Submitted:
21 December 2024
Posted:
24 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
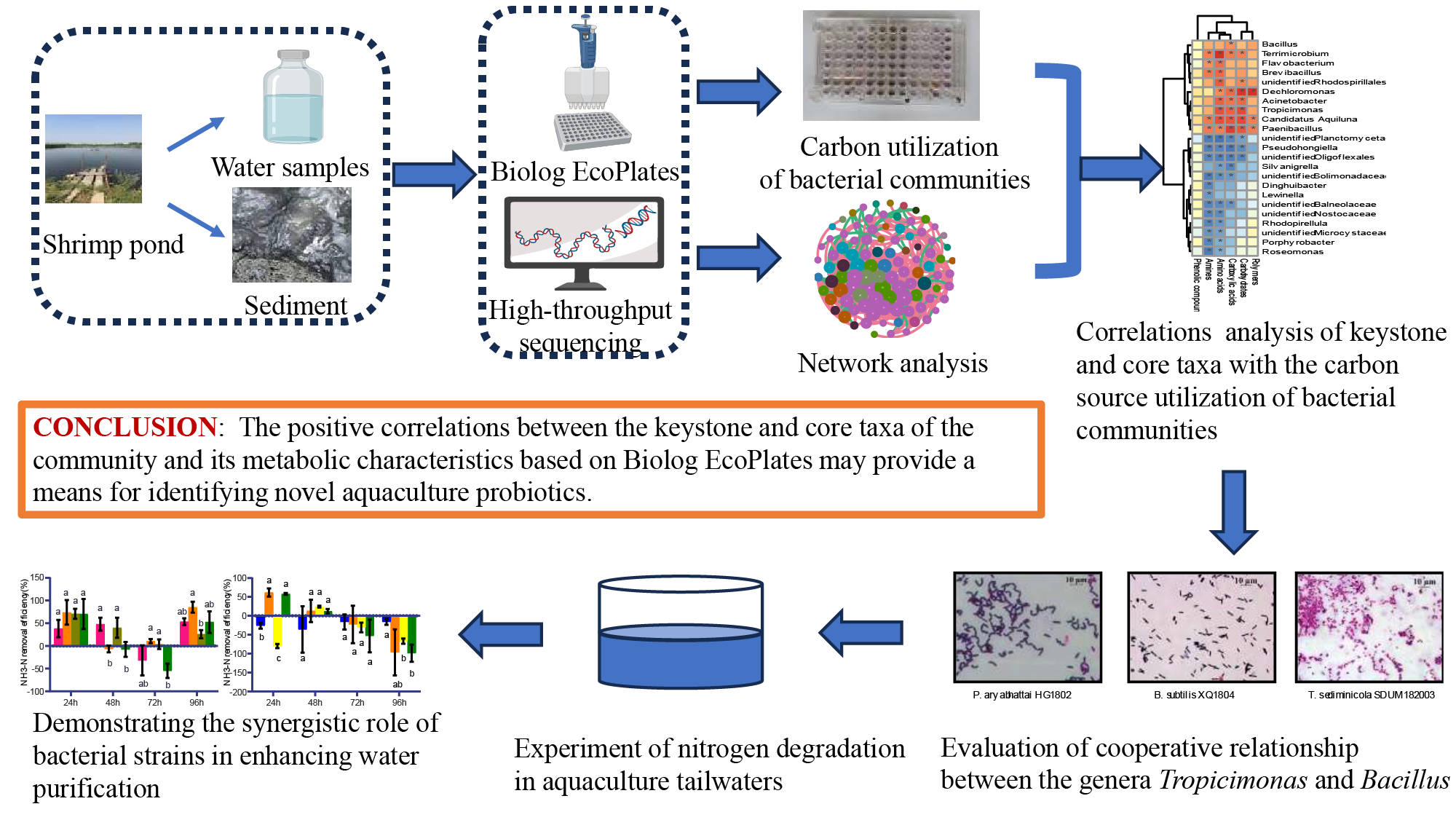
2. Materials and Methods
2.1. Water and Sediment Samples
2.2. CLPP Analysis Using Biolog EcoPlates
2.3. DNA Extraction, Bacterial 16S rRNA Gene Amplification, and Illumina Sequencing Analysis
2.4. Network Analysis and Identification of Potential Keystone Taxa and Core Taxa
2.5. Application of Tropicimonas in Nitrogen Degradation Experiments
2.6. Statistical Analysis
3. Results
3.1. Bacterial Community Network at the Genus Level
3.2. Correlation Analyses of the Relationships Between Keystone and Core Taxa and the Utilization of Grouped Carbon Sources in Bacterial Communities
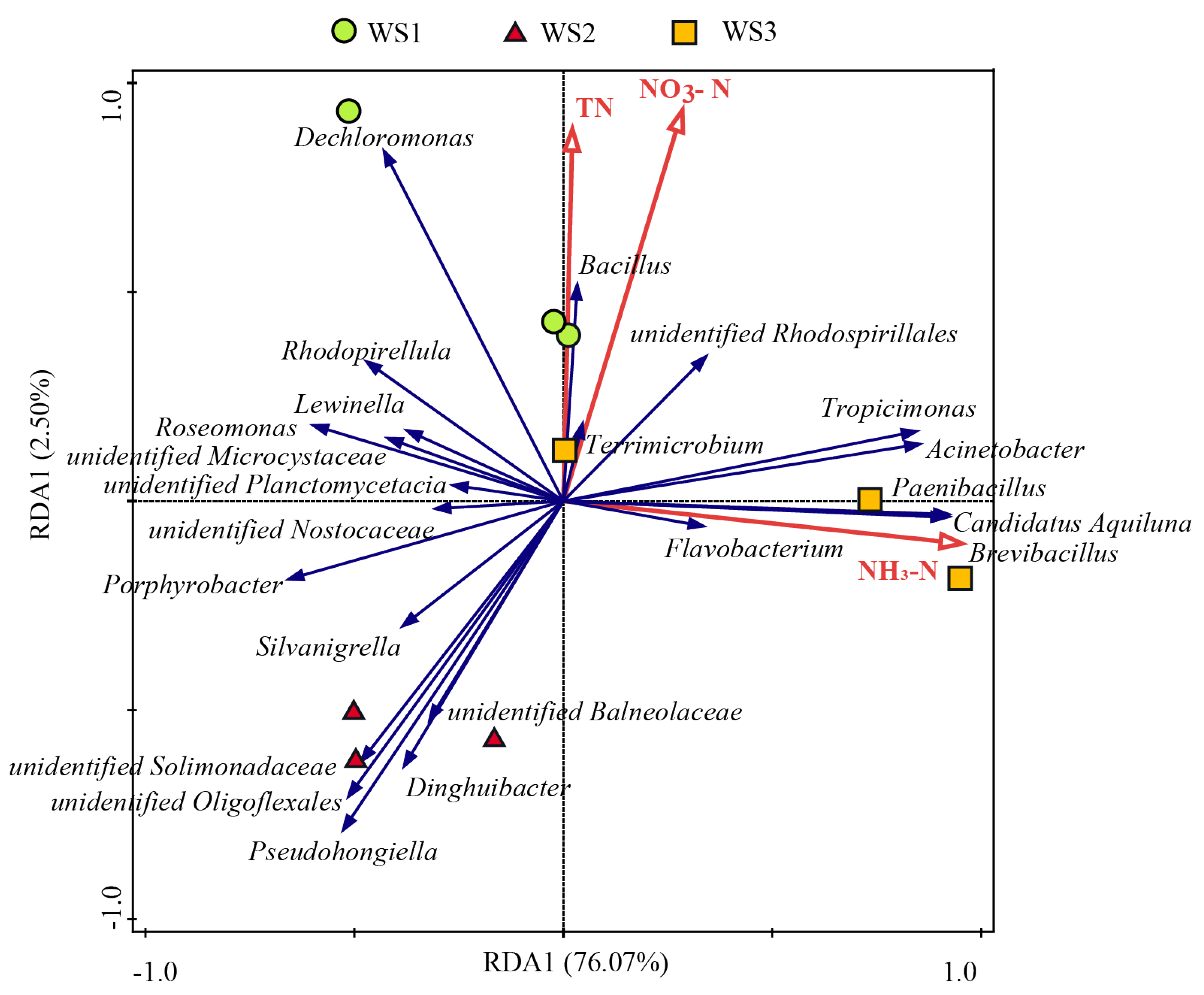
3.3. Correlations Between Keystone and Core Taxa of Bacterial Communities and Nitrogen Indices in the Water Samples
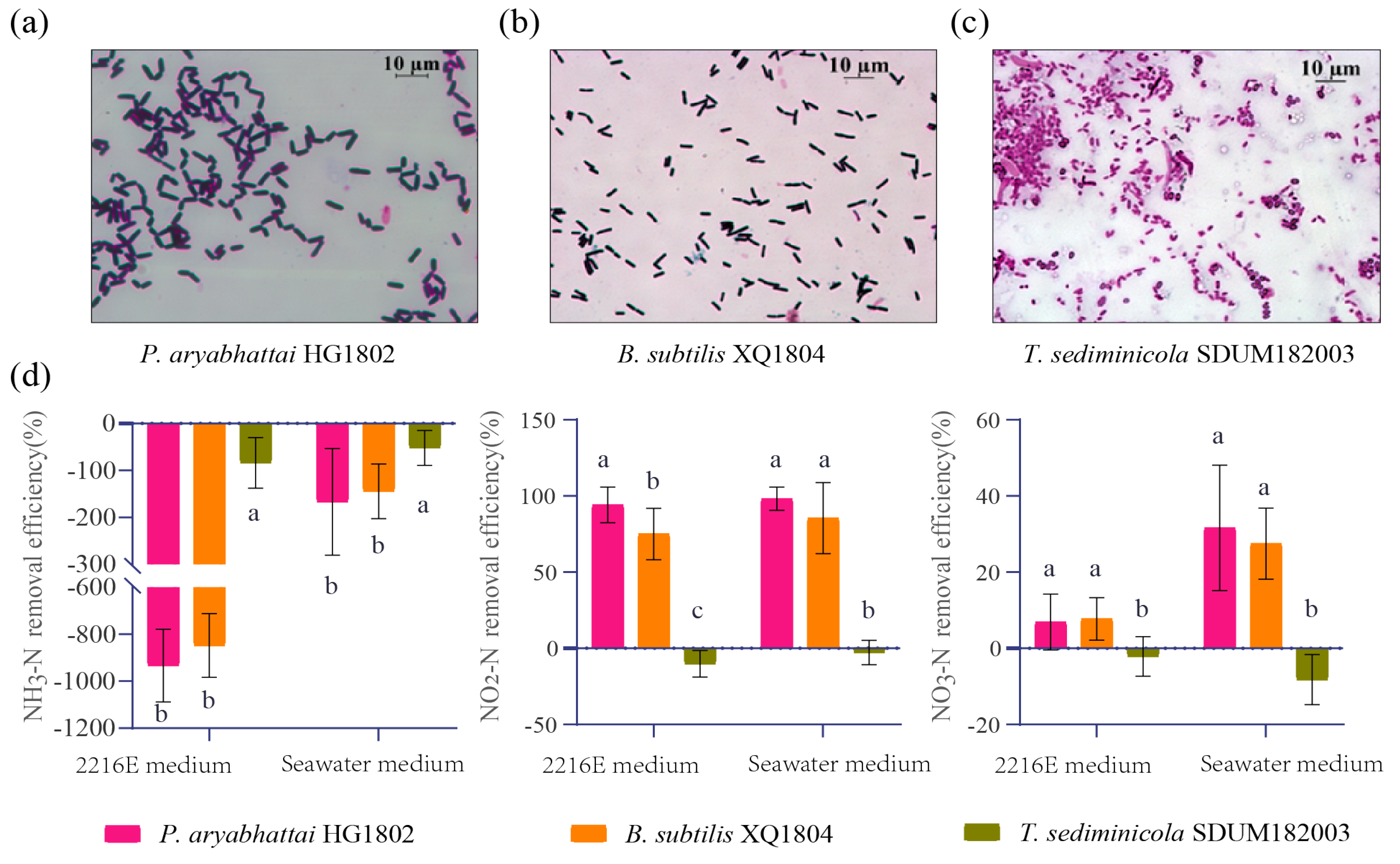
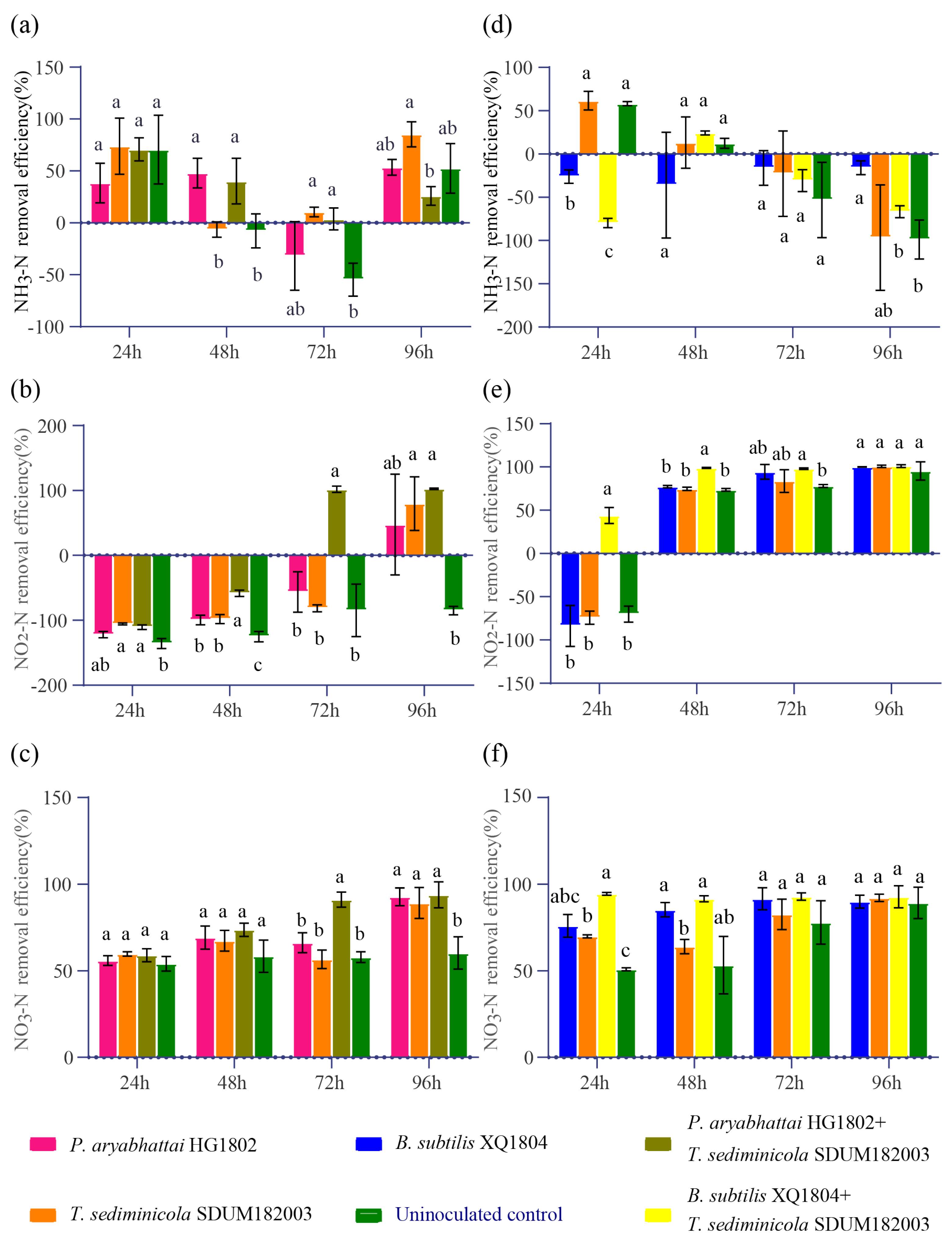
3.4. Application of T. sediminicola in Nitrogen Degradation Experiments
4. Discussion
4.1. Correlations Between Keystone and Core Taxa and Carbon Source Utilization Capacity at the Community Level
4.2. Relationship Between the Core Species in Shrimp Pond Water and the Major Aquatic Probiotics Currently in Use
4.3. Application of Tropicimonas to Aquaculture Tailwater
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Ethics approval
Consent to participate
Consent for publication
Availability of data and material
Code availability
References
- Abdelhamed, H.; Nho, S.W.; Karsi, A.; Lawrence, M.L. The role of denitrification genes in anaerobic growth and virulence of Flavobacterium columnare. J Appl Microbiol 2021, 130, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.; Kumala, R.R.C.; Mukti, A.T.; Lamid, M.; Nindarwi, D.D. Metagenomic profiles of core and signature bacteria in the guts of white shrimp, Litopenaeus vannamei, with different growth rates. Aquaculture 2022, 550, 737849. [Google Scholar] [CrossRef]
- Amoah K, Huang Q, Dong X, Tan B, Zhang S, Chi S, Yang Q, Liu H, Yang Y. Paenibacillus polymyxa improves the growth, immune and antioxidant activity, intestinal health, and disease resistance in Litopenaeus vannamei challenged with Vibrio parahaemolyticus. Aquaculture 2020, 518, 734563. [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. Bmc Bioinformatics 2003, 4, 2. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biology and Biochemistry 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Bastian, M. HSJM. (2009). Gephi: An Open Source Software for Exploring and Manipulating Networks. Paper presented at the International AAAI Conference on Weblogs and Social Media. [Google Scholar]
- Bazar, K.K.; Pemmineti, N.J.; Mohammad, S.A. Effect of soil probiotic on water quality and soil quality maintenance and growth of freshwater fish Pangasius hypophthalmus. Letters in Applied NanoBioScience 2022, 11, 3291–3304. [Google Scholar] [CrossRef]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 2014, 5, 219. [Google Scholar] [CrossRef]
- Buerger, P.; Vanstone, R.T.; Maire, J.; van Oppen, M. Long-term heat selection of the coral endosymbiont Cladocopium C1acro (Symbiodiniaceae) stabilizes associated bacterial communities. Int J Mol Sci 2022, 23, 4913. [Google Scholar] [CrossRef]
- Cao, X.; Zheng, H.; Liao, Y.; Feng, L.; Jiang, L.; Liu, C.; Mao, Y.; Shen, Q.; Zhang, Q.; Ji, F. Effects of iron-based substrate on coupling of nitrification, aerobic denitrification and Fe(II) autotrophic denitrification in tidal flow constructed wetlands. Bioresource Technol 2022, 361, 127657. [Google Scholar] [CrossRef]
- Choi, K.; Dobbs, F.C. Comparison of two kinds of Biolog microplates (GN and ECO) in their ability to distinguish among aquatic microbial communities. J Microbiol Meth 1999, 36, 203–213. [Google Scholar] [CrossRef]
- Classen, A.E.T.; Boyle, S.I.; Haskins, K.E.; Overby, S.T.; Hart, S.C. Community-level physiological profiles of bacteria and fungi: plate type and incubation temperature influences on contrasting soils. Fems Microbiol Ecol 2003, 44, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-Granados, F.; Gallardo-Becerra, L.; Leonardo-Reza, M.; Ochoa-Romo, J.P.; Ochoa-Leyva, A. A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. Peerj 2018, 6, e5382. [Google Scholar] [CrossRef] [PubMed]
- Deng Q, Luo X, Li S, Li J, Wang P, Yuan Y, Yang Z, Li W. Alsobacter ponti sp. nov., A novel denitrification and sulfate reduction bacterium isolated from Pearl River sediment. Anton Leeuw Int J G 2023, 116, 987–994. [CrossRef]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. Bmc Bioinformatics 2012, 13, 113. [Google Scholar] [CrossRef]
- Dong, P.; Guo, H.; Huang, L.; Zhang, D.; Wang, K. Glucose addition improves the culture performance of Pacific white shrimp by regulating the assembly of Rhodobacteraceae taxa in gut bacterial community. Aquaculture 2023, 567, 739254. [Google Scholar] [CrossRef]
- Elsadek, M.M.; Wang, S.; Wu, Z.; Wang, J.; Wang, X.; Zhang, Y.; Yu, M.; Guo, Z.; Wang, Q.; Wang, G.; Chen, Y.; Zhang, D. Characterization of Bacillus spp. isolated from the intestines of Rhynchocypris lagowskii as a potential probiotic and their effects on fish pathogens. Microb Pathogenesis 2023, 180, 106163. [Google Scholar] [CrossRef]
- Enke, T.N.; Datta, M.S.; Schwartzman, J.; Cermak, N.; Schmitz, D.; Barrere, J.; Pascual-García, A.; Cordero, O.X. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol 2019, 29, 1528–1535. [Google Scholar] [CrossRef]
- FAO (2024) The state of world fisheries and aquaculture 2024 – blue transformation in action, Rome. [CrossRef]
- Feng, K.; Zhang, Y.; He, Z.; Ning, D.; Deng, Y. Interdomain ecological networks between plants and microbes. Molucular ecology resources 2019, 19, 1565–1577. [Google Scholar] [CrossRef]
- Garland, J.L. Analysis and interpretation of community-level physiological profiles in microbial ecology. Fems Microbiol Ecol 1997, 24, 289–300. [Google Scholar] [CrossRef]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int J Syst Evol Micr 2020, 70, 5753. [Google Scholar] [CrossRef]
- Hou, D.; Huang, Z.; Zeng, S.; Liu, J.; Weng, S.; He, J. Comparative analysis of the bacterial community compositions of the shrimp intestine, surrounding water and sediment. J Appl Microbiol 2018, 125, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhou, R.; Zeng, S.; Wei, D.; Deng, X.; Xing, C.; Weng, S.; He, J.; Huang, Z. Stochastic processes shape the bacterial community assembly in shrimp cultural pond sediments. Appl Microbiol Biot 2021, 105, 5013–5022. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ye, J.; Jiang, K.; Wang, Y.; Li, Y. Oil contamination drives the transformation of soil microbial communities: Co-occurrence pattern, metabolic enzymes and culturable hydrocarbon-degrading bacteria. Ecotox Environ Safe 2021, 225, 112740. [Google Scholar] [CrossRef]
- Kasozi, N.; Kaiser, H.; Wilhelmi, B. Effect of Bacillus spp. on lettuce growth and root associated bacterial community in a small-scale aquaponics system. Agronomy 2021, 11, 947. [Google Scholar] [CrossRef]
- Kiersztyn, B.; Chróst, R.; Kaliński, T.; Siuda, W.; Bukowska, A.; Kowalczyk, G.; Grabowska, K. Structural and functional microbial diversity along a eutrophication gradient of interconnected lakes undergoing anthropopressure. Sci Rep 2019, 9, 11144. [Google Scholar] [CrossRef]
- Kolde R. (2019). Pretty Heatmaps (Version 1.0.12). https://CRAN.R-project.org/package=pheatmap.
- Kong Y, Shi G, Wu R, Zhang Y. k -core: Theories and applications. Physics Reports 2019, 832, 1–32. [CrossRef]
- Kviatkovski, I.; Minz, D. A member of the Rhodobacteraceae promotes initial biofilm formation via the secretion of extracellular factor(s). Aquat Microb Ecol 2015, 75, 155–167. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Fu, B.; Mu, X. Improvement of aquaculture water quality by mixed Bacillus and its effects on microbial community structure. Environ Sci Pollut R 2022, 29, 69731–69742. [Google Scholar] [CrossRef]
- Liu, X.; Li, H. Nitrogen removal performance and microorganism community of an A/O-MBBR system under extreme hydraulic retention time. Desalin Water Treat 2019, 158, 105–113. [Google Scholar] [CrossRef]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proceedings of the National Academy of Sciences 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Yu, X.; Gu, H.; Liu, F.; Fan, Y.; Wang, C.; He, Q.; Tian, Y.; Peng, Y.; Shu, L.; Wang, S.; Huang, Z.; Yan, Q.; He, J.; Liu, G.; Tu, Q.; He, Z. Vertically stratified methane, nitrogen and sulphur cycling and coupling mechanisms in mangrove sediment microbiomes. Microbiome 2023, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Revelle W. (2020). Psych: procedures for personality and psychological research. Retried from https://CRAN.R-project.org/p ackage=psych. Version=2.0.8.
- Ringø, E.; Li, X.; Doan, H.V.; Ghosh, K. Interesting probiotic bacteria other than the more widely used lactic acid bacteria and bacilli in finfish. Front Mar Sci 2022, 9, 848037. [Google Scholar] [CrossRef]
- Sangwan, N.; Zarraonaindia, I.; Hampton-Marcell, J.T.; Ssegane, H.; Eshoo, T.W.; Rijal, G.; Negri, M.C.; Gilbert, J.A. Differential functional constraints cause strain-level endemism in Polynucleobacter populations. Msystems 2016, 1, e3–e16. [Google Scholar] [CrossRef]
- Shin N, Roh SW, Kim M, Yun B, Whon TW, Kim Y, Bae J. Tropicimonas sediminicola sp. nov., isolated from marine sediment. Int J Syst Evol Micr 2012, 62, 2424–2429. [CrossRef]
- Team RC. (2022). R: A language and environment for statistical computing. Vienna. Retried from https:// www.R-project.org/.
- Wang, A.; Ran, C.; Wang, Y.; Zhang, Z.; Ding, Q.; Yang, Y.; Olsen, R.E.; Ringø, E.; Bindelle, J.; Zhou, Z. Use of probiotics in aquaculture of China—a review of the past decade. Fish Shellfish Immun 2019, 86, 734–755. [Google Scholar] [CrossRef]
- Wang, M.; Chen, X.; Liu, X.; Fang, Y.; Zheng, X.; Huang, T.; Tang, Y.; Ackermann, M.; Nie, Y.; Wu, X. Even allocation of benefits stabilizes microbial community engaged in metabolic division of labor. Cell Rep 2022, 40, 111410. [Google Scholar] [CrossRef]
- Wang M, Fan Z, Wang R, Liu Z, Gao F, Zhang Z, Yi M, Lu M (2022b) Nitrogen removal performance, and microbial community structure of water and its association with nitrogen metabolism of an ecological engineering pond aquaculture system. Aquacult Rep 25:101258. [CrossRef]
- Wang, Y.; Wang, K.; Huang, L.; Dong, P.; Wang, S.; Chen, H.; Lu, Z.; Hou, D.; Zhang, D. Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome 2020, 8, 106–116. [Google Scholar] [CrossRef]
- Wang, Z.; Cui, T.; Wang, Q. Optimization of degradation conditions and analysis of degradation mechanism for nitrite by Bacillus aryabhattai 47. Sci Total Environ 2024, 921, 171096. [Google Scholar] [CrossRef]
- Woźniak, M.; Gałązka, A.; Marzec-Grządziel, A.; Frąc, M. Microbial community, metabolic potential and seasonality of endosphere microbiota associated with leaves of the bioenergy tree Paulownia elongata × fortunei. International Journal of Molecular Sciences 2022, 23, 8978. [Google Scholar] [CrossRef]
- Wu, L.; Yang, P.; Zhang, L.; Luo, L.; Hong, Y.; Zhu, W.; Zheng, L.; Zhao, G.; Tong, C.; Peñuelas, J. Sediment sulfate content determines assembly processes and network stability of bacteria communities of coastal land-based shrimp aquaculture ponds. Aquaculture 2023, 563, 738953. [Google Scholar] [CrossRef]
- Xie, F.; Zhu, T.; Zhang, F.; Zhou, K.; Zhao, Y.; Li, Z. Using Bacillus amyloliquefaciens for remediation of aquaculture water. Springerplus 2013, 2, 119. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wang, Z.; Dai, X.; Liu, M.; Zhang, D.; Zeng, Y.; Zeng, D.; Ni, X.; Pan, K. Addition of Brevibacillus laterosporus to the rearing water enhances the water quality, growth performance, antioxidant capacity, and digestive enzyme activity of crucian carp Carassius auratus. Fisheries Sci 2023, 89, 659–670. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, C.; Zheng, Z.; Wei, Y.; Lu, K.; Zhu, J. Nutrient enrichment during shrimp cultivation alters bacterioplankton assemblies and destroys community stability. Ecotox Environ Safe 2018, 156, 366–374. [Google Scholar] [CrossRef]
- Yang, W.; Zhu, J.; Zheng, C.; Qiu, H.; Zheng, Z.; Lu, K. Succession of bacterioplankton community in intensive shrimp (Litopenaeus vannamei) aquaculture systems. Aquaculture 2018, 497, 200–213. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Hu, Z.; Chen, D.; Li, F.; Huang, X.; Li, C. Transcriptome analysis reveals the algicidal mechanism of Brevibacillus laterosporus against Microcystis aeruginosa through multiple metabolic pathways. Toxins 2022, 14, 492. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, X.; Yan, J.; Zou, W.; Wang, E.; Lu, X.; Chen, X. Keystone microbiomes revealed by 14 years of field restoration of the degraded agricultural soil under distinct vegetation scenarios. Front Microbiol 2020, 11, 1915. [Google Scholar] [CrossRef]
- Zhang M, Zhang Y, Yang F, Yao Q, Zhu H. Gemmobacter denitrificans sp. nov., a denitrifying bacterium, isolated from pond water for Litopenaeus vannamei. Int J Syst Evol Micr 2024, 74. [CrossRef]
- Zhao, Q.; Xie, F.; Zhang, F.; Zhou, K.; Sun, H.; Zhao, Y.; Yang, Q. Analysis of bacterial community functional diversity in late-stage shrimp (Litopenaeus vannamei) ponds using Biolog EcoPlates and PICRUSt2. Aquaculture 2022, 546, 737288. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, K.; Zhang, F.; Zhao, Y.; Sun, H.; Xie, F. Effect of aquaculture water eutrophication on color development in Biolog EcoPlates. Aquacult Int 2021, 29, 373–386. [Google Scholar] [CrossRef]
- Zhao, W.; Bi, X.; Peng, Y.; Bai, M. Research advances of the phosphorus-accumulating organisms of Candidatus Accumulibacter, Dechloromonas and Tetrasphaera: Metabolic mechanisms, applications and influencing factors. Chemosphere 2022, 307, 135675. [Google Scholar] [CrossRef]
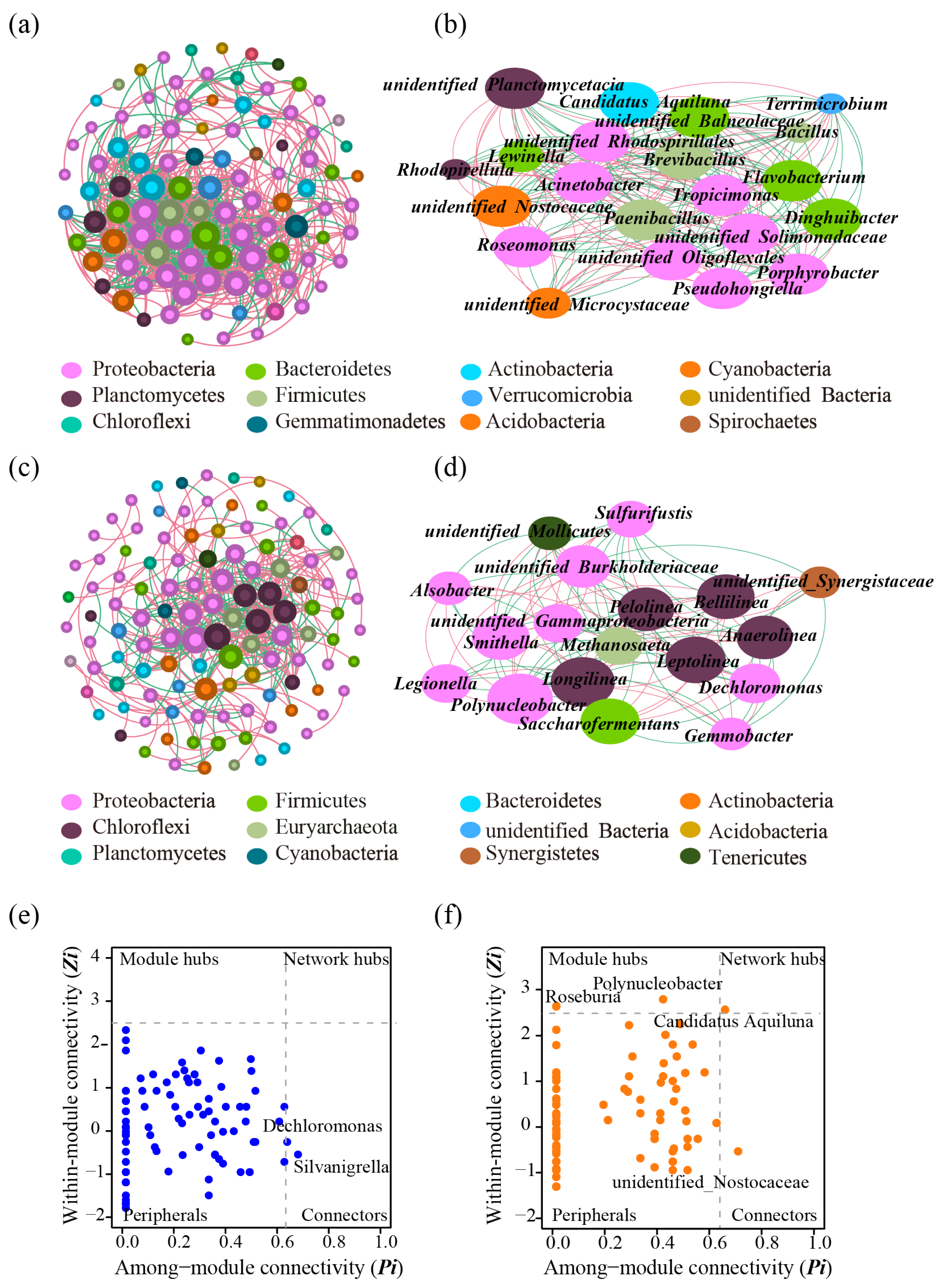

| Topological properties | Water samples | Sediment |
| Number of nodes | 99 | 115 |
| Number of edges | 782 | 398 |
| Network Density | 0.161 | 0.061 |
| Average Degree | 15.798 | 6.922 |
| Network Diameter | 6 | 9 |
| Positive/negative association (%) | 57.8/42.2 | 60.8/39.2 |
| Average Path length | 2.457 | 3.388 |
| Average Clustering Coefficient | 0.679 | 0.551 |
| Modularity | 0.318 | 0.440 |
| Genus | Interaction | correlation |
| Brevibacillus | positive | 0.9849 |
| Paenibacillus | positive | 0.9547 |
| unidentified_Rhodospirillales | positive | 0.9529 |
| Flavobacterium | positive | 0.8802 |
| Bacillus | positive | 0.8721 |
| Rhodopirellula | negative | -0.7601 |
| unidentified_Microcystaceae | negative | -0.7736 |
| unidentified_Planctomycetacia | negative | -0.7772 |
| unidentified_Oligoflexales | negative | -0.8079 |
| Pseudohongiella | negative | -0.8147 |
| Roseomonas | negative | -0.8355 |
| unidentified_Solimonadaceae | negative | -0.8476 |
| Dinghuibacter | negative | -0.8594 |
| Porphyrobacter | negative | -0.9075 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





