Submitted:
23 March 2025
Posted:
26 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
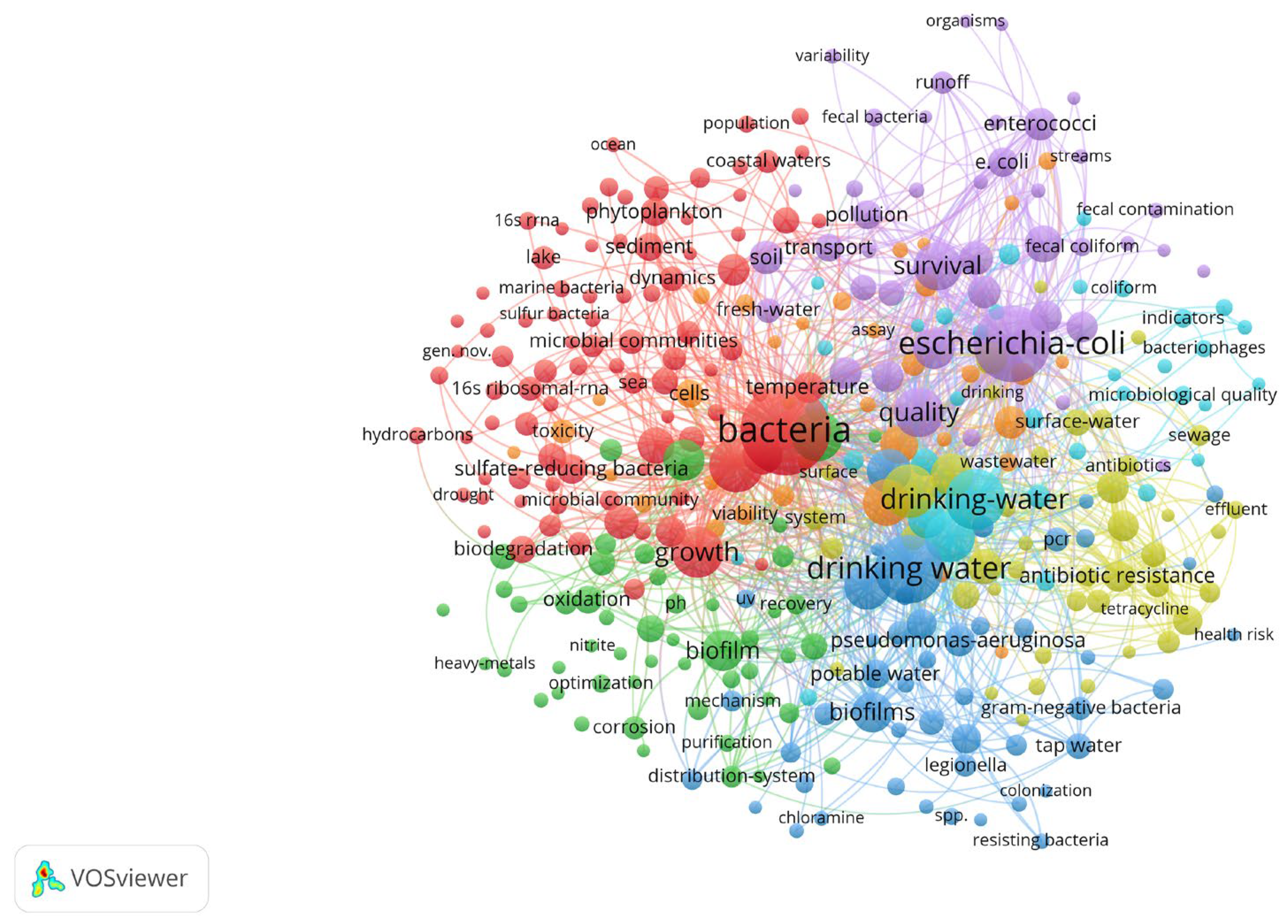
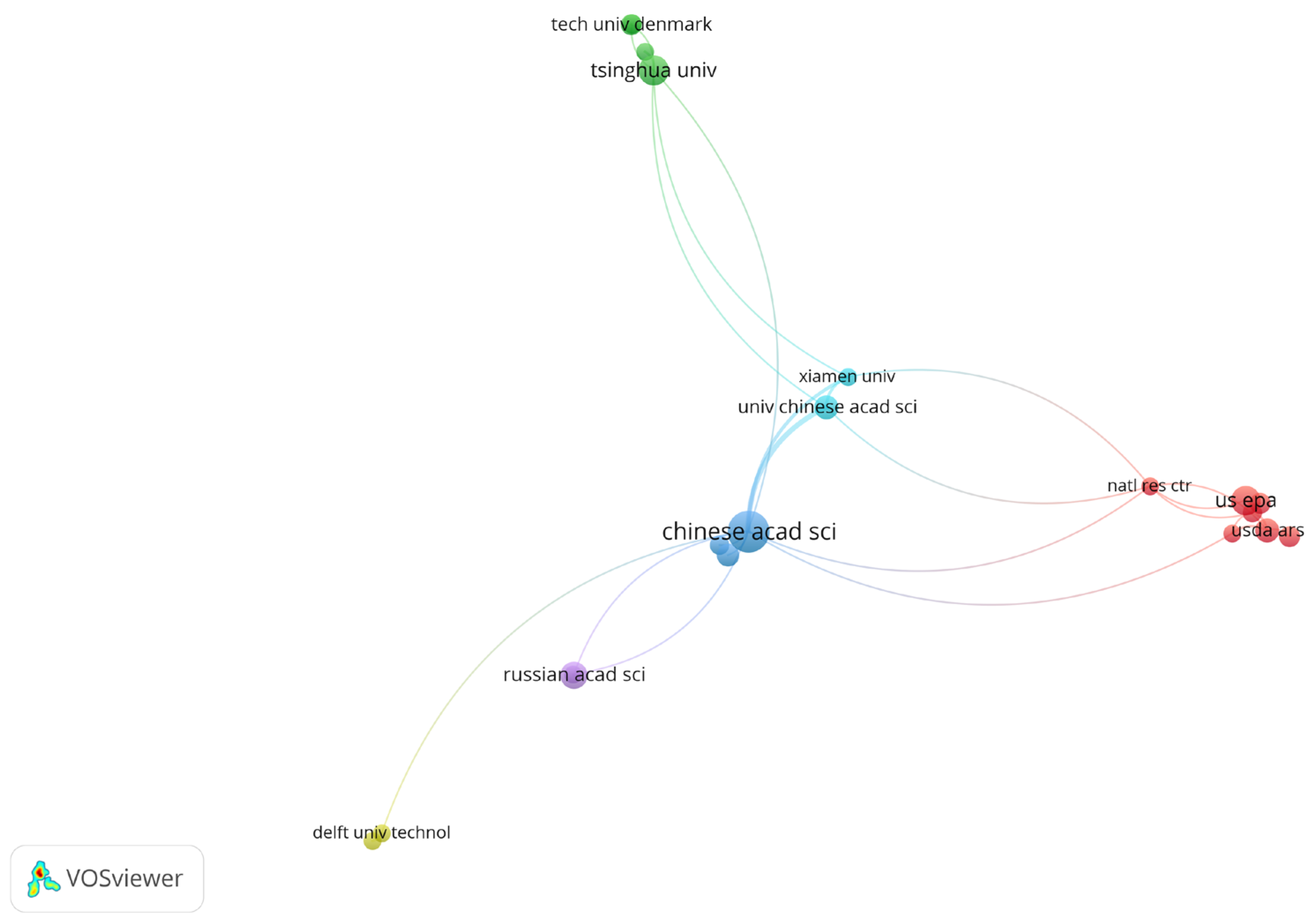
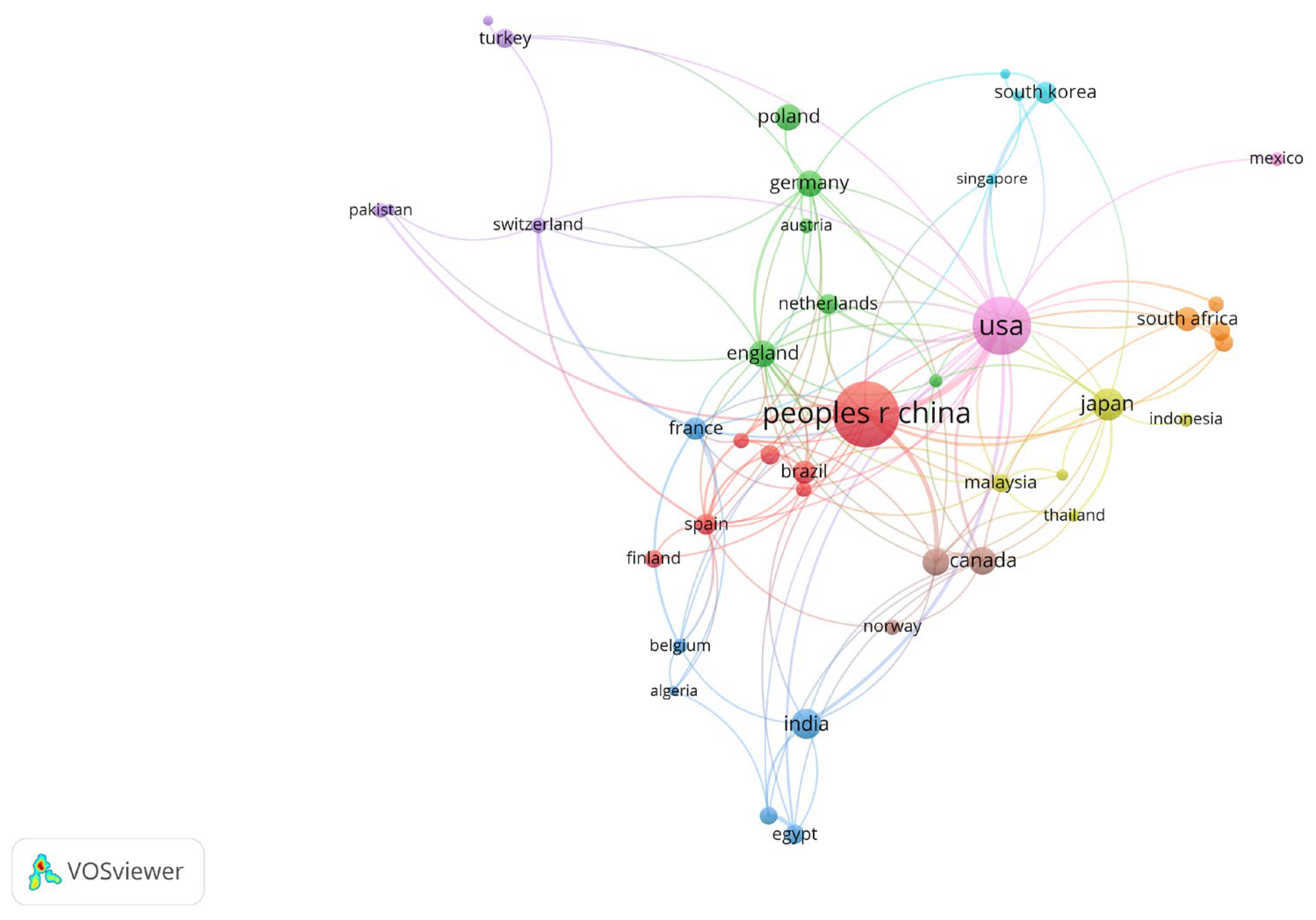
3. Results
4. Discussion
4.1. Advances in Water Bacteria Applications
4.2. Transforming Water Bacteria Research with Big Data and Machine Learning
5. Conclusions
References
- Cabral, J.P.S. Water microbiology. Bacterial pathogens and water. International journal of environmental research and public health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Hammes, F.; Egli, T. Cytometric methods for measuring bacteria in water: advantages, pitfalls and applications. Analytical and bioanalytical chemistry 2010, 397, 1083–1095. [Google Scholar] [CrossRef]
- Ding, Y.; Peng, N.; Du, Y.; Ji, L.; Cao, B. Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr (VI) immobilization. Applied and environmental microbiology 2014, 80, 1498–1506. [Google Scholar] [CrossRef]
- Yeferni, M.; Saïd, O.B.; Mahmoudi, E.; Essid, N.; Hedfi, A.; Almalki, M.; Pacioglu, O.; Dervishi, A.; Boufahja, F. Effectiveness of Shewanella oneidensis bioaugmentation in the bioremediation of phenanthrene-contaminated sediments and possible consortia with omnivore-carnivore meiobenthic nematodes. Environmental Pollution 2022, 292, 118281. [Google Scholar] [CrossRef] [PubMed]
- Yam, H.M.; Leong, S.K.W.; Qiu, X.; Zaiden, N. Bioremediation of Arsenic-contaminated water through application of bioengineered Shewanella oneidensis. 2021; pp. 559-574.
- Cabrera-Sosa, L.; Ochoa, T.J. Escherichia coli diarrhea. In Hunter's tropical medicine and emerging infectious diseases; Elsevier: 2020; pp. 481-485.
- Aydin, O.; Ulas, N.; Genc, A.; Baysal, S.; Kandemir, O.; Aktas, M.S. Investigation of hemogram, oxidative stress, and some inflammatory marker levels in neonatal calves with escherichia coli and coronavirus diarrhea. Microbial Pathogenesis 2022, 173, 105802. [Google Scholar] [CrossRef] [PubMed]
- Robins-Browne, R.M.; Hartland, E.L. Escherichia coli as a cause of diarrhea. Journal of gastroenterology and hepatology 2002, 17, 467–475. [Google Scholar] [CrossRef]
- Zhou, S.-X.; Wang, L.-P.; Liu, M.-Y.; Zhang, H.-Y.; Lu, Q.-B.; Shi, L.-S.; Ren, X.; Wang, Y.-F.; Lin, S.-H.; Zhang, C.-H. Characteristics of diarrheagenic Escherichia coli among patients with acute diarrhea in China, 2009‒2018. Journal of Infection 2021, 83, 424–432. [Google Scholar] [CrossRef]
- Nnachi, R.C.; Sui, N.; Ke, B.; Luo, Z.; Bhalla, N.; He, D.; Yang, Z. Biosensors for rapid detection of bacterial pathogens in water, food and environment. Environment international 2022, 166, 107357. [Google Scholar] [CrossRef]
- Qu, W.; Zhang, C.; Chen, X.; Ho, S.-H. New concept in swine wastewater treatment: development of a self-sustaining synergetic microalgae-bacteria symbiosis (ABS) system to achieve environmental sustainability. Journal of hazardous materials 2021, 418, 126264. [Google Scholar] [CrossRef]
- Bukar, U.A.; Sayeed, M.S.; Razak, S.F.A.; Yogarayan, S.; Amodu, O.A.; Mahmood, R.A.R. A method for analyzing text using VOSviewer. MethodsX 2023, 11, 102339. [Google Scholar] [CrossRef]
- Kirby, A. Exploratory bibliometrics: using VOSviewer as a preliminary research tool. Publications 2023, 11, 10. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, D.; Zhao, Y.; Liu, D.; Guo, X.; Chen, Y.; Zhang, C.; Sun, X.; Guo, J.; Yuan, D. Engineering Shewanella oneidensis to efficiently harvest electricity power by co-utilizing glucose and lactate in thin stillage of liquor industry. Science of The Total Environment 2023, 855, 158696. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, J.; Zou, L.; Shi, C.; Li, X.; Shi, Y.; Liu, M.; Duan, Y.; Wang, Q.; Ding, C. Electricity-powered cryptic CO2 fixation pathway in heterotrophic Shewanella oneidensis for acetate synthesis. Bioresource Technology 2025, 132324. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.J.; Kuzel, T.M.; Shafikhani, S.H. Pseudomonas aeruginosa: infections, animal modeling, and therapeutics. Cells 2023, 12, 199. [Google Scholar] [CrossRef]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa biofilm: a review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Nair, J.P.; Vijaya, M.S. Predictive models for river water quality using machine learning and big data techniques-a Survey. 2021; pp. 1747-1753.
- Kamyab, H.; Khademi, T.; Chelliapan, S.; SaberiKamarposhti, M.; Rezania, S.; Yusuf, M.; Farajnezhad, M.; Abbas, M.; Jeon, B.H.; Ahn, Y. The latest innovative avenues for the utilization of artificial Intelligence and big data analytics in water resource management. Results in Engineering 2023, 20, 101566. [Google Scholar] [CrossRef]
- Wang, C.; Mao, G.; Liao, K.; Ben, W.; Qiao, M.; Bai, Y.; Qu, J. Machine learning approach identifies water sample source based on microbial abundance. Water Research 2021, 199, 117185. [Google Scholar] [CrossRef]
- Kang, J.; Zhang, Z.; Chen, Y.; Zhou, Z.; Zhang, J.; Xu, N.; Zhang, Q.; Lu, T.; Peijnenburg, W.; Qian, H. Machine learning predicts the impact of antibiotic properties on the composition and functioning of bacterial community in aquatic habitats. Science of the Total Environment 2022, 828, 154412. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. From bibliography to understanding: water microbiology and human health. Journal of Water and Health 2024, 22, 1911–1921. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. Bibliographic Insights into Biofilm Engineering. Acta Microbiologica Hellenica 2024, 69, 3–13. [Google Scholar] [CrossRef]
- Boudry, C. Availability of ORCIDs in publications archived in PubMed, MEDLINE, and Web of Science Core Collection. Scientometrics 2021, 126, 3355–3371. [Google Scholar] [CrossRef]
- Ge, M.; Zhang, Y.; Li, Y.; Feng, C.; Tian, J.; Huang, Y.; Zhao, T. Publication Trends and hot spots in subacromial impingement syndrome research: A bibliometric analysis of the Web of Science Core Collection. Journal of Pain Research 2022, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Saiz-Alvarez, J.M. Innovation Management: A Bibliometric Analysis of 50 Years of Research Using VOSviewer® and Scopus. World 2024, 5, 901–928. [Google Scholar] [CrossRef]
- Ding, Y.; Zhou, Y.; Yao, J.; Szymanski, C.; Fredrickson, J.; Shi, L.; Cao, B.; Zhu, Z.; Yu, X.-Y. In situ molecular imaging of the biofilm and its matrix. Analytical chemistry 2016, 88, 11244–11252. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhou, Y.; Yao, J.; Xiong, Y.; Zhu, Z.; Yu, X.-Y. Molecular evidence of a toxic effect on a biofilm and its matrix. Analyst 2019, 144, 2498–2503. [Google Scholar] [CrossRef]
- Zhao, C.e.; Wu, J.; Ding, Y.; Wang, V.B.; Zhang, Y.; Kjelleberg, S.; Loo, J.S.C.; Cao, B.; Zhang, Q. Hybrid conducting biofilm with built-in bacteria for high-performance microbial fuel cells. ChemElectroChem 2015, 2, 654–658. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, Y.; Hu, Y.; Cao, B.; Rice, S.A.; Kjelleberg, S.; Song, H. Enhancing bidirectional electron transfer of Shewanella oneidensis by a synthetic flavin pathway. ACS synthetic biology 2015, 4, 815–823. [Google Scholar] [CrossRef]
- Dai, H.N.; Nguyen, T.-A.D.; Le, L.-P.M.; Van Tran, M.; Lan, T.-H.; Wang, C.-T. Power generation of Shewanella oneidensis MR-1 microbial fuel cells in bamboo fermentation effluent. International Journal of Hydrogen Energy 2021, 46, 16612–16621. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, C.; Zhao, G.; Chen, Y. Versatile mechanisms and enhanced strategies of pollutants removal mediated by Shewanella oneidensis: A review. Journal of Hazardous Materials 2022, 440, 129703. [Google Scholar] [CrossRef]
- Ng, C.K.; Sivakumar, K.; Liu, X.; Madhaiyan, M.; Ji, L.; Yang, L.; Tang, C.; Song, H.; Kjelleberg, S.; Cao, B. Influence of outer membrane c-type cytochromes on particle size and activity of extracellular nanoparticles produced by Shewanella oneidensis. Biotechnology and Bioengineering 2013, 110, 1831–1837. [Google Scholar] [CrossRef]
- Ng, C.K.; Mohanty, A.; Cao, B. Biofilms in Bio-Nanotechnology: Opportunities and Challenges. Bio-Nanoparticles: Biosynthesis and Sustainable Biotechnological Implications, 2015; 83–100. [Google Scholar]
- Zhang, Z.; Ding, Y.; Qian, S. Influence of bacterial incorporation on mechanical properties of engineered cementitious composites (ECC). Construction and Building Materials 2019, 196, 195–203. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, D.; Ding, Y.; Wang, S. Mechanical performance of strain-hardening cementitious composites (SHCC) with bacterial addition. Journal of Infrastructure Preservation and Resilience 2022, 3, 3. [Google Scholar] [CrossRef]
- Zhang, Z.; Weng, Y.; Ding, Y.; Qian, S. Use of genetically modified bacteria to repair cracks in concrete. Materials 2019, 12, 3912. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, N.; Huang, T.Y.; Zhong, F.; Peng, G. Pseudomonas aeruginosa: A typical biofilm forming pathogen and an emerging but underestimated pathogen in food processing. Frontiers in Microbiology 2023, 13, 1114199. [Google Scholar] [CrossRef]
- Faure, E.; Kwong, K.; Nguyen, D. Pseudomonas aeruginosa in chronic lung infections: how to adapt within the host? Frontiers in immunology 2018, 9, 2416. [Google Scholar] [CrossRef]
- van Heeckeren, A.M.; Schluchter, M.D. Murine models of chronic Pseudomonas aeruginosa lung infection. Laboratory animals 2002, 36, 291–312. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, H.; Zhou, Z.; Miao, Y.; Li, R.; Yang, B.; Cao, C.; Xiao, S.; Wang, X.; Liu, H. Characterization of extended-spectrum β-Lactamase-producing Escherichia coli isolates that cause Diarrhea in Sheep in Northwest China. Microbiology Spectrum 2022, 10, e01595–01522. [Google Scholar] [CrossRef]
- Manhique-Coutinho, L.; Chiani, P.; Michelacci, V.; Taviani, E.; Bauhofer, A.F.L.; Chissaque, A.; Cossa-Moiane, I.; Sambo, J.; Chilaúle, J.; Guimarães, E.L. Molecular characterization of diarrheagenic Escherichia coli isolates from children with diarrhea: A cross-sectional study in four provinces of Mozambique: Diarrheagenic Escherichia coli in Mozambique. International Journal of Infectious Diseases 2022, 121, 190–194. [Google Scholar] [CrossRef]
- Pakbin, B.; Brück, W.M.; Rossen, J.W.A. Virulence factors of enteric pathogenic Escherichia coli: A review. International journal of molecular sciences 2021, 22, 9922. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, S.; Zhang, W.; Wu, D.; Yang, G. Probiotic Escherichia coli NISSLE 1917 for inflammatory bowel disease applications. Food & Function 2022, 13, 5914–5924. [Google Scholar]
- Niveditha, A.; Pandiselvam, R.; Prasath, V.A.; Singh, S.K.; Gul, K.; Kothakota, A. Application of cold plasma and ozone technology for decontamination of Escherichia coli in foods-a review. Food control 2021, 130, 108338. [Google Scholar] [CrossRef]
- Bharti, B.; Li, H.; Ren, Z.; Zhu, R.; Zhu, Z. Recent advances in sterilization and disinfection technology: A review. Chemosphere 2022, 308, 136404. [Google Scholar] [CrossRef] [PubMed]
- Hamdany, A.H.; Ding, Y.; Qian, S. Mechanical and antibacterial behavior of photocatalytic lightweight engineered cementitious composites. Journal of Materials in Civil Engineering 2021, 33, 04021262. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Cementitious Composite Materials for Self-Sterilization Surfaces. ACI Materials Journal 2022, 119, 197–210. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Visible light antibacterial potential of graphene-TiO2 cementitious composites for self-sterilization surface. Journal of Sustainable Cement-Based Materials 2023, 12, 972–982. [Google Scholar] [CrossRef]
- Hamdany, A.H.; Ding, Y.; Qian, S. Graphene-based TiO2 cement composites to enhance the antibacterial effect of self-disinfecting surfaces. Catalysts 2023, 13, 1313. [Google Scholar] [CrossRef]
- Kotsiri, Z.; Vidic, J.; Vantarakis, A. Applications of biosensors for bacteria and virus detection in food and water–A systematic review. journal of environmental sciences 2022, 111, 367–379. [Google Scholar] [CrossRef]
- Sreedevi, P.R.; Suresh, K.; Jiang, G. Bacterial bioremediation of heavy metals in wastewater: a review of processes and applications. Journal of Water Process Engineering 2022, 48, 102884. [Google Scholar] [CrossRef]
- Bachute, M.R.; Subhedar, J.M. Autonomous driving architectures: insights of machine learning and deep learning algorithms. Machine Learning with Applications 2021, 6, 100164. [Google Scholar] [CrossRef]
- Kiran, B.R.; Sobh, I.; Talpaert, V.; Mannion, P.; Al Sallab, A.A.; Yogamani, S.; Pérez, P. Deep reinforcement learning for autonomous driving: A survey. IEEE transactions on intelligent transportation systems 2021, 23, 4909–4926. [Google Scholar] [CrossRef]
- Khan, A.R. Facial emotion recognition using conventional machine learning and deep learning methods: current achievements, analysis and remaining challenges. Information 2022, 13, 268. [Google Scholar] [CrossRef]
- Tabassum, F.; Islam, M.I.; Khan, R.T.; Amin, M.R. Human face recognition with combination of DWT and machine learning. Journal of King Saud University-Computer and Information Sciences 2022, 34, 546–556. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. Machine learning and its applications in studying the geographical distribution of ants. Diversity 2022, 14, 706. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. A machine learning approach to predicting academic performance in Pennsylvania’s schools. Social Sciences 2023, 12, 118. [Google Scholar] [CrossRef]
- Chen, S.; Ding, Y. The feasibility of using machine learning to predict COVID-19 cases. International Journal of Medical Informatics 2025, 105786. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, J.; Yang, X.; Zhang, Y.; Zhang, L.; Ren, H.; Wu, B.; Ye, L. A review of the application of machine learning in water quality evaluation. Eco-Environment & Health 2022, 1, 107–116. [Google Scholar]
- Nasir, N.; Kansal, A.; Alshaltone, O.; Barneih, F.; Sameer, M.; Shanableh, A.; Al-Shamma'a, A. Water quality classification using machine learning algorithms. Journal of Water Process Engineering 2022, 48, 102920. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Beaglehole, D.; Pandit, P.; Belkin, M. Mechanism for feature learning in neural networks and backpropagation-free machine learning models. Science 2024, 383, 1461–1467. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




