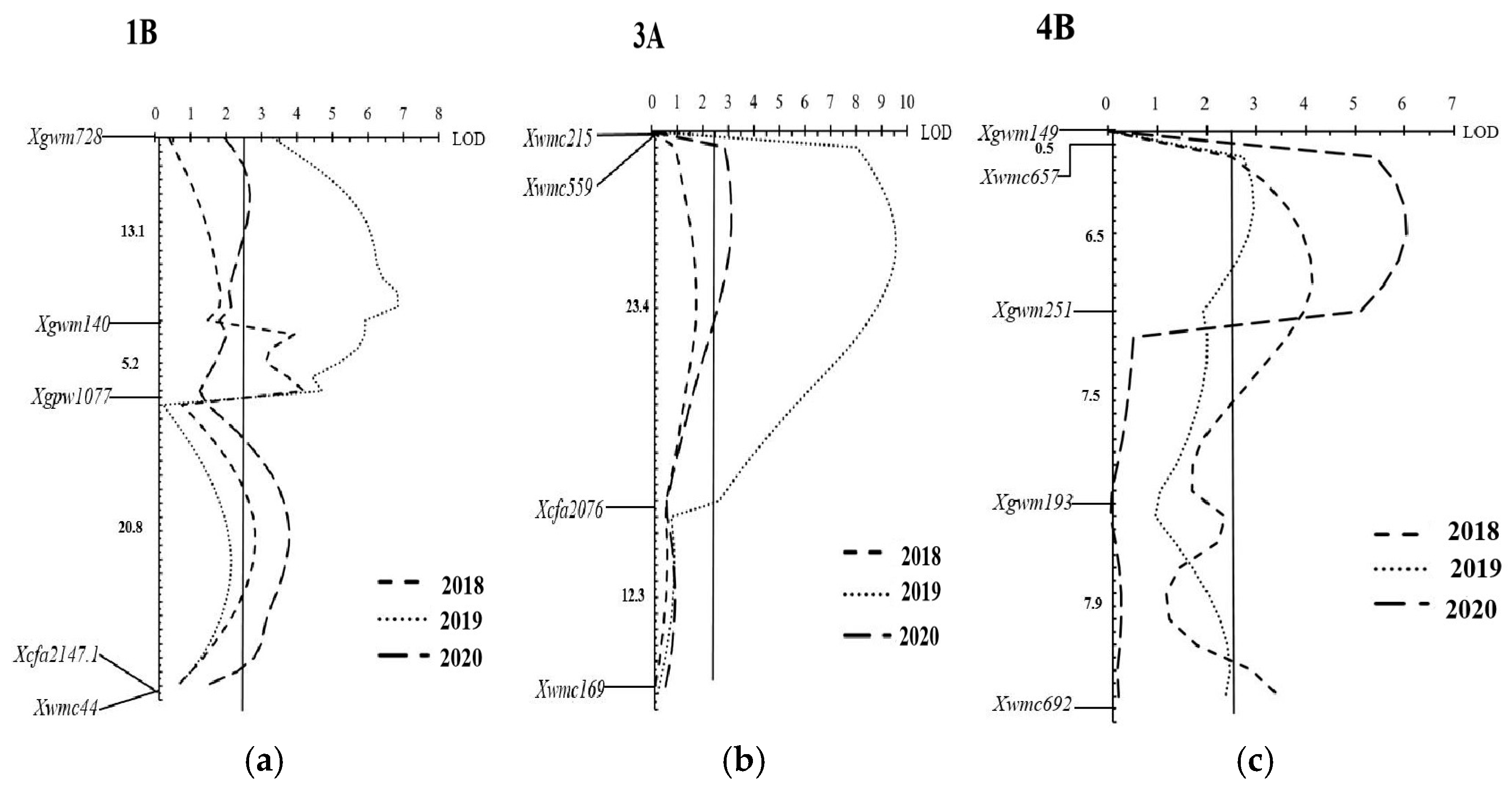
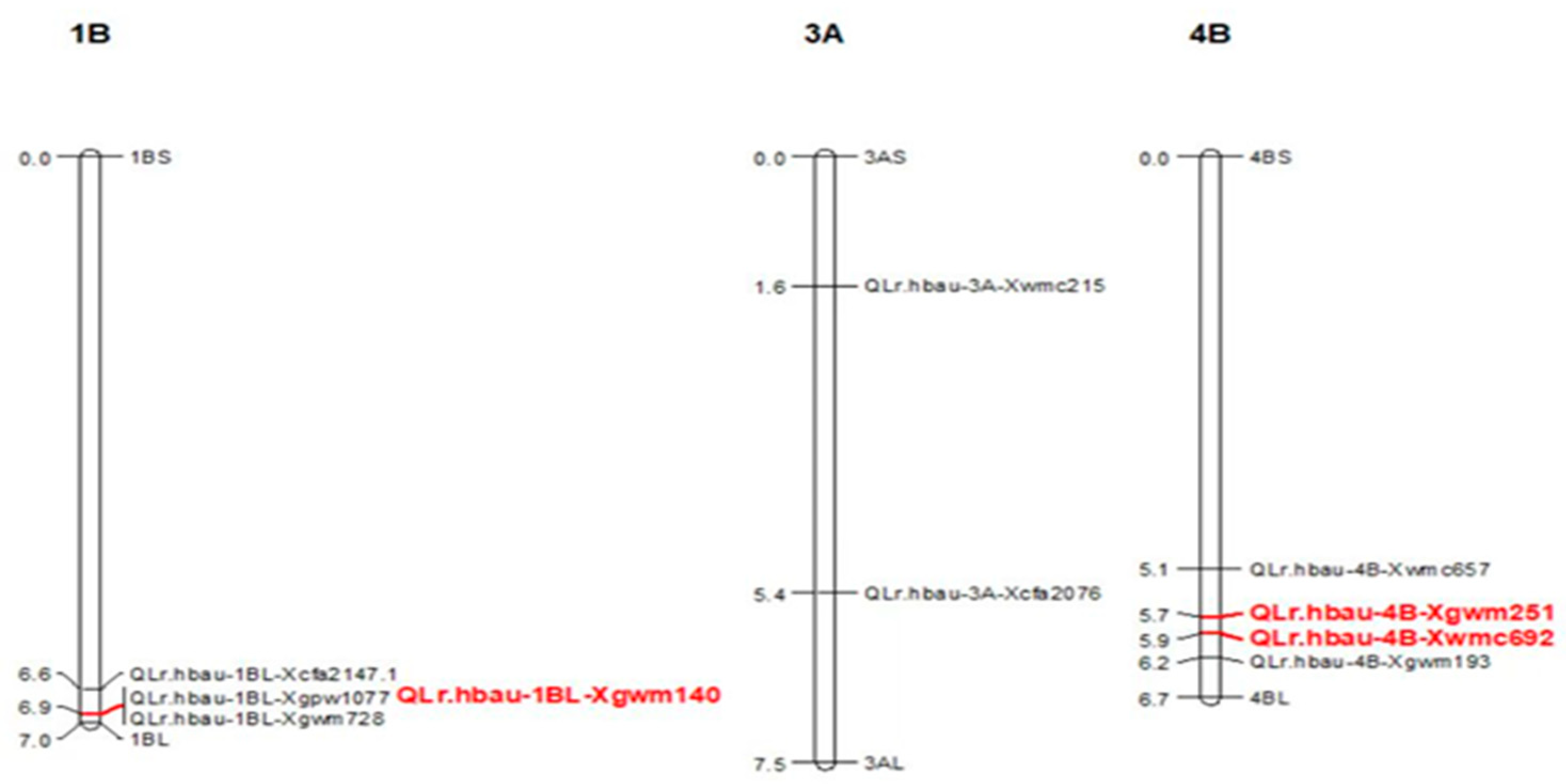
Six QTLs for leaf rust APR were identified on chromosomes 1B, 3A, and 4B (
Figure 2 and
Table 3). The
QLr.hbau-1BL.1 was detected in the 2019 and 2018,
QLr.hbau-1BL.2 was detected in the 2018 and 2020, and
QLr.hbau-1BL.3 was detected in the 2019 and 2020 season, and data experiments for all three traits explained 10.66%, 3.37–5.44% and 2.82–3.79% of the phenotypic variation for leaf rust, respectively (
Figure 2 and
Table 3). A major QTL for leaf rust resistance located on chromosome 1BL, was identified as
Lr46 based on the presence of marker
Xgwm728-Xgwm140. The fourth QTL,
QLr.hbau-3A was detected in the 2019 and 2020 experiments, with phenotypic variation explained (PVE) of 24.95–6.63%,for leaf rust, respectively. It was flanked by markers
Xwmc215-Xcfa2076. A major QTL for leaf rust resistance,
QLr.hbau-4BL was detected in all the three experiments, with a PVE of 8.13%, 5.31%, and 12.23% for leaf rust, respectively. It was flanked by markers
Xwmc657-Xgwm251. Among the Six QTLs, the resistance were from Xinong1163-4 (
Table 3).
3.4. csLV46G22 Molecular Marker Identification
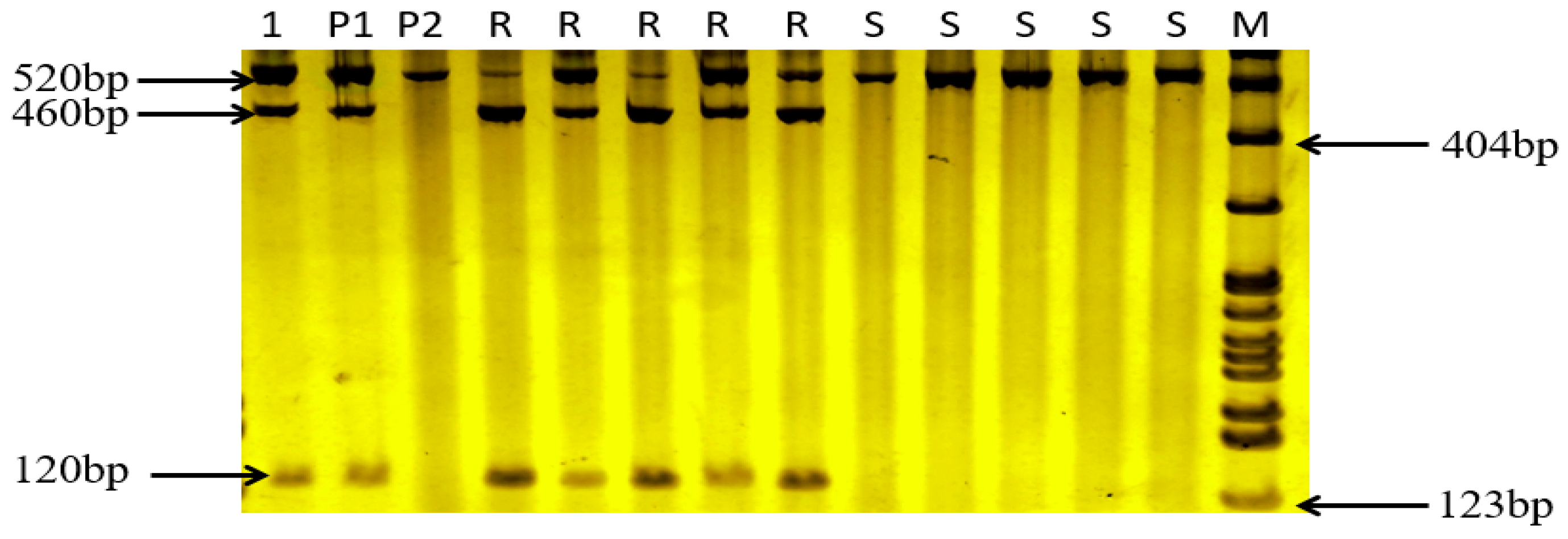
The gene
Lr46, which has now been identified at 1BL, was validated against two parents, five disease-resistant and five disease-susceptible lines using the CAPs marker
csLV46G22, which is co-segregated with the gene (
Figure 4). The near-isogenic lines containing
Lr46 could detect a 460 bp and a 120 bp DNA fragment, and the cultivars without
Lr46 could only detect a fragment of about 520 bp. In this test, both disease-resistant parents and disease-resistant subpopulations were able to detect two bands of 460 bp and 120 bp, while disease-susceptible parents and disease- susceptible subpopulations were able to detect only a single band of 520 bp, demonstrating the presence of
Lr46.
4. Discussion
Six QTLs for leaf rust resistance were detected in the current study. The total phenotypic variance across environments for leaf rust and 3.37–24.95% across environments for leaf rust, indicates their significant effects in reducing disease severity. The QTLs detected in the study were compared with the known genes or QTLs based on the chromosome position, molecular marker, pedigree, and resistance to rusts.
QLr.hbau-1BL
Based on the positions of closely linked markers,
QLr.hebau-1BL appeared to be the pleiotropic gene
Lr46/Yr29/Pm39/Ltn2. The QTLs identified on chromosome 1BL(
QLr.hbau-1BL.1,QLr.hbau-1BL.2 and
QLr.hbau-1BL.3) were predicted to correspond to the gene
Lr46/Yr29/Pm39/Ltn2.Lr46, a slow rust gene from common wheat that is located at the end of chromosome 1BL and confers resistance to leaf rust in adult plants.
Lr46 is widely used in CIMMYT wheat cultivars and has provided stable improvements in disease resistance for more than 30 years [
31].
Lr46 was linked to the SSR marker
Xwmc44 [
32], which was detected in the 1BL linkage map in all three experiments. This suggests that the three QTLs on chromosome 1BL (
QLr.hbau-1BL.1,QLr.hbau-1BL.2, and
QLr.hbau-1BL.3) are equivalent to
Lr46. Interestingly, the
Xgpw1077 marker was detected for the first time in the
Lr46 marker interval, providing a new marker reference for gene location and an additional option for molecular marker-assisted breeding. Genes associated with powdery mildew, stripe rust and stem rust resistance were also detected at this location. Therefore, Xinong1163-4 may also be resistant to powdery mildew, stripe rust and stem rust, although this possibility requires further testing.
QLr.hbau-3AL
To date, three APR QTLs have been identified on wheat chromosome 3A:
QLr.ubo-3A,QLr.sfrf-3A and
QLr.fcu-3AL [
33].
QLr.ubo-3A is located at 45 cM on chromosome 3AS, near the centromere, while
QLr.sfrf-3A and
QLr.fcu-3AL are located at 51 cM and 82 cM, respectively, and the marker interval
Xgwm666-Xcfa2183 on chromosome 3AL. According to the Chinese spring wheat reference sequence, this QTL should be different from
QLr.hbau-3AL, so,
QLr.hbau-3AL may be a new QTL.
QLr.hbau-4BL
To date, the APR genes
Lr12 [
34] and
Lr49 [
35]and three QTLs.
QLr.cimmyt-4BL, QLr.pbi-4BL and
QLr.zh-4B [
20,
29,
36]have been identified. These genes and loci are of great significance in the study of wheat leaf rust resistance, and their specific characteristics and roles will be described in detail below The
Lr12, has been mapped on 4BL, and is flanked by the markers
Xgwm251-Xgwm149, and the
Lr49 flanked by the markers
Xbarc163-Xwmc349. Both
Lr49 and
Lr12 were vulnerable to wheat leaf rust, but the slight species specificity of
Lr49 differed from that of
Lr12.
QLr.cimmyt-4BL flanked by the markers
Xgwm495-Xgwm368, and the marker linked to
QLr.pbi-4BL is wPt-1708 [
37]. Kang and colleagues identified
QLr.zh-4B, flanked by
Xwmc692 and
Xwmc657 in wheat cultivar Zhoumai 22×Chinese Spring F2:3 population [
38]. The QTL was also found to be consistent with
Lr12.
QLr.hbau-4BL flanked by the markers
Xwmc657 and
Xgwm251 was detected in all tests.
Xgwm149 also appears in the all tested linkage maps at this locus and is only 0.5 cM away from
Xwmc657, indicating a very close distance between them, since it closely corresponds to the reported markers in
Lr12.
Lr12 is a significant resistant gene that does not confer disease resistance during the seedling stage but has been previously identified as highly resistant at maturity. Here,
QLr.hbau-4BL was mapped very close to
Lr12, thereby confirming this QTL as
Lr12.
Author Contributions
“Conceptualization, J.Z. and Z.K.; methodology, J.Z.; software, J.Z.; validation, J.Z., X.L.(Xue Li) and L.X.; formal analysis, M.L.; investigation, J.Z.; resources, Z.K.; data curation, X.L.(Xue Li); writing—original draft preparation, J.Z.; writing—review and editing, X.L.(Xing Li ) and Z.K.; visualization, L.X.; supervision, X.L.(Xing Li ) and Z.K.; project administration, J.Z.; funding acquisition, Z.K. All authors have read and agreed to the published version of the manuscript.”
Funding
This study was sponsored by the National Natural Science Foundation of China (32001890), 2023 Scholarship Program for Introducing Overseas Students (C20230107).
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| Lr |
Wheat leaf rust |
| Pt |
Puccinia triticina Eriks |
| RILs |
Recombinant Inbred L ines |
| QTLs |
Quantitative trait loci |
| APR |
Adult plant resistance |
| ASR |
All-stage resistance |
| SSR |
Simple sequence repeat |
| SNP |
Single nucleotide polymorphism |
| GWAS |
Genome-wide association studies |
| MDS |
Maximum disease severity |
| ANOVA |
Analysis of variance |
| LOD |
Logarithm of the odds |
| PVE |
Phenotypic variation explained |
| CIMMYT |
International Maize and Wheat Improvement Center |
| MDPI |
Multidisciplinary Digital Publishing Institute |
| DOAJ |
Directory of open access journals |
| TLA |
Three letter acronym |
| LD |
Linear dichroism |
References
- Bolton, M.D.; Kolmer, J.A. and Garvin, D. F. Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol,2008,9(5), 563-75. [CrossRef]
- Huerta-Espino, J.; Singh, R.P.; Germán, S.; McCallum, B.D.; Park, R.F.; Chen, W.Q.; Bhardwaj, S.C. and Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179(1), 143-160. [CrossRef]
- Xu, X.; Duan, Z.; Su, J.; Li, X.; Wu, J. and Yao, Z. QTL mapping of adult - plant resistance to leaf rust based on SSR markers and SNP sequencing of Chinese wheat landrace Xu’ai (Triticum aestivum L.). Genet. Resour. Crop Evol. 2021, 68(4), 1359-1373. [CrossRef]
- Khan, M.; Bukhari, A.; Dar, Z.and Rizvi, S. Status and strategies in breeding for rust resistance in wheat. Agric. Sci. 2013, 4, 292-301. [CrossRef]
- Peng, H.; Lv, G.Q.; Wang, J.R. Hénán shěng 2015 nián xiǎomài zhǔyào bìnghài fāshēng tèdiǎn jí yuányīn fēnxī [Occurrence characteristics and causes of major wheat diseases in Henan Province in 2015]. China Plant Protection Guide 2016, 3, 91-93.
- Chen, X. Review Article: High - temperature adult - plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 2013, 04, 608 - 627. [CrossRef]
- Mapuranga, J.; Zhang, N.; Zhang, L.; Liu, W.; Chang, J.and Yang, W. Harnessing genetic resistance to rusts in wheat and integrated rust management methods to develop more durable resistant cultivars. Front Plant Sci 2022, 13, 951095. [CrossRef]
- Lagudah, E.S. Molecular genetics of race non-specific rust resistance in wheat. Euphytica 2011, 179(1), 81-91. [CrossRef]
- Lin, F.; Chen, X. M. Genetics and molecular mapping of genes for race - specific all - stage resistance and non - race - specific high - temperature adult - plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 2007, 114(7), 1277-1287. [CrossRef]
- Zhang, P.; Lan, C.; Asad, M.A.; Gebrewahid, T.W.; Xia, X.; He, Z.; Li, Z.and Liu, D. QTL mapping of adult-plant resistance to leaf rust in the Chinese landraces Pingyuan 50/Mingxian 169 using the wheat 55K SNP array. Mol. Breed. 2019, 39(7), 98. [CrossRef]
- Kolmer, J. A.; Bajgain, P.; Rouse, M. N.; Li, J.and Zhang, P. Mapping and characterization of the recessive leaf rust resistance gene Lr83 on wheat chromosome arm 1DS. Theor Appl Genet 2023, 136(5), 115. [CrossRef]
- Dyck, P. Genetics of leaf rust reaction in three introductions of common wheat. Can. J. Genet. Cytol. 1977, 19, 711-716. [CrossRef]
- Singh, R. P.; Mujeeb-Kazi, A.; Huerta-Espino, J. Lr46: a gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 1998, 88(9), 890 - 894. [CrossRef]
- Herrera-Foessel, S. A.; Singh, R. P.; Lillemo, M.; Huerta-Espino, J.; Bhavani, S.; Singh, S.; Lan, C.and Calvo-Salazar, V.; Lagudah, E. S. Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor Appl Genet 2014, 127(4), 781 - 789. [CrossRef]
- Herrera - Foessel, S.A.; Singh, R.P.; Huerta-Espino, J.; Rosewarne, G.M.; Periyannan, S.K.; Viccars, L.; Calvo - Salazar, V.; Lan, C.; Lagudah, E. S. Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theoret. Appl. Genet. 2012, 124(8), 1475 - 1486. [CrossRef]
- Kolmer, J.A.; Chao, S.; Brown-Guedira, G.; Bansal, U. and Bariana, H. Adult plant leaf rust resistance derived from the soft red winter wheat cultivar ‘Caldwell’ maps to chromosome 3BS. Crop Sci. 2018b, 58(1), 152-158. [CrossRef]
- Kolmer, J.A.; Su, Z.; Bernardo, A.; Bai, G.and Chao, S. Mapping and characterization of the new adult plant leaf rust resistance gene Lr77 derived from Santa Fe winter wheat. Theor Appl Genet 2018c, 131(7), 1553-1560. [CrossRef]
- Kolmer, J. A.; Bernardo, A.; Bai, G.; Hayden, M. J.and Chao, S. Adult plant leaf rust resistance derived from Toropi wheat is conditioned by Lr78 and three minor QTL. Phytopathology 2018a, 108(2), 246-253. [CrossRef]
- Singla, J.; Lüthi, L.; Wicker, T.; Bansal, U.; Krattinger, S.G.and Keller, B. Characterization of Lr75: a partial, broad - spectrum leaf rust resistance gene in wheat. Theor Appl Genet 2017, 130(1), 1-12. [CrossRef]
- Kolmer, J.A.; Rouse, M.N. Adult plant leaf rust resistance QTL derived from wheat line CI13227 maps to chromosomes 2AL, 4BS, and 7AL. Plant Genome 2022, 15(3), e20215. [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta - Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.and Keller, B.A. putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323(5919), 1360 - 1363. [CrossRef]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta - Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; Kong, X.; Spielmeyer, W.; Talbot, M.; Bariana, H.; Patrick, J. W.; Dodds, P.; Singh, R.and Lagudah, E. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 2015, 47(12), 1494 - 1498. [CrossRef]
- Krattinger, S.G.; Kang, J.; Bräunlich, S.; Boni, R.; Chauhan, H.; Selter, L.L.; Robinson, M.D.; Schmid, M.W.; Wiederhold, E.; Hensel, G.; Kumlehn, J.; Sucher, J.; Martinoia, E.and Keller, B. Abscisic acid is a substrate of the ABC transporter encoded by the durable wheat disease resistance gene Lr34. New Phytol. 2019, 223(2), 853-866. [CrossRef]
- Pinto da Silva, G.B.; Zanella, C.M.; Martinelli, J.A.; Chaves, M.S.; Hiebert, C.W.; McCallum, B.D.; Boyd, L.A. Quantitative trait loci conferring leaf rust resistance in hexaploid wheat. Phytopathology 2018, 108(12), 1344-1354. [CrossRef]
- Singh, R.; Huerta-Espino, J.; Rajaram, S. Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol. Entomol. Hung. 2000, 35, 133-139.
- Semagn, K.; Babu, R.and Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33(1), 1-14. [CrossRef]
- Peterson, R.F.; Campbell, A.B.and Hannah, A.E. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 1948, 26, 496-500. [CrossRef]
- Sharp, P.J.; Kreis, M.; Shewry, P.R.; Gale, M.D. Location of β-amylase sequences in wheat and its relatives. Theor. Appl. Genet. 1988, 75(2), 286-290. [CrossRef]
- Zhang, P.; Yin, G.; Zhou, Y.; Qi, A.; Gao, F.; Xia, X.; He, Z.; Li, Z.and Liu, D. QTL mapping of adult-plant resistance to leaf rust in the wheat cross Zhou 8425B/Chinese spring using high - density SNP Markers. Front Plant Sci 2017, 8, 793. [CrossRef]
- Singh, D.; Simmonds, J.; Park, R. F.; Bariana, H. S.; Snape, J. W. Inheritance and QTL mapping of leaf rust resistance in the European winter wheat cultivar ‘Beaver’. Euphytica 2009, 169(2), 253 - 261. [CrossRef]
- Lillemo, M.; Joshi, A.K.; Prasad, R.; Chand, R.; Singh, R.P. QTL for spot blotch resistance in bread wheat line Saar co-locate to the biotrophic disease resistance loci Lr34 and Lr46. Theor Appl Genet 2013, 126(3), 711-719. [CrossRef]
- Rosewarne, G.M.; Singh, R.P.; Huerta-Espino, J.; William, H. M.; Bouchet, S.; Cloutier, S.; McFadden, H.and Lagudah, E.S. Leaf tip necrosis, molecular markers and β1-proteasome subunits associated with the slow rusting resistance genes Lr46/Yr29. Theor. Appl. Genet. 2006, 112(3), 500-508. [CrossRef]
- Lan, C.X.; Singh, R.P.; Huerta - Espino, J.; Calvo - Salazar, V.; Herrera - Foessel, S.A. Genetic analysis of resistance to leaf rust and stripe rust in wheat cultivar Francolin#1. Plant Dis 2014, 98(9), 1227-1234. [CrossRef]
- Singh, S.; Bowden, R.L. Molecular mapping of adult-plant race-specific leaf rust resistance gene Lr12 in bread wheat. Mol. Breed. 2011, 28(2), 137-142. [CrossRef]
- Bansal, U.K.; Hayden, M.J.; Venkata, B.P.; Khanna, R.; Saini, R.G.; Bariana, H.S. Genetic mapping of adult plant leaf rust resistance genes Lr48 and Lr49 in common wheat. Theor. Appl. Genet. 2008, 117(3), 307-312. [CrossRef]
- William, H.M.; Singh, R.P.; Huerta-Espino, J.; Palacios, G.; Suenaga, K. Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 2006, 49(8), 977-990. [CrossRef]
- Yu, H.; Zhiying, D.; Xiang, C.; Tian, J. Analysis of diversity and linkage disequilibrium mapping of agronomic traits on B - genome of wheat. J. Genom. 2014, 2, 20-30. [CrossRef]
- Kang, L.; Qin, J.; Yu, T.; Gebrewahid, T.W.; Liu, J.; Chu, Z.; Xi, J.; Li, Z.; Yan, X.; Yao, Z. QTL mapping of adult plant resistance for leaf rust in F2:3 wheat pedigrees derived from Zhou mai 22/Chinese spring. Euphytica 2024, 220(11), 177. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).