Submitted:
07 April 2023
Posted:
07 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant husbandry
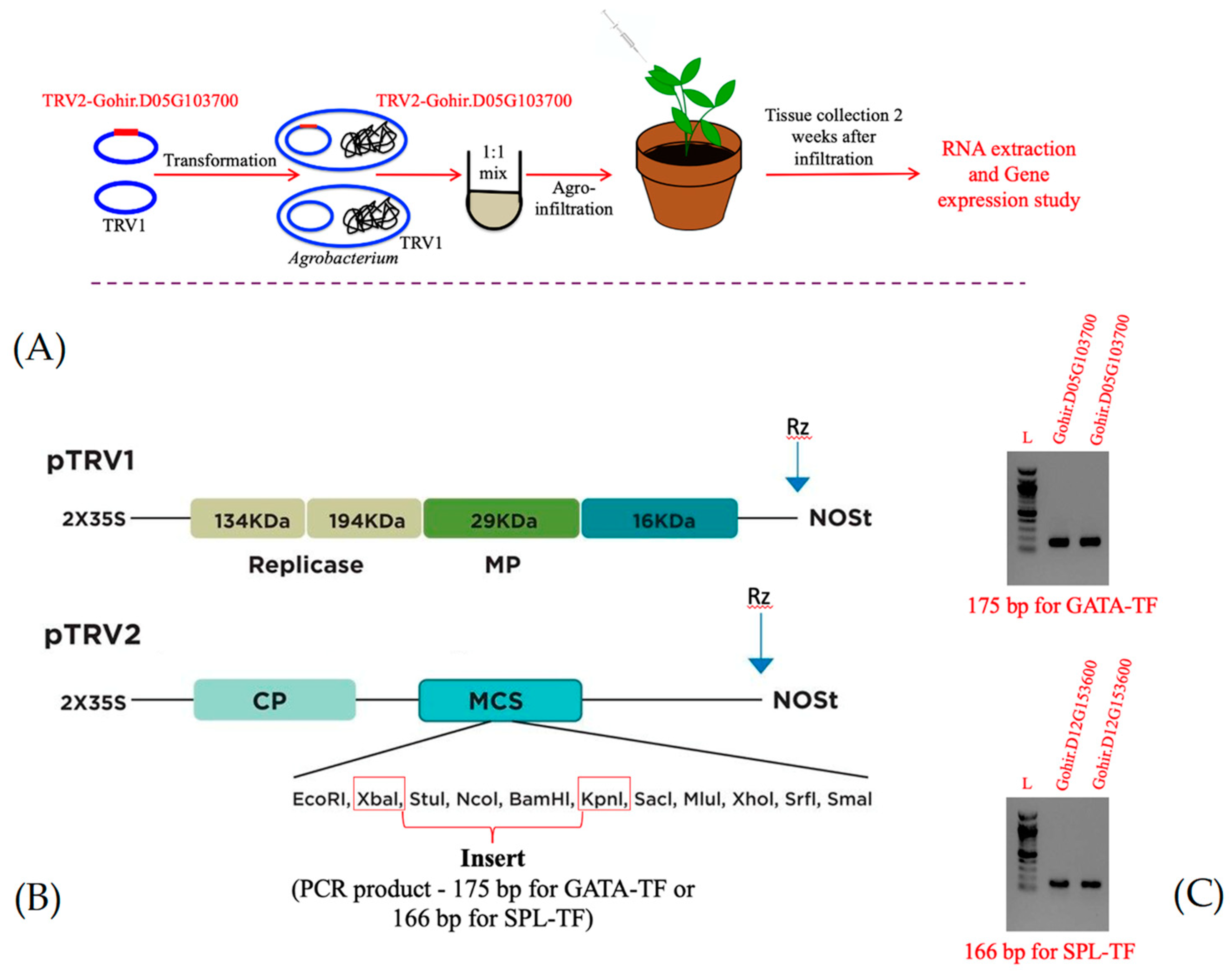
2.2. Construction of the gene silencing constructs
2.3. Agrobacterium tumefaciens transfection with TRV1 and TRV2 constructs and plant inoculation
2.4. RNA extraction and cDNA conversion
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Phenotypic data analysis
3. Results
3.1. Sequence analysis for the antisense construct development
3.2. Agrobacterium mediated-VIGS of Gohir.D05G103700 and Gohir.D12G153600 genes in the upland Cotton Cultivar ‘Coker-201’
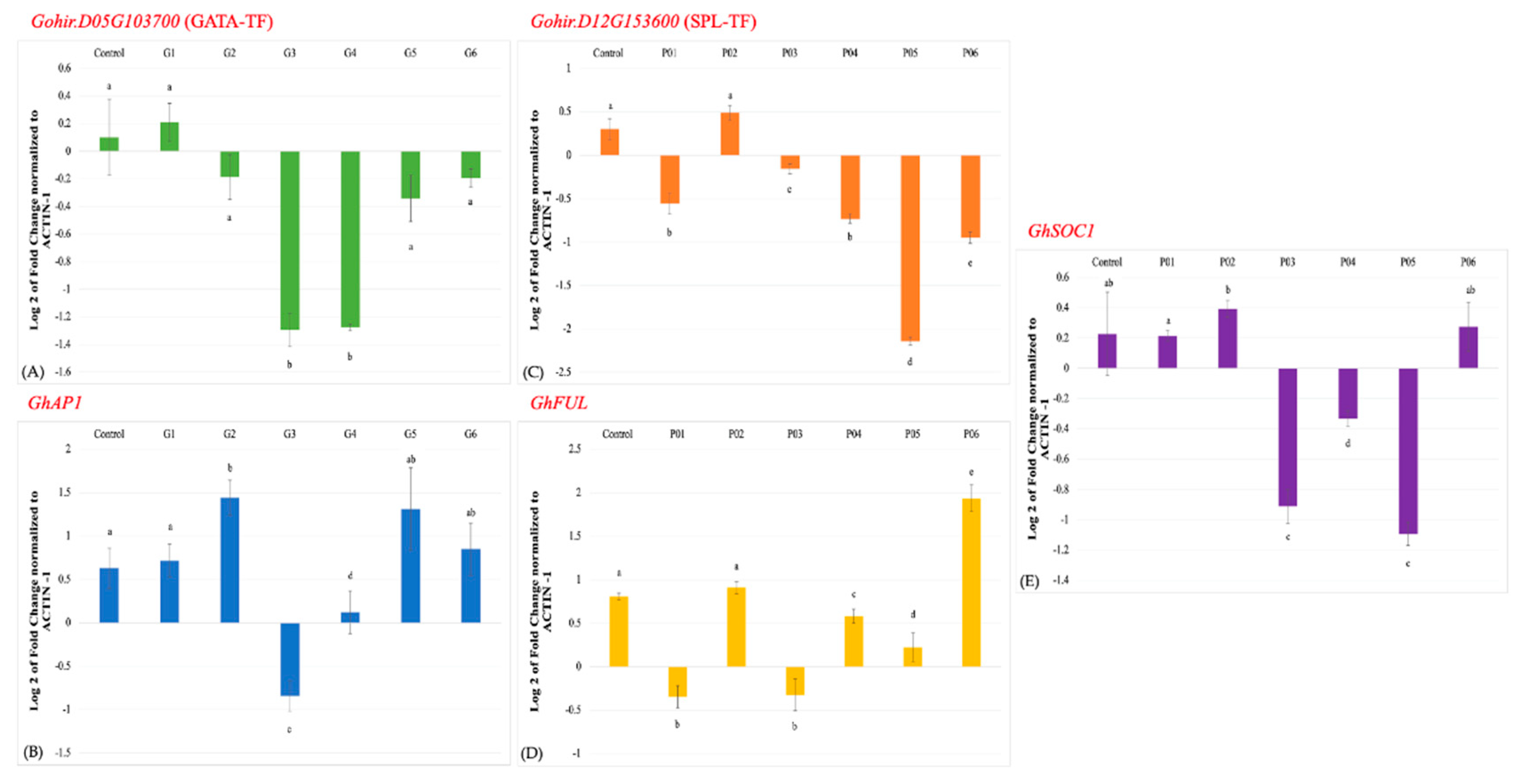
3.3. Expression profiling of the cotton AP1 genes in TRV2-GATA-TF inoculated plants and the FUL and SOC1 genes in TRV2-SPL-TF inoculated plants
3.4. Phenotypic observations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paterson, A. H.; Wendel, J. F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K. C.; Shu, S.; Udall, J.; Yoo, M.; Byers, R.; Chen, W.; Doron-Faigenboim, A.; Duke, M. V.; Gong, L.; Grimwood, J.; Grover, C.; Grupp, K.; Hu, G. Repeated Polyploidization of Gossypium Genomes and the Evolution of Spinnable Cotton Fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Page, J. T.; Udall, J. A.; Sanders, W. S.; Peterson, D. G.; Arick, M. A.; Grover, C. E.; Wendel, J. F. Independent Domestication of Two Old World Cotton Species. Genome Biology and Evolution 2016, 8, 1940–1947. [Google Scholar] [CrossRef]
- Zhang, H.-B.; Li, Y.; Wang, B.; Chee, P. W. Recent Advances in Cotton Genomics. International Journal of Plant Genomics 2008, 2008, 1–20. [Google Scholar] [CrossRef]
- Ahmad, S. (2014). Cotton Production and Uses Agronomy. Crop Protection and Postharvest Technologies. Springer Nature Singapore 2014. [Google Scholar] [CrossRef]
- Sussex, I. M.; Kerk, N. M. The Evolution of Plant Architecture. Current Opinion in Plant Biology 2001, 4, 33–37. [Google Scholar] [CrossRef]
- Borello, U.; Ceccarelli, E.; Giuliano, G. Constitutive, Light-Responsive and Circadian Clock-Responsive Factors Compete for the Different I Box Elements in Plant Light-Regulated Promoters. The Plant Journal 1993, 4, 611–619. [Google Scholar] [CrossRef]
- Terzaghi, W. B.; Cashmore, A. R. Light-Regulated Transcription. Annual Review of Plant Physiology and Plant Molecular Biology 1995, 46, 445–474. [Google Scholar] [CrossRef]
- Luo, X.-M.; Lin, W.-H.; Zhu, S.; Zhu, J.-Y.; Sun, Y.; Fan, X.-Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M.; Liu, L.; Zhang, M.; Xie, Q.; Chong, K.; Wang, Z.-Y. Integration of Light- and Brassinosteroid-Signaling Pathways by a GATA Transcription Factor in Arabidopsis. Developmental Cell 2010, 19, 872–883. [Google Scholar] [CrossRef]
- Daniel-Vedele, F.; Caboche, M. A Tobacco CDNA Clone Encoding a GATA-1 Zinc Finger Protein Homologous to Regulators of Nitrogen Metabolism in Fungi. Molecular and General Genetics MGG 1993, 240, 365–373. [Google Scholar] [CrossRef]
- Reyes J., C.; Muro-Pastor, M. I.; Florencio, F. J. The GATA Family of Transcription Factors in Arabidopsis and Rice. Plant Physiology 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hou, Y.; Hao, Q.; Chen, H.; Chen, L.; Yuan, S.; Shan, Z.; Zhang, X.; Yang, Z.; Qiu, D.; Zhou, X.; Huang, W. Genome-Wide Survey of the Soybean GATA Transcription Factor Gene Family and Expression Analysis under Low Nitrogen Stress. PLOS ONE 2015, 10, e0125174. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, X.; Huang, Z.; Fan, S.; Qun, G.; Liu, A.; Gong, J.; Li, J.; Gong, W.; Shi, Y.; Fan, L.; Zhang, Z.; Liu, R.; Jiang, X.; Lei, K.; Shang, H.; Xu, A.; Yuan, Y. Genome-Wide Identification and Analysis of the Evolution and Expression Patterns of the GATA Transcription Factors in Three Species of Gossypium Genus. Gene 2019, 680, 72–83. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A New Family of DNA Binding Proteins Includes Putative Transcriptional Regulators of TheAntirrhinum Majus Floral Meristem Identity GeneSQUAMOSA. Molecular and General Genetics MGG 1996, 250, 7–16. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative Study of SBP-Box Gene Family in Arabidopsis and Rice. Gene 2007, 407, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H. The MiR156/SPL Module, a Regulatory Hub and Versatile Toolbox, Gears up Crops for Enhanced Agronomic Traits. Molecular Plant 2015, 8, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K. W.; Wu, G.; Yang, L.; Poethig, R. S. Developmental Functions of MiR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-like (SPL) Genes in Arabidopsis Thaliana. PLOS Genetics 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Silva, G. F. F. e; Silva, E. M.; da Silva Azevedo, M.; Guivin, M. A. C.; Ramiro, D. A.; Figueiredo, C. R.; Carrer, H.; Peres, L. E. P.; Nogueira, F. T. S. MicroRNA156-Targeted SPL/SBP Box Transcription Factors Regulate Tomato Ovary and Fruit Development. The Plant Journal 2014, 78, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xu, W.; Liu, X.; Mao, X.; Li, A.; Wang, J.; Chang, X.; Zhang, X.; Jing, R. Functional Conservation and Divergence among Homoeologs of TaSPL20 and TaSPL21, Two SBP-Box Genes Governing Yield-Related Traits in Hexaploid Wheat1[OPEN]. Plant Physiology 2017, 174, 1177–1191. [Google Scholar] [CrossRef]
- Liu, N.; Tu, L.; Wang, L.; Hu, H.; Xu, J.; Zhang, X. MicroRNA 157-Targeted SPL Genes Regulate Floral Organ Size and Ovule Production in Cotton. BMC Plant Biology 2017, 17. [Google Scholar] [CrossRef]
- Hayward, A.; Padmanabhan, M.; Dinesh-Kumar, S. P. Virus-Induced Gene Silencing in Nicotiana Benthamiana and Other Plant Species. Methods in Molecular Biology (Clifton, N.J.) 2011, 678, 55–63. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA Silencing in Plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T. M.; Anderson, J. C.; Martin, G. B.; Dinesh-Kumar, S. P. Applications and Advantages of Virus-Induced Gene Silencing for Gene Function Studies in Plants. The Plant Journal 2004, 39, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Dasgupta, I. Gene Silencing Approaches through Virus-Based Vectors: Speeding up Functional Genomics in Monocots. Plant Molecular Biology 2019, 100, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S. P. Tobacco Rar1, EDS1 and NPR1/NIM1 like Genes Are Required for N-Mediated Resistance to Tobacco Mosaic Virus. The Plant Journal 2002, 30, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Gedling, C. R.; Ali, E. M.; Gunadi, A.; Finer, J. J.; Xie, K.; Liu, Y.; Yoshikawa, N.; Qu, F.; Dorrance, A. E. Improved Apple Latent Spherical Virus-Induced Gene Silencing in Multiple Soybean Genotypes through Direct Inoculation of Agro-Infiltrated Nicotiana Benthamiana Extract. Plant Methods 2018, 14. [Google Scholar] [CrossRef]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A.; Yaegashi, H.; Yamagishi, N.; Takahashi, T.; Isogai, M.; Takahashi, H.; Yoshikawa, N. Apple Latent Spherical Virus Vectors for Reliable and Effective Virus-Induced Gene Silencing among a Broad Range of Plants Including Tobacco, Tomato, Arabidopsis Thaliana, Cucurbits, and Legumes. Virology 2009, 386, 407–416. [Google Scholar] [CrossRef]
- Lentz, E. M.; Kuon, J.-E.; Alder, A.; Mangel, N.; Zainuddin, I. M.; McCallum, E. J.; Anjanappa, R. B.; Gruissem, W.; Vanderschuren, H. Cassava Geminivirus Agroclones for Virus-Induced Gene Silencing in Cassava Leaves and Roots. Plant Methods 2018, 14. [Google Scholar] [CrossRef]
- Tzean, Y.; Lee, M.-C.; Jan, H.-H.; Chiu, Y.-S.; Tu, T.-C.; Hou, B.-H.; Chen, H.-M.; Chou, C.-N.; Yeh, H.-H. Cucumber Mosaic Virus-Induced Gene Silencing in Banana. Scientific Reports 2019, 9. [Google Scholar] [CrossRef]
- Rustgi, S.; Naveed, S.; Windham, J.; Zhang, H.; Demirer, G. S. Plant Biomacromolecule Delivery Methods in the 21st Century. Frontiers in Genome Editing 2022, 4. [Google Scholar] [CrossRef]
- MacFarlane, S. A. Molecular Biology of the Tobraviruses. Journal of General Virology 1999, 80, 2799–2807. [Google Scholar] [CrossRef]
- Ziegler-Graff, V.; Guilford, P. J.; Baulcombe, D. C. Tobacco Rattle Virus RNA-1 29K Gene Product Potentiates Viral Movement and Also Affects Symptom Induction in Tobacco. Virology 1991, 182, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Visser, P. B.; Bol, J. F. Nonstructural Proteins of Tobacco Rattle Virus Which Have a Role in Nematode-Transmission: Expression Pattern and Interaction with Viral Coat Protein. Journal of General Virology 1999, 80, 3273–3280. [Google Scholar] [CrossRef]
- Ratcliff, F.; Martin-Hernandez, A. M.; Baulcombe, D. C. Technical Advance: Tobacco Rattle Virus as a Vector for Analysis of Gene Function by Silencing. The Plant Journal 2001, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Senthil-Kumar, M.; Mysore, K. S. Tobacco Rattle Virus–Based Virus-Induced Gene Silencing in Nicotiana Benthamiana. Nature Protocols 2014, 9, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Saedler, R. Virus-Induced Gene Silencing of Jasmonate-Induced Direct Defences, Nicotine and Trypsin Proteinase-Inhibitors in Nicotiana Attenuata. Journal of Experimental Botany 2003, 55, 151–157. [Google Scholar] [CrossRef]
- Galis, I.; Schuman, M. C.; Gase, K.; Hettenhausen, C.; Hartl, M.; Dinh, S. T.; Wu, J.; Bonaventure, G.; Baldwin, I. T. The Use of VIGS Technology to Study Plant-Herbivore Interactions. Methods in Molecular Biology (Clifton, N.J.) 2013, 975, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Atamian, H.S. Virus induced gene silencing optimization in plants: things to be considered. Virus 2014, 2. [Google Scholar] [CrossRef]
- Jin, S.; Zhang, X.; Nie, Y.; Guo, X.; Liang, S.; Zhu, H. Identification of a Novel Elite Genotype for in Vitro Culture and Genetic Transformation of Cotton. Biologia plantarum 2006, 50, 519–524. [Google Scholar] [CrossRef]
- Ye, K.; Teng, T.; Yang, T.; Zhao, D.; Zhao, Y. Transcriptome Analysis Reveals the Effect of Grafting on Gossypol Biosynthesis and Gland Formation in Cotton. BMC Plant Biology 2023, 23. [Google Scholar] [CrossRef]
- Mauney, J. Chapter 2 VEGETATIVE GROWTH AND DEVELOPMENT OF FRUITING SITES. Available online: https://www.semanticscholar.org/paper/Chapter-2-VEGETATIVE-GROWTH-AND-DEVELOPMENT-OF-Mauney/7f0dcf43b3d71eaac0317c6690f8a9aedcb3989c.
- Ribeiro, T. P.; Lourenço-Tessutti, I. T.; de Melo, B. P.; Morgante, C. V.; Filho, A. S.; Lins, C. B. J.; Ferreira, G. F.; Mello, G. N.; Macedo, L. L. P.; Lucena, W. A.; Silva, M. C. M.; Oliveira-Neto, O. B.; Grossi-de-Sa, M. F. Improved Cotton Transformation Protocol Mediated by Agrobacterium and Biolistic Combined-Methods. Planta 2021, 254, 20. [Google Scholar] [CrossRef]
- Rajasekaran, K. Biolistic Transformation of Cotton Embryogenic Cell Suspension Cultures. Methods in Molecular Biology 2018, 55–66. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Zhang, C.; Zhang, J.; Chen, Y.; Liu, G.; Zhao, Y.; Hao, F.; Zhang, J. A Novel VIGS Method by Agroinoculation of Cotton Seeds and Application for Elucidating Functions of GhBI-1 in Salt-Stress Response. Plant Cell Reports 2018, 37, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Bennypaul, H. S.; Mutti, J. S.; Rustgi, S.; Kumar, N.; Okubara, P. A.; Gill, K. S. Virus-Induced Gene Silencing (VIGS) of Genes Expressed in Root, Leaf, and Meiotic Tissues of Wheat. Functional & Integrative Genomics 2012, 12, 143–156. [Google Scholar] [CrossRef]
- Chaudhary, B.; Flagel, L.; Stupar, R. M.; Udall, J. A.; Verma, N.; Springer, N. M.; Wendel, J. F. Reciprocal Silencing, Transcriptional Bias and Functional Divergence of Homeologs in Polyploid Cotton (Gossypium). Genetics 2009, 182, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Coate, J. E.; Doyle, J. J. Quantifying Whole Transcriptome Size, a Prerequisite for Understanding Transcriptome Evolution across Species: An Example from a Plant Allopolyploid. Genome Biology and Evolution 2010, 2, 534–546. [Google Scholar] [CrossRef]
- Gao, X.; Britt Jr., R.C.; Shan, L.; He, P. Agrobacterium-Mediated Virus-Induced Gene Silencing Assay in Cotton. Journal of Visualized Experiments 2011, 54. [Google Scholar] [CrossRef]
- Tuttle, J. R.; Idris, A. M.; Brown, J. K.; Haigler, C. H.; Robertson, D. Geminivirus-Mediated Gene Silencing from Cotton Leaf Crumple Virus Is Enhanced by Low Temperature in Cotton. Plant Physiology 2008, 148, 41–50. [Google Scholar] [CrossRef]
- Mustafa, R.; Shafiq, M.; Mansoor, S.; Briddon, R. W.; Scheffler, B. E.; Scheffler, J.; Amin, I. Virus-Induced Gene Silencing in Cultivated Cotton (Gossypium Spp.) Using Tobacco Rattle Virus. Molecular Biotechnology 2015, 58, 65–72. [Google Scholar] [CrossRef]
- Lloyd, A.; Blary, A.; Charif, D.; Charpentier, C.; Tran, J.; Balzergue, S.; Delannoy, E.; Rigaill, G.; Jenczewski, E. Homoeologous Exchanges Cause Extensive Dosage-Dependent Gene Expression Changes in an Allopolyploid Crop. New Phytologist 2017, 217, 367–377. [Google Scholar] [CrossRef]
- Glover, N. M.; Redestig, H.; Dessimoz, C. Homoeologs: What Are They and How Do We Infer Them? Trends in Plant Science 2016, 21, 609–621. [Google Scholar] [CrossRef]
- Zhao, Y.; Medrano, L.; Ohashi, K.; Fletcher, J. C.; Yu, H.; Sakai, H.; Meyerowitz, E. M. HANABA TARANU Is a GATA Transcription Factor That Regulates Shoot Apical Meristem and Flower Development in Arabidopsis[W]. The Plant Cell 2004, 16, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yin, H.; Qian, Q.; Yang, J.; Huang, C.; Hu, X.; Luo, D. NECK LEAF 1, a GATA Type Transcription Factor, Modulates Organogenesis by Regulating the Expression of Multiple Regulatory Genes during Reproductive Development in Rice. Cell Research 2009, 19, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Whipple, C. J.; Hall, D. H.; DeBlasio, S.; Taguchi-Shiobara, F.; Schmidt, R. J.; Jackson, D. P. A Conserved Mechanism of Bract Suppression in the Grass Family. The Plant Cell 2010, 22, 565–578. [Google Scholar] [CrossRef] [PubMed]


| Sr. No. | Molecular markers | Gene ID | Sub-genome | Gene Name | Position on genome (Mb) | Annotation |
|---|---|---|---|---|---|---|
| 1 | i02927Gh | Gohir.A01G208700 | A01 | AP1, FT, LFY | 117.19592 | Trihelix transcription factor PTL |
| 2 | i43992Gh | Gohir.A08G034500 | A08 | FT | 4.35425 | MYB3-like transcription factor |
| 3 | i13158Gh | Gohir.A13G050400 | A13 | FT, LFY | 7.06423 | GATA transcription factor 28-like |
| 4* | i09222Gh; i00443Gh | Gohir.D05G103700 | D05 | AP1 | 8.73032 | GATA transcription factor 11-like |
| 5* | i08185Gh | Gohir.D12G153600 | D12 | FUL, SOC1 | 48.44348 | SQUAMOSA promoter binding-like transcription factor |
| 6 | i13848Gh; i13851Gh | Gohir.D13G236200 | D13 | LFY | 64.67230 | Homeobox-leucine zipper protein REVOLUTA-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





