Submitted:
29 September 2024
Posted:
01 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
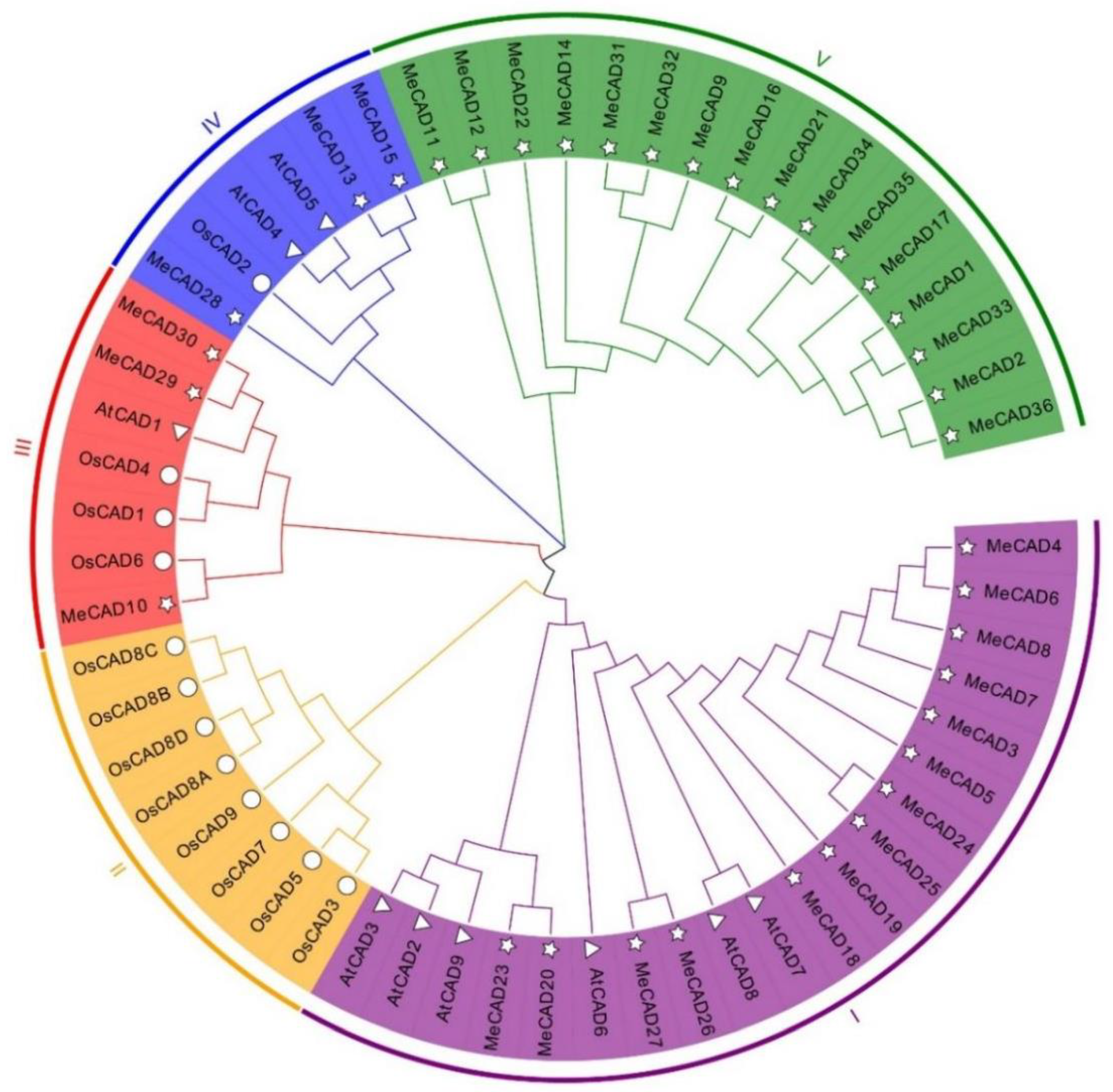
2.1. Identification and Phylogenetic Analysis of CAD in M. esculenta
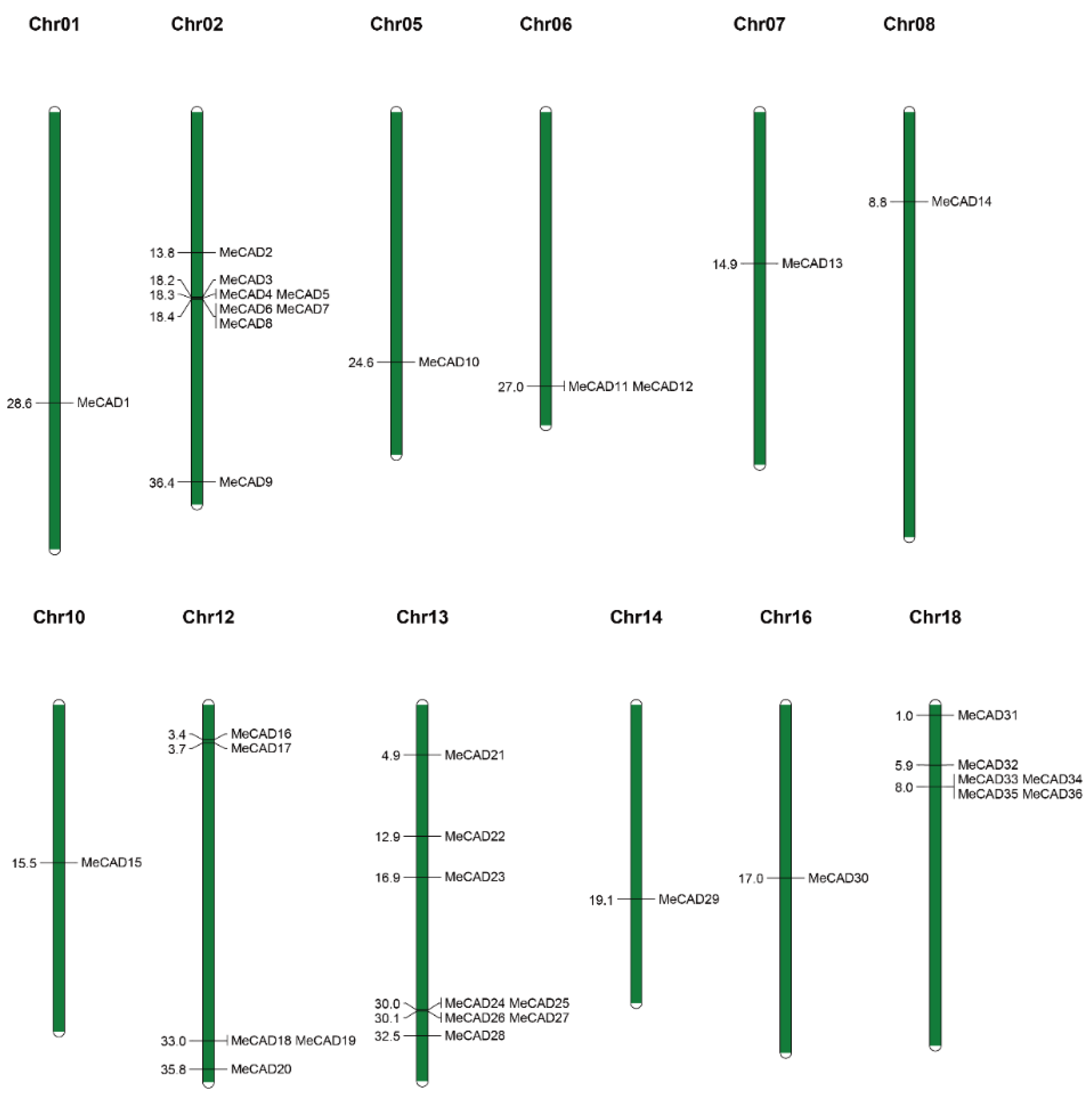
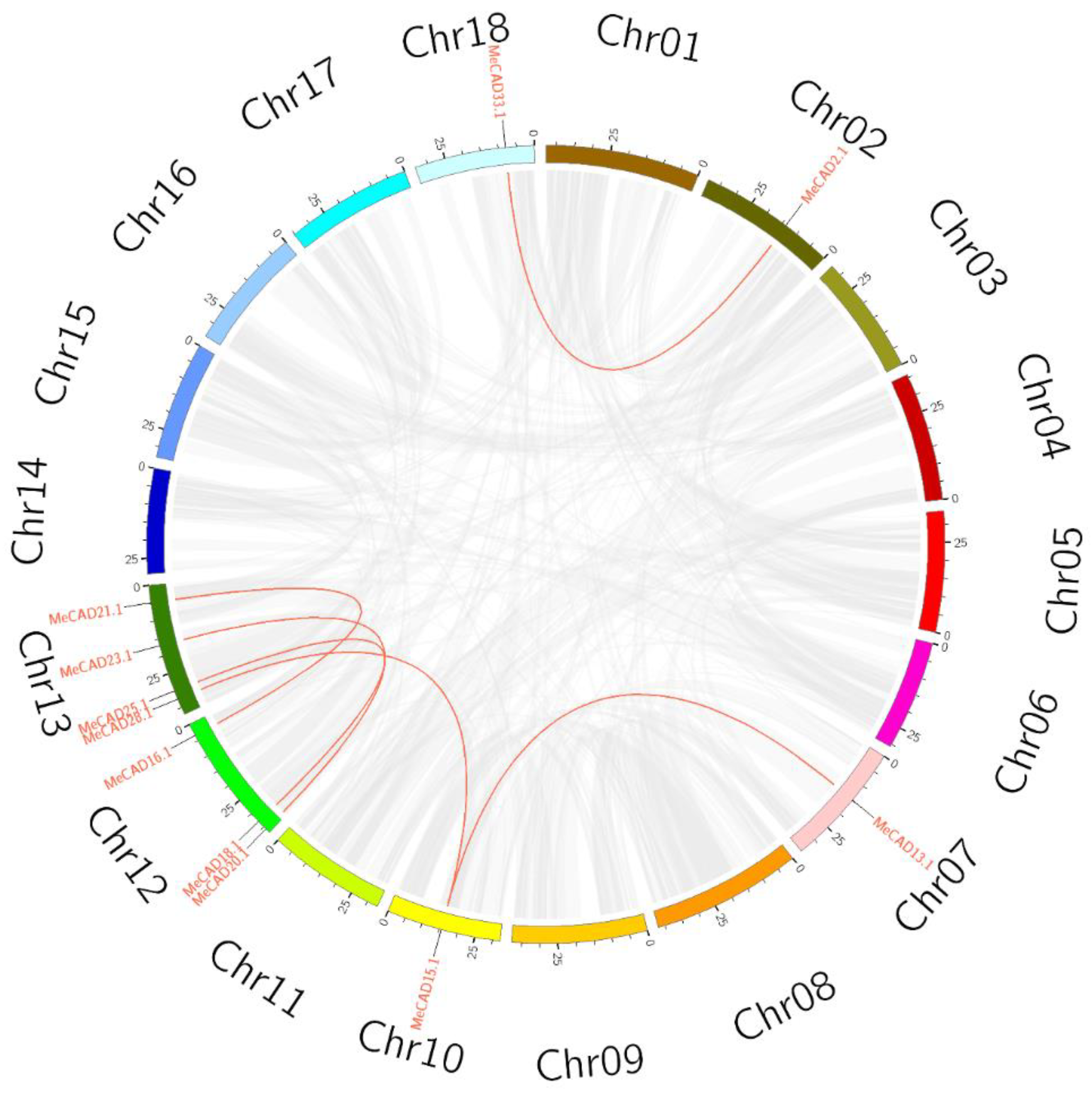
2.2. Chromosomal Locations, Duplications, and Synteny Analysis of the MeCAD Genes
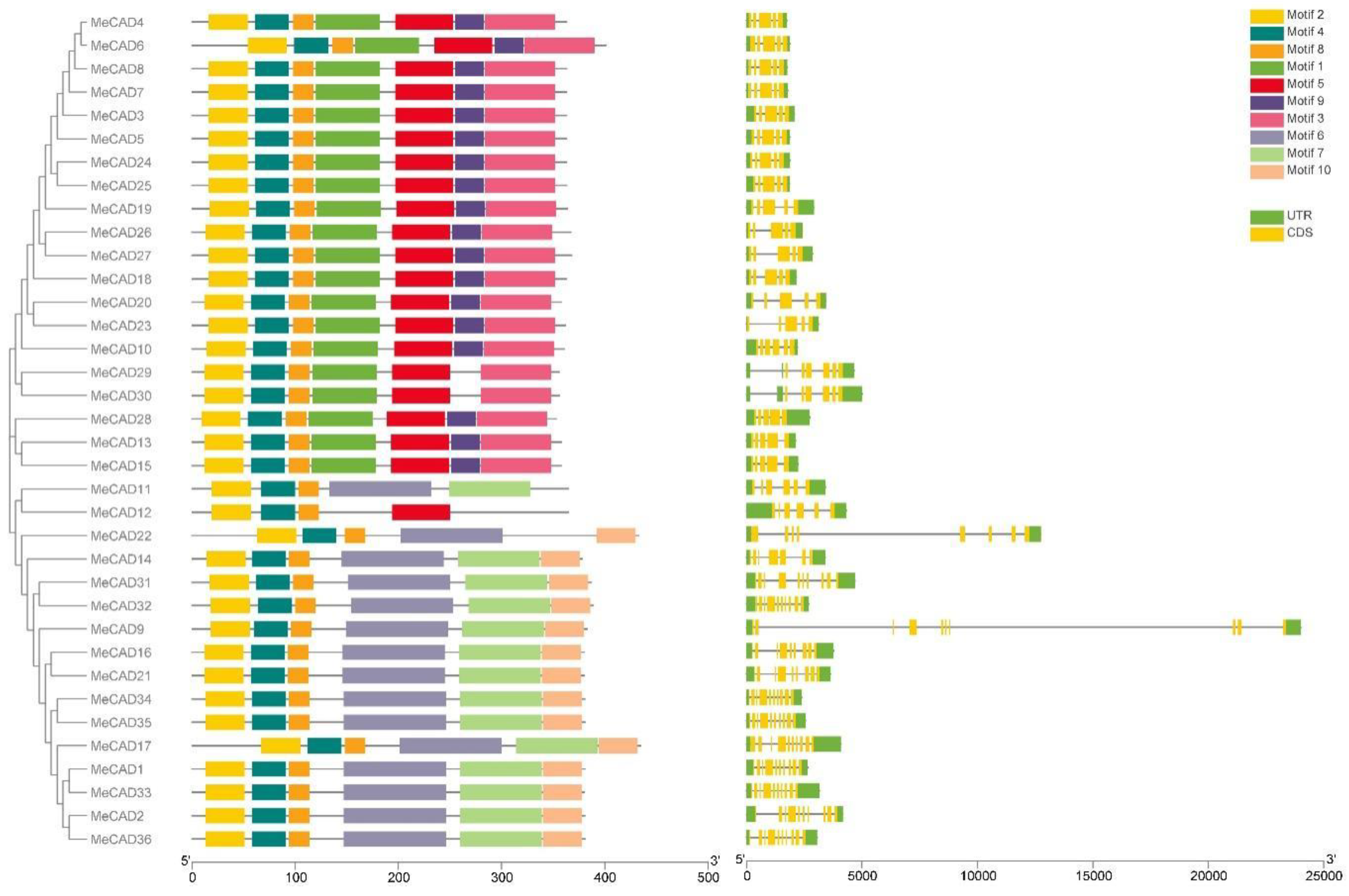
2.3. MeCAD Structure, Conserved Motifs and Cis-Acting Regulatory Element
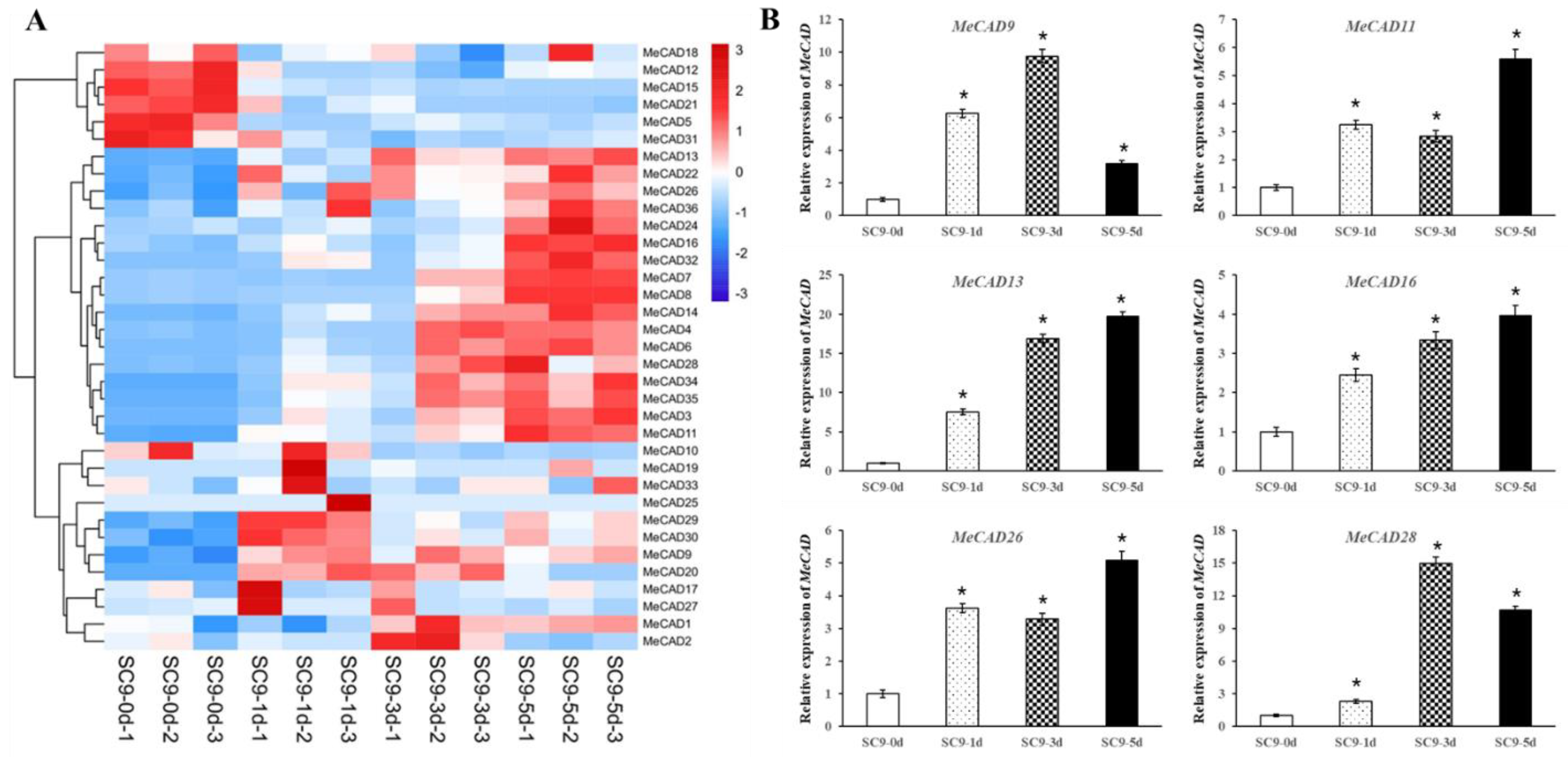
2.4. Expression Profile of MeCADs and Lignin Changes in Response to PPD
2.5. MeCAD13 and MeCAD28 Were Necessary for Lignin Biosynthesis and Wound Response
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of MeCAD Genes in Cassava Genome
4.3. Chromosomal Mapping, Gene Structure, Conserved Motif, and Cis-Acting Regulatory Element Analysis
4.4. Phylogenetic Analysis, Gene Duplication, Multiple Alignments, and Synteny Analysis
4.5. RNA-seq Analysis and qRT-PCR Verification
4.6. Gene Cloning, Expression and Subcellular Localization of MeCAD13 and MeCAD28
4.7. Virus-Induced Gene Silencing (VIGS) in Cassava and qRT-PCR Verification
4.8. Quantitative Analysis of Lignin
4.9. Stress of Yeast W303
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability
Conflicts of Interest
References
- Mohidin: S. R. N. S. P., Moshawih, S., Hermansyah, A., Asmuni, M. I., Shafqat, N., Ming, L. C. Cassava (Manihot esculenta Crantz): A systematic review for the pharmacological activities, traditional uses, nutritional values, and phytochemistry. J. Evid. Based Integr. Med. 2023, 28, 2515690X231206227.
- FAOSTAT. FAOSTAT statistical database, agriculture data. 2022. http://apps.fao.org.
- Lebot, V., Lawac, F., Muñoz-Cuervo, I., Mercier, P. E., Legendre, L. Metabolite fingerprinting of cassava (Manihot esculenta Crantz) landraces assessed for post-harvest physiological deterioration (PPD). Food Chem. 2023, 421, 136217.
- Li, R., Yuan, S., Zhou, Y., Wang, S., Zhou, Q., Ding, Z., Wang, Y., Yao, Y., Liu, J., Guo, J. Comparative transcriptome profiling of cassava tuberous roots in response to postharvest physiological deterioration. Int. J. Mol. Sci. 2023, 24, 246.
- Zainuddin, I. M., Fathoni, A., Sudarmonowati, E., Beeching, J. R., Gruissem, W., Vanderschuren, H. Cassava post–harvest physiological deterioration: From triggers to symptoms. Postharvest Biol. Technol. 2018, 142, 115–123.
- An, F., Xue, J., Luo, X., Chen, T., Wei, Z., Zhu, W., Ou, W., Li, K., Cai, J., Chen, S. MePOD12 participates the regulation to postharvest physiological deterioration by ROS scavenging and lignin accumulation in cassava tuberous roots. Postharvest Biol. Technol. 2024, 207, 112609.
- Wahengbam, E. D., Devi, C. P., Sharma, S. K., Roy, S. S., Maibam, A., Dasgupta, M., Luikham, S., Chongtham, T., Ningombam, A., Bhupenchandra, I., Singh, L. K., Devi, Y. P., Thokchom, S., Khaba, C. I., Singh, N. B., Rajashekar, Y., Das, S., Mohanty, S., Sahoo, M. R. Reactive oxygen species turnover, phenolics metabolism, and some key gene expressions modulate postharvest physiological deterioration in cassava tubers. Front. Microbiol. 2023, 14, 1148464.
- Djabou, A. S. M., Carvalho, L. J. C. B., Li, Q., Niemenak, N., Chen, S. Cassava postharvest physiological deterioration: a complex phenomenon involving calcium signaling, reactive oxygen species and programmed cell death. Acta. Physiol. Plant. 2017, 39, 91.
- Zidenga, T., Leyva-Guerrero, E., Moon, H., Siritunga, D., Sayre, R. Extending cassava root shelf life via reduction of reactive oxygen species production. Plant Physiol. 2012, 159, 1396–1407.
- Xu, J., Duan, X., Yang, J., Beeching, J. R., Zhang, P. Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 2013, 161, 1517–1528.
- Oirschot, Q. E. A. v., Rees, D., Aked, J., Kihurani, A. Sweetpotato cultivars diffier in efficiency of wound healing. Postharvest Biol. Technol. 2006, 42, 65–74.
- Wang, B., Jiang, H., Bi, Y., He, X., Wang, Y., Li, Y., Zheng, X., Prusky, D. Preharvest multiple sprays with sodium nitroprusside promote wound healing of harvested muskmelons by activation of phenylpropanoid metabolism. Postharvest Biol. Technol. 2019, 158, 110988.
- Zhao, Y., Deng, L., Zhou, Y., Ming, J., Yao, S., Zeng, K. Wound healing in citrus fruit is promoted by chitosan and Pichia membranaefaciens as a resistance mechanism against Colletotrichum gloeosporioides. Postharvest Biol. Technol. 2018, 145, 134–143.
- Reilly, K., G´omez-V´asquez, R., Buschmann, H., Tohme, J., Beeching, J. R. Oxidative stress responses during cassava post-harvest physiological deterioration. Plant Mol. Biol. 2003, 53, 669–685. [CrossRef]
- Weng, J. and Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 2010, 187, 273–285.
- Ge, X., Zhu, Y., Li, Z., Bi, Y., Yang, J., Zhang, J., Prusky, D. Preharvest multiple fungicide stroby sprays promote wound healing of harvested potato tubers by activating phenylpropanoid metabolism. Postharvest Biol. Technol. 2021, 171, 111328.
- Kim, Y. H., Bae, J. M., Huh, G. H. Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweetpotato in response to plant developmental stage and environmental stress. Plant Cell Rep 2010, 29, 779–791.
- Goujon, T., Sibout, R., Eudes, A., MacKay, J., Jouanin, L. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol. Biochem. 2003, 41, 677–687.
- Zhu, Y., Wang, Y., Jiang, H., Liu, W., Zhang, S., Hou, X., Zhang, S., Wang, N., Zhang, R., Chen, X. Transcriptome analysis reveals that PbMYB61 and PbMYB308 are involved in the regulation of lignin biosynthesis in pear fruit stone cells. Plant J. 2023, 116, 217–233.
- Dong, N. Q. and Lin, H. X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209.
- Yan, Y., Wang, P., Lu, Y., Bai, Y., Wei, Y., Liu, G., Shi, H. MeRAV5 promotes drought stress resistance in cassava by modulating hydrogen peroxide and lignin accumulation. Plant J. 2021, 107, 847–860.
- Liu, Q., Luo, L., Zheng, L. Lignins: biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335.
- Kim, S., Kim, M., Bedgar, D. L., Moinuddin, S. G. A., Cardenas, C. L. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. PNAS 2004, 101, 1455–1460.
- Liu, X., Acker, R. V., Voorend, W., Pallidis, A., Goeminne, G., Pollier, J., Morreel, K., Kim, H., Muylle, H., Bosio, M., Ralph, J., Vanholme, R., Boerjan, W. Rewired phenolic metabolism and improved saccharification efficiency of a Zea mays cinnamyl alcohol dehydrogenase 2 (zmcad2) mutant. Plant J. 2021, 105, 1240–1257.
- Ibrahim, W., Zhu, Y. M., Chen, Y., Qiu, C. W., Zhu, S., Wu, F. Genotypic differences in leaf secondary metabolism, plant hormones and yield under alone and combined stress of drought and salinity in cotton genotypes. Physiol. Plantarum 2019, 165, 343–355.
- Jiao, F., Luo, R., Dai, X., Liu, H., Yu, G., Han, S., Lu, X., Su, C., Chen, Q., Song, Q., Liu, W., Jiang, Y., Jin, Y., Wang, C., Yang, J., Qi, H. Drought-induced ABA, H2O2 and JA positively regulate CmCAD genes and lignin synthesis in melon stems. BMC Plant Biol. 2021, 21, 83.
- Tobias, C. M. and Chow, E. K. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 2005, 220, 678–688.
- Barakat, A., Bagniewska-Zadworna, A., Choi, A., Plakkat, U., DiLoreto, D., Yellanki, P., Carlson, J. E. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol. 2009, 9, 26.
- Chao, N., Huang, S., Kang, X., Yidilisi, K., Dai, M., Liu, L. Systematic functional characterization of cinnamyl alcohol dehydrogenase family members revealed their functional divergence in lignin biosynthesis and stress responses in mulberry. Plant Physiol. Bioch. 2022, 186, 145–156. [CrossRef]
- Qi, K., Song, X., Yuan, Y., Bao, J., Gong, X., Huang, X., Khanizadeh, S., Zhang, S., Tao, S. CAD genes: genome-wide identification, evolution, and their contribution to lignin biosynthesis in Pear (Pyrus bretschneideri). Plants 2021, 10, 1444.
- Sibout, R., Eudes, A., Mouille, G., Pollet, B., Lapierre, C., Jouanin, L., Séguin, A. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 2005, 17, 2059–2076.
- Jin, Y., Zhang, C., Liu, W., Qi, H., Chen, H., Cao, S. The cinnamyl alcohol dehydrogenase gene family in Melon (Cucumis melo L.): bioinformatic analysis and expression patterns. PLOS ONE 2014, 9, e101730.
- Cheng, H., Li, L., Xu, F., Cheng, S., Cao, F., Wang, Y., Yuan, H., Jiang, D., Wu, C. Expression patterns of a cinnamyl alcohol dehydrogenase gene involved in lignin biosynthesis and environmental stress in Ginkgo biloba. Mol. Biol. Rep. 2013, 40, 707–721.
- Ma, Q. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 2010, 10, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Saballos, A., Ejeta, G., Sanchez, E., Kang, C., Vermerris, W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the Brown midrib6 gene. Genetics 2009, 181, 783–795.
- Deng, W., Zhang, M., Wu, J., Jiang, Z., Tang, L., Li, Y., Wei, C., Jiang, C., Wan, X. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant, Camellia sinensis. J. Plant Physiol. 2013, 170, 272–282.
- Zhang, K., Qian, Q., Huang, Z., Wang, Y., Li, M., Hong. L., Zeng, D., Gu, M., Chu, C., Cheng, Z. Gold Hull and internode2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 2006, 140, 972–983.
- Bukh, C., Nord-Larsen, P. H., Rasmussen, S. K. Phylogeny and structure of the cinnamyl alcohol dehydrogenase gene family in Brachypodium distachyon. J. Exp. Bot. 2012, 63, 6223–6236.
- Tronchet, M., Balague, C., Kroj, T., Jouanin, L., Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92.
- An, F., Xiao, X., Chen, T., Xue, J., Luo, X., Ou, W., Li, K., Cai, J., Chen, S. Systematic analysis of bHLH transcription factors in cassava uncovers their roles in postharvest physiological deterioration and cyanogenic glycosides biosynthesis. Front. Plant Sci. 2022, 13, 901128.
- Bano, N., Patel, P., Chakrabarty, D., Bag, S. K. Genome-wide identification, phylogeny, and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum). Physiol. Mol. Biol. Plants 2021, 27, 1747–1764.
- Moura, J. C. M. S., Bonine, C. A. V., Viana, J. O. F., Dornelas, M. C., Mazzafera, P. Abiotic and biotic stresses and changes in lignin content and composition in plants. J. Integ. Plant Biol. 2010, 52, 360–376. [CrossRef]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M. A., Zhao, Q., Gallego-Giraldo, L., Dai, X., Zhao, P. X., Dixon, R. A. Genome-wide analysis of phenylpropanoid defense pathways. Mol. Plant Pathol. 2010, 11, 829–846.
- Wang, C., Wu, J., Tang, Y., Min, Y., Wang, D., Ma, X., Li, H., Li, J., Chen, Y., Chen, S., Liu, Z. Understanding the changes of phenylpropanoid metabolism and lignin accumulation in wounded cassava root during postharvest storage. Sci. Hortic. 2023, 310, 111765.
- Li Z, Xue, S., Xu, X., Wang, B., Zheng, X., Li, B., Xie, P., Bi, Y., Prusky, D. Preharvest multiple sprays with chitosan accelerate the deposition of suberin polyphenolic at wound sites of harvested muskmelons. Postharvest Biol. Tec. 2021, 179, 111565.
- Rahmawati, R., Khumaida, N., Ardie, S., Sukma, D., Sudarsono. Effects of harvest period, storage, and genotype on postharvest physiological deterioration response in cassava. Biodiversitas 2022, 23, 100–109.
- Yan, P., Zeng, Y., Shen, W., Tuo, D., Li, X., Zhou, P. Nimble cloning: A simple, versatile, and efficient system for standardized molecular cloning. Front. Bioeng. Biotech. 2020, 7, 460.
- Tuo, D., Zhou, P., Yan, P., Cui, H., Liu, Y., Wang, H., Yang, X., Liao, W., Sun, D., Li, X., Shen, W. A cassava common mosaic virus vector for virus-induced gene silencing in cassava. Plant Methods 2021, 17, 74. [CrossRef]
- Foster, C., Martin, T., Pauly, M. Comprehensive compositional analysis of plant cell wall (Lignocellulosic biomass) Part II: carbohydrates. J. Vis. Exp. 2010, 37, e1837.
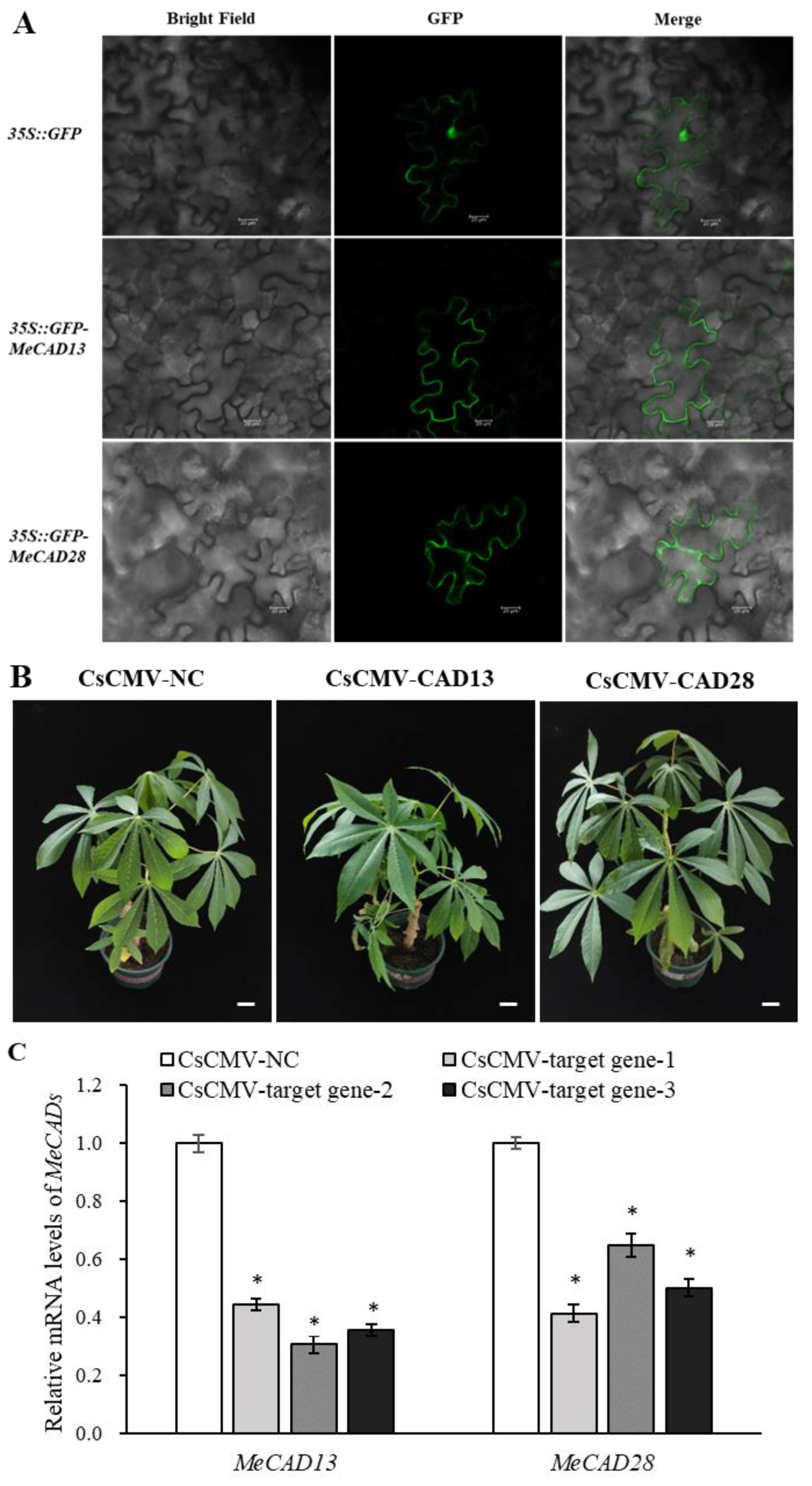
| Content (μg/mg) |
Tuberous roots | Leaves | |||||
|---|---|---|---|---|---|---|---|
| SC9-0d | SC9-1d | SC9-3d | SC9-5d | CsCMV-NC | CsCMV-CAD13 | CsCMV-CAD28 | |
| Lignin | 67.45 ± 0.43 d | 69.22 ± 0.20 c | 70.16 ± 0.44 b | 72.22 ± 0.48 a | 25.39 ± 0.47 a | 10.14 ± 0.20 c | 14.89 ± 0.27 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





