1. Introduction
Cotton is a pivotal economic crop and the primary source of natural fiber for the textile industry [
1]. However, it is severely affected by Verticillium wilt caused by Verticillium dahlia. In China, this disease affects over 50% of cotton fields annually, resulting in substantial economic losses [
2].
V. dahliae is particularly difficult to control due to its persistence in soil as dormant microsclerotia [
3]. Currently, there is no effective chemical control for cotton Verticillium wilt [
4]. The most cost-effective and efficient strategy involves breeding and deployment of disease-resistant cotton varieties [
5]. The scarcity of available resistance genes for genetic engineering has significantly constrained cotton molecular breeding. GTs can link sugar molecules to specific receptors and produce glycosidic bonds, which are present in most organisms and can be particularly important in plants.
Some natural glycosides or sugar esters with more stable structure were usually formed by glycosylation modification [
6]. Glycosylation is crucial for promoting plant storage, intracellular transport, and regulating the balance of growth agents within plants [
7]. One important role of
GTs is responsible to catalyze plant glycosylation [
8].
GTs transfer activated sugars to various plant molecules, including monosaccharides, oligosaccharides, polysaccharides, and non-carbohydrate substrates such as proteins, antibiotics, and lipids, leading to the glycosylation of these compounds [
9].
GTs are essential for plant growth, development, and responses to biotic and abiotic stresses.
UGT73C7 is a glycosyltransferase that regulates disease resistance by redirecting the phenylpropanoid pathway and activating the TIR-type NLR protein SNC1 in
Arabidopsis thaliana [
10]. UGT76B1 can regulate the basic defense and SAR by coordinating the glycosylation of three defense activators SA, ILA, and NHP [
11]. CsUGT87E7 encodes a SA carboxyl-glucosyltransferase and enhances the disease resistance of tea by regulating SA homeostasis [
12].
TaUGT6 is a novel UDP-glycosyltransferase gene, which can enhance the resistance of wheat to Fusarium head blight by converting deoxynivalenol (DON) into DON-3-glucoside [
13]. However, few reports have reported on the role of
GTs in cotton against
V. dahliae.
In our previous study, we found that GTs are involved in the regulation of cotton's defense response to V. dahliae. In order to clarify the molecular function of GTs in cotton, the GTs were analyzed in G. arboreum, G. raimondii, G. hirsutum, and G. barbadense genomes, respectively. And the characteristics of GTs were comprehensively analyzed in G. hirsutum, including their chromosomal locations, motif distributions, gene structures, and cis-elements in the promoter regions. Additionally, the impact of GhGT61 on V. dahliae was verified by VIGS. This study will preliminarily clarify the function of GTs and provide gene resources for molecular breeding of upland cotton resistant to Verticillium wilt.
2. Results
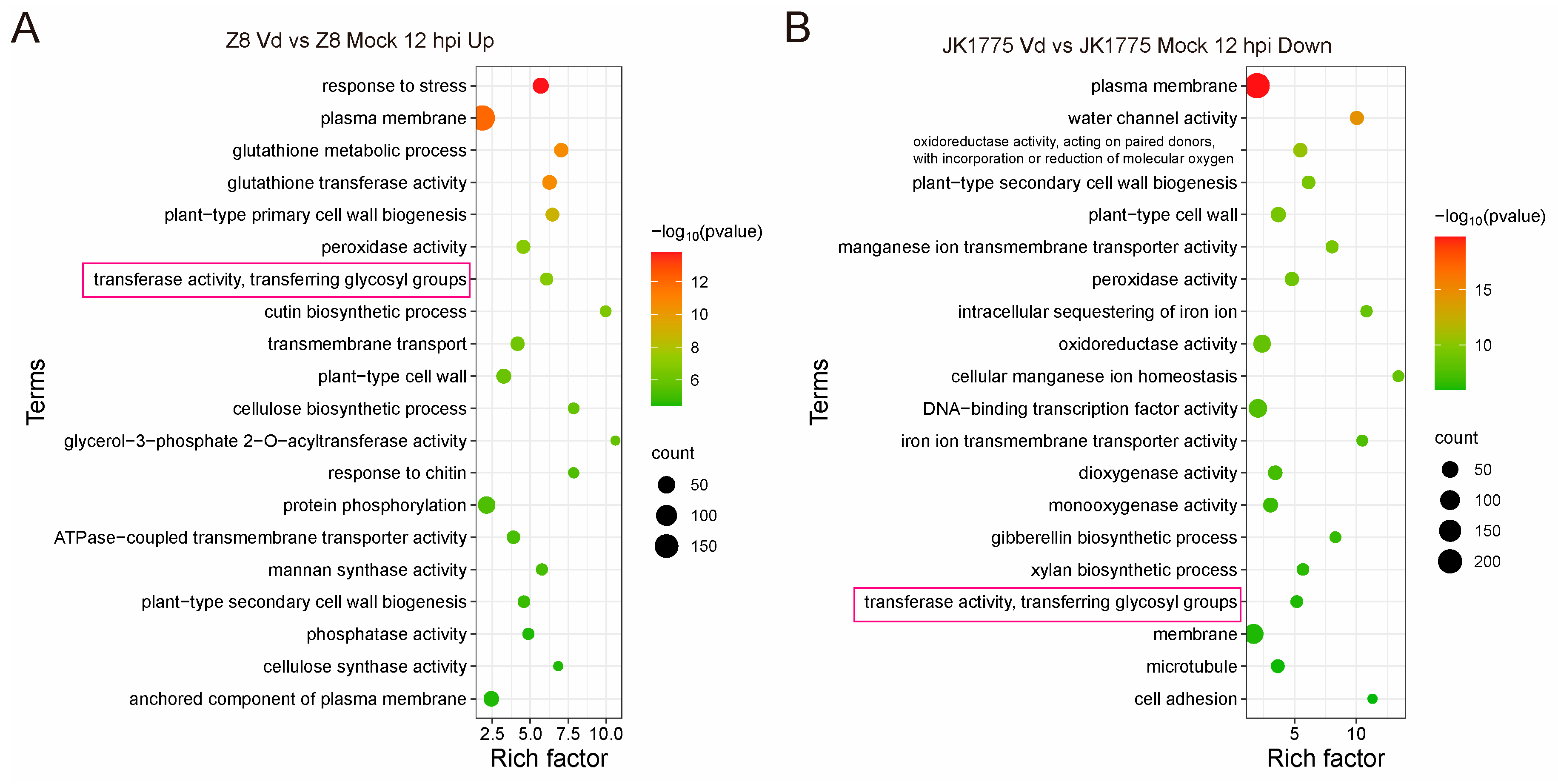
2.1. GTs Are Involved in the Response of Cotton to V. dahliae Infection
In order to screen the key resistance- associated genes of upland cotton responding to
V. dahliae infection, we analyzed the differentially expressed genes (DEGs) in both resistant and susceptible upland cotton varieties after inoculation with
V. dahliae. GO enrichment analysis revealed that the transferase activity and transferring glycosyl groups pathway were significantly enriched in both cotton varieties at 12 h post-inoculation. However, the expression patterns of these DEGs were completely opposite because of that the related DEGs were up-regulated and down-regulated in susceptible and resistant upland cotton, respectively (
Figure 1A, B).
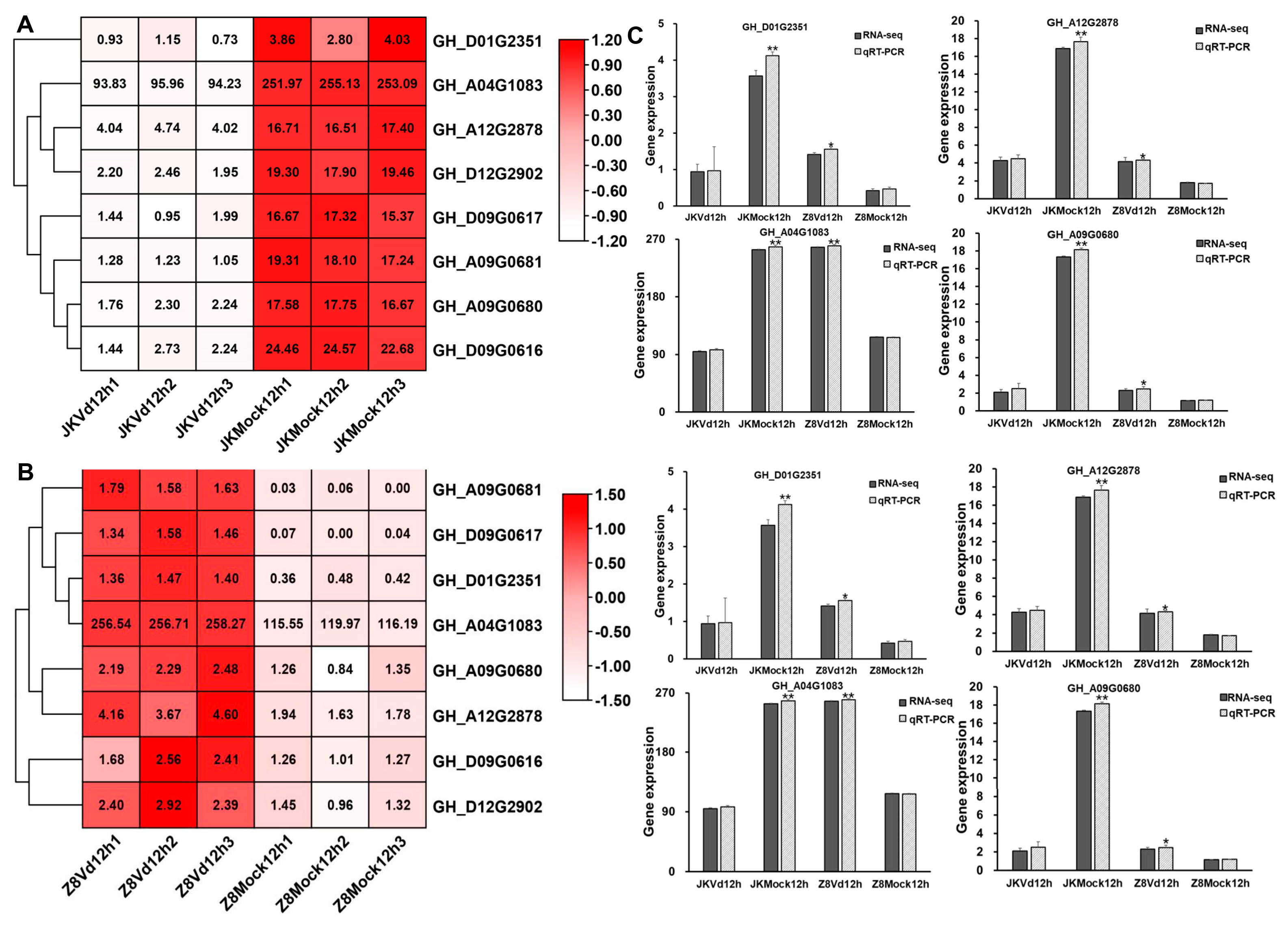
There were eight identical
GTs involved in the transferase activity and transferring glycosyl groups pathway in both resistant and susceptible upland cotton (
Figure 2A, B). These
GTs may play a crucial role in the immune response of upland cotton to
V. dahliae In order to verify the reliability of the selected
GTs data, the differential expression levels of
GTs were analyzed by qRT-PCR in the early stage of
V. dahliae infection in both resistant and susceptible upland cotton. The results indicated that the expression level of the
GTs were basically consistent with the results of transcriptome data (
Figure 2 C). Notably,
GH_A04G1083 exhibited high expression in both inoculated and uninoculated upland cotton, with expression levels higher than those of other
GTs (Figure2A, B, C). These results suggested that
GH_A04G1083 may play a role not only in the immune response to
V. dahliae but also in other vital activities of upland cotton.
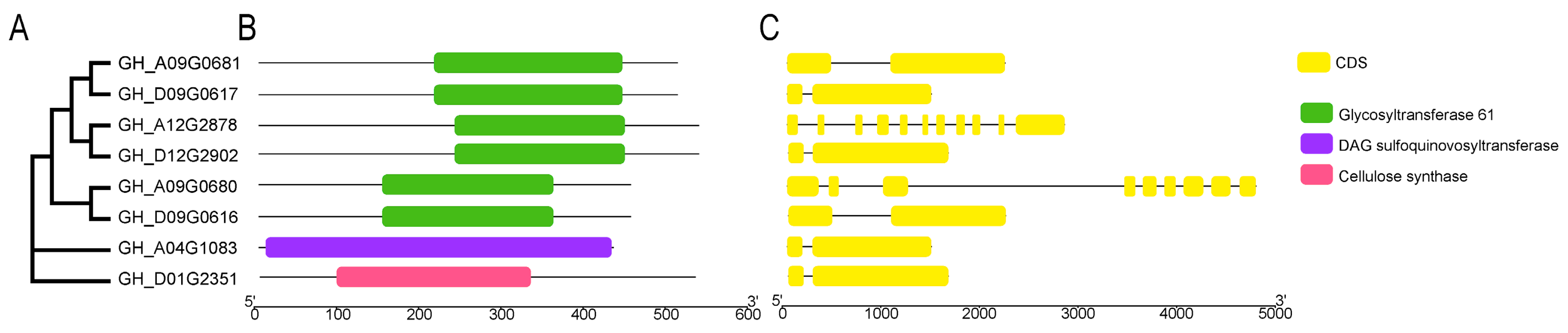
2.2. Protein Structure and Genetic Relationship of the Eight GTs in Cotton
To eclucidate the biological function of the eight
GTs furtherly, we analyzed their protein structure and genetic relationship. The results revealed that although the distribution of CDS sequences on the eight
GTs showed certain differences (
Figure 3C),
GH_A09G0681 and
GH_D09G0617,
GH_A12G2878 and
GH_D12G2902,
GH_A09G0680 and
GH_D09G0616 were clustered together, respectively, indicating a relatively close relationship (
Figure 3A). And this six
GTs were identified as members of GT family 61 based on their domain analysis (
Figure 3B). Additionally, the
GH_D01G2351 and
GH_AO4G1083 were classified under GT family 1 and GT family 2, respectively (
Figure 3A).
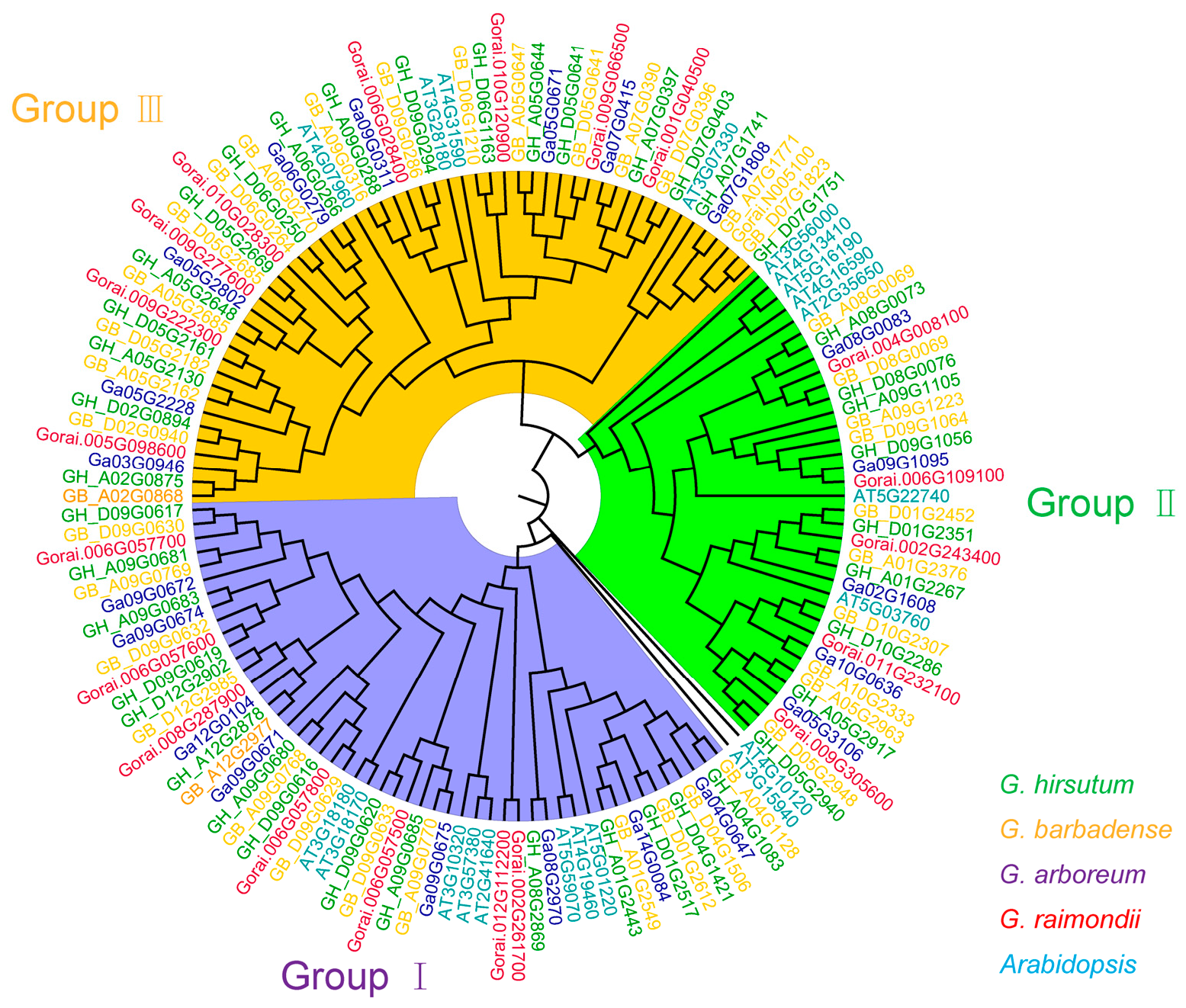
To investigate the evolutionary relationship of the eight
GTs in cotton, we obtained their homologous genes in
G. arboreum,
G. raimondii,
G. hirsutum,
G. barbadense and
A. thaliana by BLAST in cotton and
A. thaliana genomes Phylogenetic analysis revealed that the homologous genes of the eight
GTs in present study were divided into three groups, consisting of 51, 36, and 54 genes, respectively. Among which seven
GTs were in the Group Ι and one
GTs (GH_D01G2351) in the Group II (
Figure 4). The number of homologous
GTs in
G. barbadense and
G. hirsutum was approximately twice than that of
G. arboreum and
G. raimondii (
Figure 4).
2.3. Analysis of Promoter Region of the Eight GTs
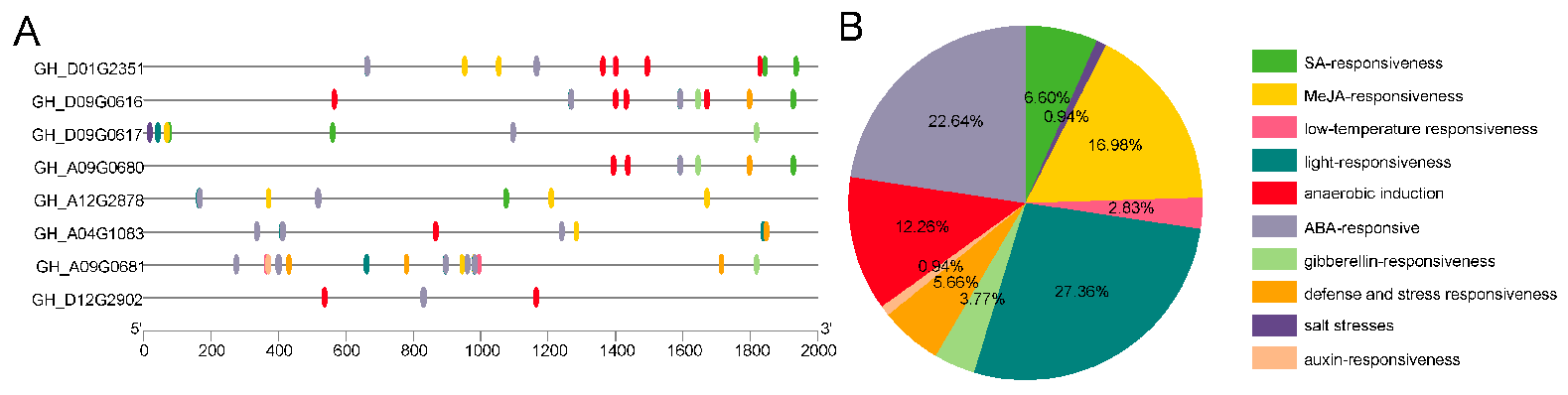
Promoter regions are crucial for determining gene expression patterns and functions. In order to predict the roles of the
GTs, we analyzed the upstream cis-acting elements within their promoter regions. Interestingly, many of these elements were associated with plant hormones, including abscisic acid (ABA), gibberellin (GA), salicylic acid (SA), and MeJA, as well as abiotic stress factors such as anaerobic conditions, defense and stress responses, light responses, and low-temperature responses (
Figure 5A). Among these elements, hormone-related elements (SA, MeJA, auxin, ABA, and gibberellin) account for a significant proportion, representing 50.93% of the total elements (
Figure 5B). More importantly, defense and stress responses elements accounted for 5.66% of the total (
Figure 5B). Notably, each GT contains at least one hormone-related cis-acting element, particularly those related to ABA. Among the eight
GTs,
GH_A09G0680,
GH_D09G0616, and
GH_D12G2902 lack MeJA-related elements. Collectively, these results indicate that
GTs are crucial for conferring resistance to upland cotton against adverse environmental conditions.
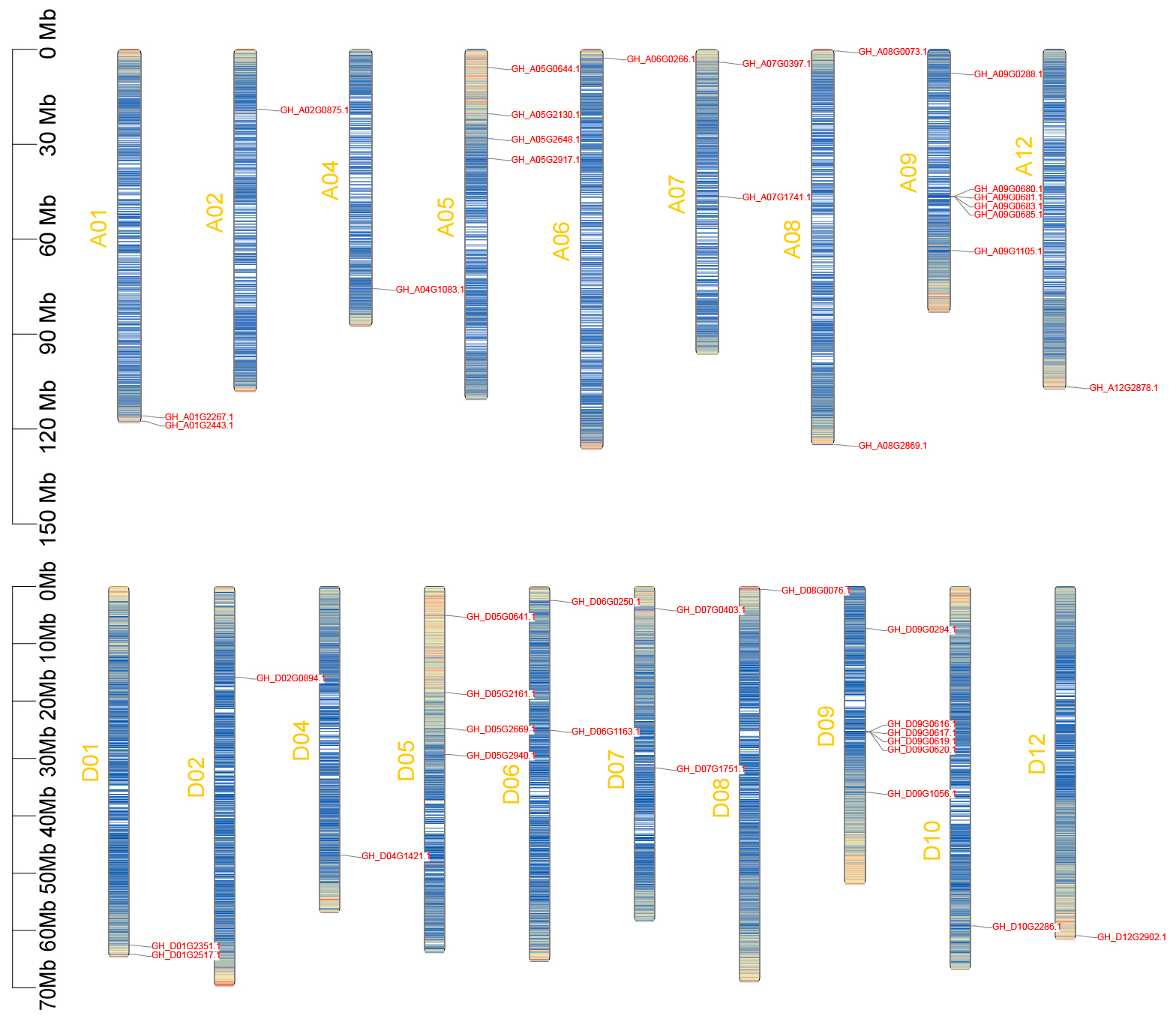
2.4. Chromosomal Localization of Cotton GTs
To investigate the genomic distribution of cotton
GTs, a chromosome map was constructed for 41
GTs in upland cotton. A total of 20
GTs were mapped to 9 chromosomes of the At subgenome, while 21
GTs were mapped to 10 chromosomes of the Dt subgenome (
Figure 6). The distribution of
GTs genes across At and Dt chromosomes is uneven, ranging from 1 to 6. Specifically, there are 6
GTs located on chromosomes A09 and D09, while 6
GTs located on chromosomes A02, A04, A06, A12, D02, D04, D06, D10, D12. Notably, the number and chromosomal location of certain
GTs on the At subgenome are the same as those on the Dt subgenome, as evidenced by pairs such as A01-D01, A02-D02, A04-D04, A05-D05, A07-D07, A09-D09, and A12-D12.
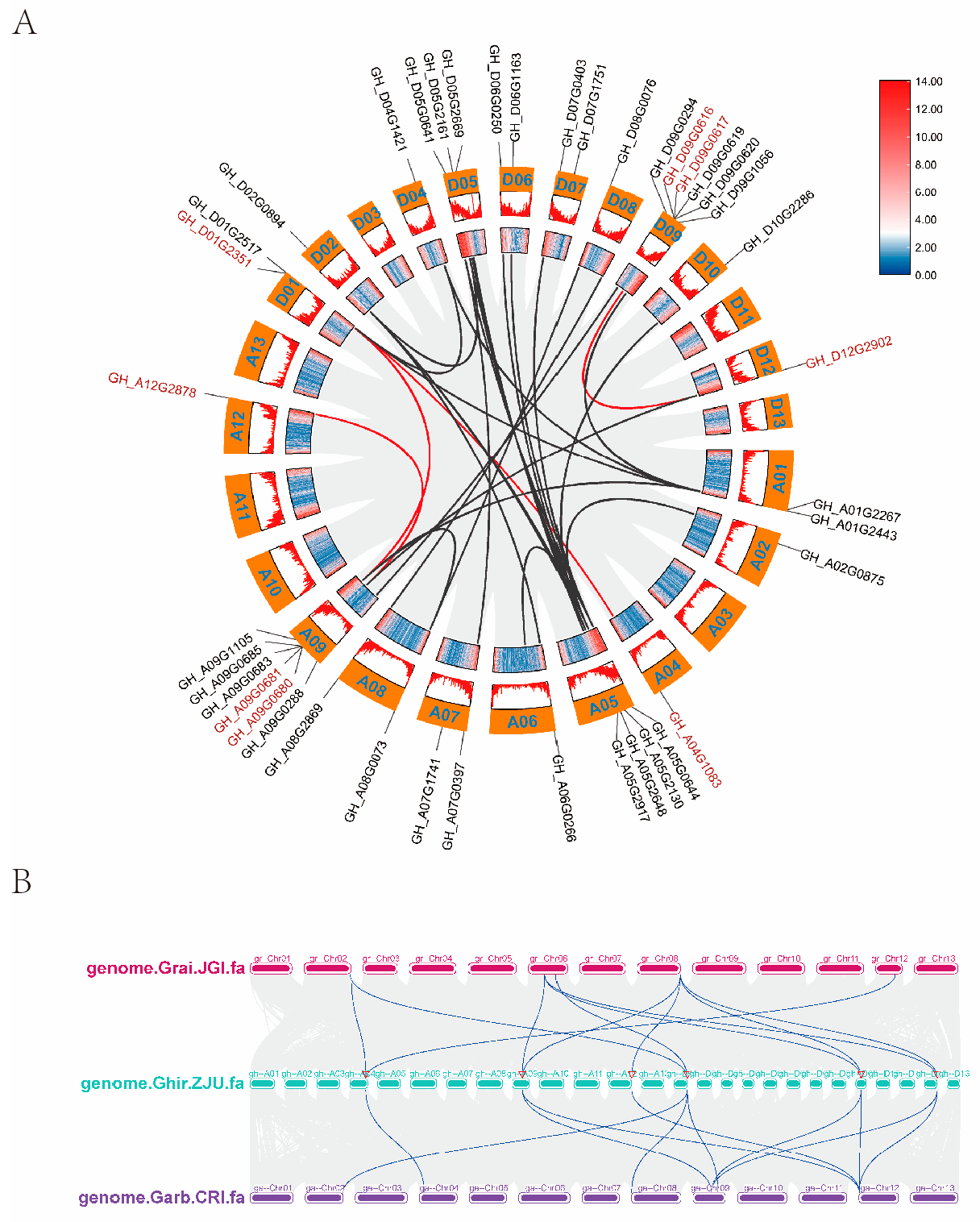
2.5. Synteny Analysis of GTs in Cotton
In order to investigate the fragment replication and tandem replication of
GTs in upland cotton, the amplification mechanism of cotton
GTs were analyzed using BLAST and MCScanX. The results suggensted that six of the eight
GTs formed gene clusters, including
GH_A04G1083-
GH_D01G2517,
GH_D01G2351-
GH_A09G0685,
GH_A12G2878-
GH_A09G0680, and
GH_D09G0616-
GH_D12G2902, which constituted four tandem clusters on chromosomes A12, A09, D09, and D12, respectively (
Figure 7A). In contrast,
GH_D09G0617 and
GH_A09G0681 did not form gene clusters (
Figure 7A). These four tandem duplications are inferred to have originated from polyploid ancestors, while
GH_D09G0617 and
GH_A09G0681 are believed to have arisen in
G. hirsutum following its divergence from
G. barbadense, as they are absent in
G. barbadense (
Figure 7B).
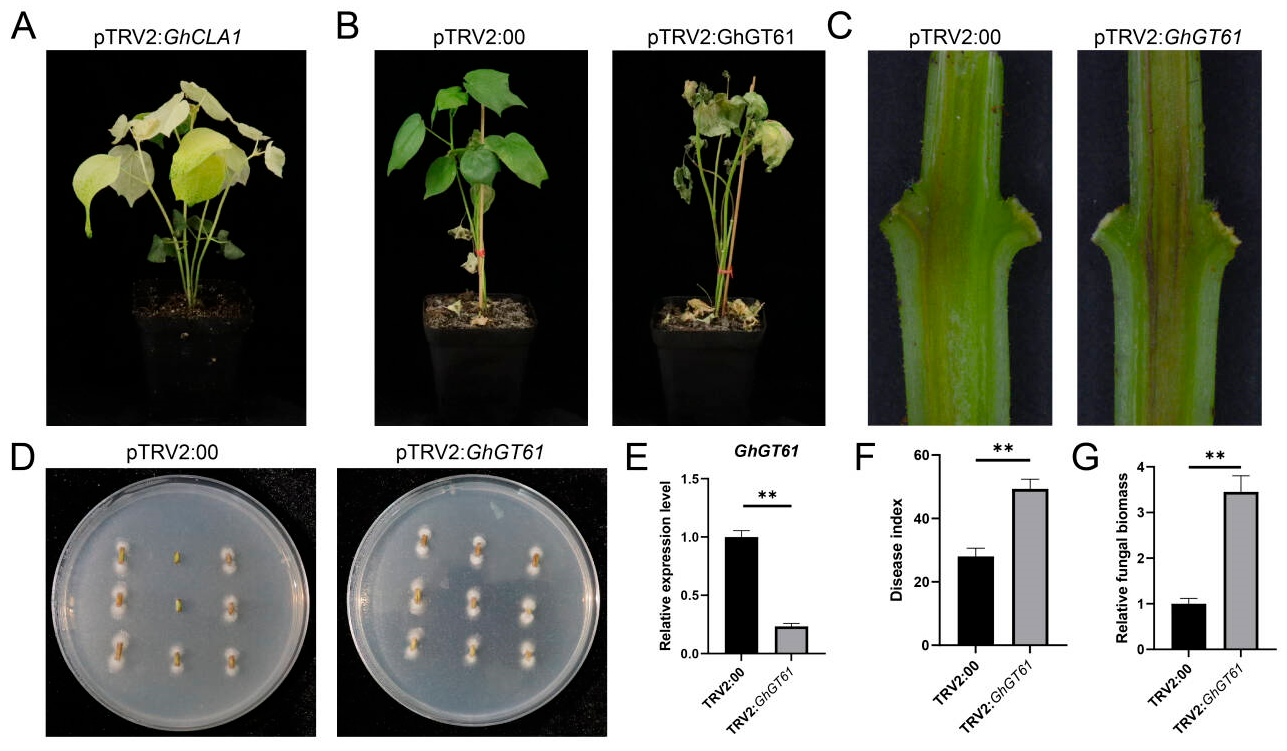
2.6. Silence of GhGT61 GhGTs Affected the Disease Resistance of Cotton
To elucidate the role of
GhGTs in cotton resistance to Verticillium wilt, VIGS and disease resistance assays were carried out. The results showed that an albino phenotype was observed at 14 dpi (
Figure 7A). Following inoculation with
V. dahliae 13 days after silencing, plants with suppressed
GhGT61 exhibited markedly reduced resistance to the
V. dahliae (
Figure 7B, F), with a significant reduction in expression level of
GhGT61 (
Figure 7E). Histological analysis of longitudinal sections of cotton cotyledonary nodes revealed pronounced browning in TRV2:
GhGT61 plants compared to TRV2:
00 controls (
Figure 7C). Fungal biomass quantification via recovery culture showed significantly higher
V. dahliae accumulation in stems of TRV2:
GhGT61 plants than in TRV2:
00 controls (
Figure 7D, G). These results collectively demonstrate that silencing
GhGT61 enhances cotton susceptibility to
V. dahliae, underscoring its critical role in mediating resistance to Verticillium wilt.
3. Discussion
In recent years, continuous cropping has resulted in a significant increase in Verticillium wilt incidence in the cotton industry, severely impacting cotton yields. The application of modern molecular techniques for identifying resistance-related genes has accelerated the breeding of disease-resistant varieties [
14]. Exploring key genes associated with cotton's resistance to Verticillium wilt and elucidating their mechanism is crucial for developing new cotton germplasm resistant to Verticillium wilt [
15]. Recent advancements in sequencing technology and decreasing costs have significantly improved and updated cotton genome sequencing efforts, providing a foundation for studying gene functions at the genomic level [
16]. Transcriptome analysis can identify potential genes and molecular mechanisms involved in various biotic stress responses, playing a vital role in elucidating gene function [
17]. A more profound comprehension of cotton genomics and genetics opens new pathways for investigating the
GTs family.
Glycosylation is a common modification of plant secondary metabolites and plays various functions, including the regulation of hormone homeostasis, detoxification of exogenous substances, and biosynthesis and storage of secondary compounds. These processes are facilitated by specific subclasses of the GT family [
18]. However, there are few studies on the biotic stress of cotton
GTs. In the present study, we identified that there were eight
GTs involved in cotton regulation of the defense response to
V. dahliae in the early infection period. The GT family has remained conserved throughout the long-term evolution of cotton, consistent with its evolutionary relationships. Notably, the number of
GTs in tetraploid cotton is approximately double that of diploid cotton species, further supporting the existence of two diploid ancestors for tetraploid cotton [
19].
Gene function is closely linked to the differentiation of promoter region, which are the combination regions of many transcription factors (TFs) [
20]. TFs usually regulate various aspects of plant growth, development, and stress resistance by regulating gene expression in responses to biotic and abiotic stress factors [
21]. Analysis of the cis-elements in the promoter regions of these eight
GTs revealed that the cis-elements with the strongest hormone response were ABA and MEJA response cis-elements. This suggests that these
GTs may be involved in the response of cotton to
V. dahliae through the ABA or MeJA pathway. Additionally, light-responsive cis-regulating elements account for a large proportion in the eight
GTs with accounting for 27.336% of the total, indicating that
GTs may participate in the regulation of light responses in cotton.
At present, the main goal of cotton breeding is to select high-yield and high-quality new varieties resistant to Verticillium wilt [
22]. In this study, we analyzed the RNA-Seq data set (PRJNA1121084) of cotton inoculated with
V. dahliae and found that eight
GTs were differentially expressed in resistant and susceptible cotton after inoculation with
V. dahliae, indicating that
GTs may be involved in the defense process of cotton against
V. dahliae. Subsequently, resistance of cotton to
V. dahliae was weakened after silencing
GhGT61. To deepen the understanding of
GTs gene family, a lot of work is still needed to produce transgenic cotton and CRISPR/Cas9-mediated knockout in the future. It is necessary to verify the function, find important genes and analyze whether there is functional redundancy between
GTs genes in the resistance to Verticillium wilt.
4. Materials and Methods
4.1. RNA-seq and qRT-PCR Analysis
The transcriptome data were obtained from were from the NCBI Sequence Read Archive database (Project ID: PRJNA1121084)[
23]. Data were filtered and quality-controlled using Fastp software [
24]. Clean data were aligned to the
G. hirsutum TM-1 genome (ZJU-improved_v2.1_a1) [
1] using HISAT2, and StringTie was used for read quantification. Gene expression was evaluated using the RPKM method [
25]. For qRT-PCR, primers were designed using Primer 5.3 software based on the cDNA sequences of eight candidate genes. Expression levels were analyzed using root tissue cDNA as a template. Each sample was replicated 7 times, and
GhUBQ7 was used as an internal reference gene. qRT-PCR was performed as described previously [
26].
4.2. Identification of GTs in Cotton
Genomic data of
G. arboreum,
G. raimondii,
G. hirsutum and
G. barbadense. were obtained from the CottonGen database (
http://www.cottongen.org/). Homologous sequences were were identified using BLAST with an e-value threshold of 1e-5. A total of 21 Ga and Gr, 41 Gh, and 40 Gb GTs were retrieved [
27].
4.3. Analysis of Gene Structure and Conserved Motifs
Gene structure analysis was performed using TBtools to identify exons, introns, and untranslated regions of GTs in cotton. Domain analysis was performed using NCBI conserved domain database (CDD) to determine the domain types and positions. Subsequently, Gene exons, introns, and conserved motifs were visualized using Tbtools (v2.149).
4.4. Phylogenetic Analysis of GTs in Cotton
Multiple sequence alignments were performed using Clustal W in MEGA X (v11.0.13) with default settings (gap opening penalty = 10, gap separation distance = 4). A phylogenetic tree with 1000 bootstrap replicates was constructed using the neighbor-joining (NJ) method. The tree was visualized and edited using FigTree (
http://tree.bio.ed.ac.uk/software/Figtree/).
4.5. Analysis of Cotton Glycosyltransferase Activity Gene Promoter Region
4.6. VIGS of GhGT61
RNA was isolated from cotton roots and reverse-transcribed into cDNA. A 300 bp CDS fragment of
GhGT61 (GH_D12G2902) was amplified using primers with BamHI and EcoRI restriction sites. The fragment was cloned into the TRV2 vector by homologous recombination and transformed into Agrobacterium GV3101. The transformed TRV2 vector was injected into Z8 with completely flattened cotyledons, and then the transformed seedlings were transferred to a growth chamber at 25℃ with a light/dark photoperiod of 16 h/ 8 h. VIGS efficiency was assessed using the TRV2:
GhCLA construct. After 15 d, the expression level of the target gene was detected, and the successfully silenced plants were infected with V592 [
30].
5. Conclusions
This study identified eight GTs that were differentially expressed in cotton varieties susceptible and resistant to Verticillium wilt through RNA-Seq analysis. The analysis of gene number, chromosome location and evolution revealed that the GTs family is conserved in cotton. The presence of ABA- and MeJA-responsive cis-acting elements suggests that these GTs may participate in the response of cotton to V. dahliae infection through these hormonal pathways. Additionally, the presence of photoresponsive elements in the cis-acting elements of each gene suggests that GTs may be involved in the regulation of cotton photoperiod. VIGS experiments further revealed that GhGT61 is involved in the resistance process of cotton to Verticillium wilt. This study provides a foundation for further research on the role of GTs in cotton and offers insights for breeding efforts aimed at developing more resistant cotton varieties.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: List of primers used in this study.
Author Contributions
Conceptualization, Y.Y and H.X.; methodology, M.Z.; software, M.Z; validation, M.Z, F.Z and G.S.; formal analysis, Y.M.; investigation, M.Z and X.Z; resources, Y.Y and H.X.; data curation, M.Z; writing—original draft preparation, M.Z; writing—review and editing, M.Z, X.Z, Y.Y, and H.X; visualization, H.X.; supervision, S,Z and H.X.; project administration, Y.Y and H.X.; funding acquisition, X.Z and H.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Shihezi University high-level talents research project(2022ZK015), Bingtuan Science and Technology Program (2022ZD056, 2023CB007-08), and the Earmarked fund for XJARS-Cotton (XJARS-03).
Data Availability Statement
The data used in this study were from the National Center for Biotechnology Information (NCBI), BioProject ID: PRJNA1121084.
Acknowledgments
We thank senior researcher Yu Yu from Xinjiang Academy of Agricultural Reclamation Sciences for his assistance with the materials used for experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (G. hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 3, 531-7. [CrossRef]
- Zhang, X.; Cheng, W.; Feng, Z.; Zhu, Q.; Sun, Y.; Li, Y.; Sun, J. Transcriptomic analysis of gene expression of V. dahliae upon treatment of the cotton root exudates. BMC Genomics. 2020, 21, 155. [CrossRef]
- Luo, X.; Xie, C.; Dong, J.; Yang, X.; Sui, A. Interactions between V. dahliae and its host: vegetative growth, pathogenicity, plant immunity. Appl. Microbiol. Biotechnol. 2014, 98, 6921-32. [CrossRef]
- Mo, H.; Wang, X.; Zhang, Y.; Zhang, G.; Zhang, J.; Ma Z. Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to V. dahliae. Plant J. 2015, 83, 962-75. [CrossRef]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant. Pathol. 2006, 7, 71-86. [CrossRef]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003, 62, 399-413. [CrossRef]
- Gilbert, M.K.; Bland, J.M.; Shockey, J.M.; Cao, H.; Hinchliffe, D.J.; Fang D.D.; Naoumkina M. A transcript profiling approach reveals an abscisic acid-specific glycosyltransferase (UGT73C14) induced in developing fiber of Ligon lintless-2 mutant of cotton (Gossypium hirsutum L.). PLoS One. 2013, 8, e75268. [CrossRef]
- Henrissat, B.; Davies, G.J. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000, 124, 1515-9. [CrossRef]
- Yu, J.; Hu, F.; Dossa, K.; Wang, Z.; Ke T. Genome-wide analysis of UDP-glycosyltransferase super family in Brassica rapa and Brassica oleracea reveals its evolutionary history and functional characterization. BMC Genomics. 2017, 18, 474. [CrossRef]
- Huang, X.; Wang, Y.; Lin, J.; Chen, L.; Li, Y.; Liu, Q.; Wang, G.; Xu, F.; Liu, L.; Hou, B. The novel pathogen-responsive glycosyltransferase UGT73C7 mediates the redirection of phenylpropanoid metabolism and promotes SNC1-dependent Arabidopsis immunity. Plant J. 2021, 107, 149-165. [CrossRef]
- Bauer, S.; Mekonnen, D.W.; Hartmann, M.; Yildiz, I.; Janowski, R.; Lange, B.; Geist, B.; Zeier, J.; Schäffner A.R. UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid, and balances plant immunity. Plant Cell. 2021, 33, 714-734. [CrossRef]
- Hu, Y.; Zhang, M.; Lu, M.; Wu, Y.; Jing, T.; Zhao, M.; Zhao, Y.; Feng, Y.; Wang, J.; Gao, T.; et al. Salicylic acid carboxyl glucosyltransferase UGT87E7 regulates disease resistance in Camellia sinensis. Plant Physiol. 2022, 188, 1507-1520. [CrossRef]
- He, Y.; Wu, L.; Liu, X.; Jiang, P.; Yu, L.; Qiu, J.; Wang, G.; Zhang, X.; Ma, H. TaUGT6, a novel UDP-glycosyltransferase gene enhances the resistance to FHB and DON accumulation in wheat. Front. Plant Sci. 2020, 11, 574775. [CrossRef]
- Mo, H.; Wang, X.; Zhang, Y.; Zhang, G.; Zhang, J.; Ma, Z. Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 2015, 83, 962-75. [CrossRef]
- Yuan, Y.; Feng, H.; Wang, L.; Li, Z.; Shi, Y.; Zhao, L.; Feng, Z.; Zhu, H. Potential of endophytic fungi isolated from cotton roots for biological control against Verticillium wilt disease. PLoS One. 2017, 12, e0170557. [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao T.; Lian J.; et al. G. barbadense and G. hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739-748. [CrossRef]
- Guo, L.; Han, L.; Yang, L.; Zeng, H.; Fan, D.; Zhu, Y.; Feng, Y.; Wang, G.; Peng, C.; Jiang, X.; et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS One. 2014, 9, e95543. [CrossRef]
- Gachon C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 2005, 10, 542-9. [CrossRef]
- Senchina, D.S.; Alvarez, I.; Cronn, R.C.; Liu, B.; Rong, J.; Noyes, R.D.; Paterson, A.H.; Wing, R.A.; Wilkins T.A.; Wendel, J.F. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 2003, 20, 633-43. [CrossRef]
- Amorim, L.L.B.; da Fonseca Dos Santos, R.; Neto, J.P.B.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A.M. Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 2017, 18, 335-351. [CrossRef]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genomics. 2021, 22, 350. [CrossRef]
- Wang, Y.; Zhao, J.; Deng, X.; Wang, P.; Geng, S.; Gao, W.; Guo, P.; Chen, Q.; Li, C.; Qu, Y. Genome-wide analysis of serine carboxypeptidase-like protein (SCPL) family and functional validation of Gh_SCPL42 unchromosome conferring cotton Verticillium der Verticillium wilt stress in Gossypium hirsutum. BMC Plant Biol. 2022, 22, 421. [CrossRef]
- Zhang, M.; Ma, Y.; Wang, Y.; Gao, H.; Zhao, S.; Yu, Y.; Zhang, X.; Xi, H. MAPK and phenylpropanoid metabolism pathways involved in regulating the resistance of upland cotton plants to Verticillium dahliae. Front. Plant Sci. 2024, 15, 1451985. [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018, 34, i884-i890. [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008, 5, 621-8. [CrossRef]
- Zhao, J.; Wang, P.; Gao, W.; Long, Y.; Wang, Y.; Geng, S.; Su, X.; Jiao, Y.; Chen, Q.; Qu, Y. Genome-wide identification of the DUF668 gene family in cotton and expression profiling analysis of GhDUF668 in Gossypium hirsutum under adverse stress. BMC Genomics. 2021, 22, 395. [CrossRef]
- Sun, Q.; Wang, G.; Zhang, X.; Zhang, X.; Qiao, P.; Long, L.; Yuan, Y.; Cai, Y. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci. Rep. 2017, 7, 42418. [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Bio.l Evol. 2016, 33, 1870-4. [CrossRef]
- Lescot, M.; Déhais, P.; Thijs. G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325-7. [CrossRef]
- Song, Y.; Liu, L.; Wang, Y.; Valkenburg, D.J.; Zhang, X.; Zhu, L.; Thomma, B.P.H.J. Transfer of tomato immune receptor Ve1 confers Ave1-dependent Verticillium resistance in tobacco and cotton. Plant Biotechnol. J. 2018, 16, 638–48. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).