Submitted:
12 September 2024
Posted:
13 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Initial Processing of Omics Data and Databases of Model and Reference Plants
2.1. De Novo Assembly of NGS Products and Other Types of Omics Information
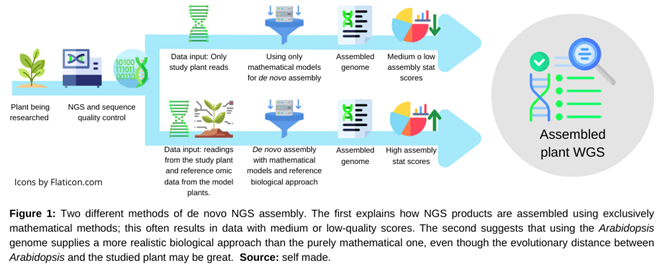
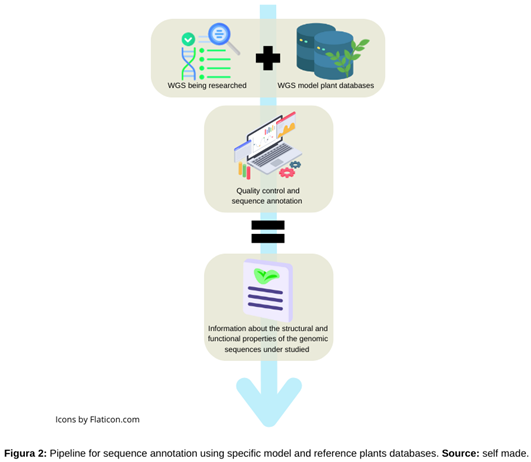
2.2. Annotations of Crops Omics Information without Reference Genome
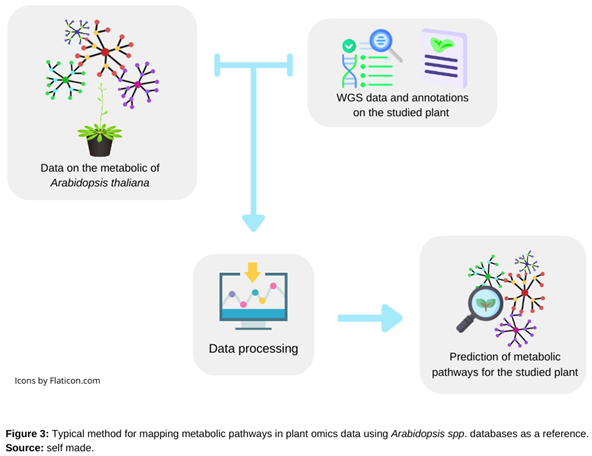
2.3. Using Data from Arabidopsis to Mapping Metabolic Pathways in Plants
- Collect information on relevant metabolites, enzymes, and pathways from a variety of sources, including literature, experimental data, and pathway databases [38].
- Using metabolic mapping tools for building a metabolic pathway map that includes all the metabolites and enzymes involved in the pathway [38]. It involves obtaining and compiling data on biochemical reactions from current sources, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG), to discover the functional annotation of genes [38,41].
- Making predictions regarding the roles of uncharacterized enzymes or metabolites while using pathway tools to examine the pathway map, identify important enzymes and metabolites, and predict the effects of genetic or environmental changes on pathway activity [41].
- Using the pathway information to develop new strategies against affections that are associated with dysregulated metabolic pathways [42].
3. Approaches to Data Integration for Plant Genetic Improvement
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO The State of Food and Agriculture 2020. Overcoming water challenges in Agriculture. 2020.
- Pazhamala, L.T.; Kudapa, H.; Weckwerth, W.; Millar, A.H.; Varshney, R.K. Systems biology for crop improvement. Plant Genome 2021, 14. [Google Scholar] [CrossRef]
- Jhansi Rani, S.; Usha, R. Transgenic plants: Types, benefits, public concerns and future. J Pharm Res 2013, 6, 879–83. [Google Scholar] [CrossRef]
- Koch, M.S.; Ward, J.M.; Levine, S.L.; Baum, J.A.; Vicini, J.L.; Hammond, B.G. The food and environmental safety of Bt crops. Front Plant Sci 2015, 06. [Google Scholar] [CrossRef]
- Rousselière, D.; Rousselière, S. Is biotechnology (more) acceptable when it enables a reduction in phytosanitary treatments? A European comparison of the acceptability of transgenesis and cisgenesis. PLoS One 2017, 12, e0183213. [Google Scholar] [CrossRef]
- Mullins, E.; Bresson, J.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; et al. Updated scientific opinion on plants developed through cisgenesis and intragenesis. EFSA Journal 2022, 20. [Google Scholar] [CrossRef]
- Lai, K.; Lorenc, M.T.; Edwards, D. Genomic Databases for Crop Improvement. Agronomy 2012, 2, 62–73. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, F.; Zhang, J.; Liu, H.; Rahman, S.; Islam, S.; et al. Application of CRISPR/Cas9 in Crop Quality Improvement. Int J Mol Sci 2021, 22, 4206. [Google Scholar] [CrossRef]
- Zhu, X.-G.; Lynch, J.P.; LeBauer, D.S.; Millar, A.J.; Stitt, M.; Long, S.P. Plants in silico : why, why now and what?-an integrative platform for plant systems biology research. Plant Cell Environ 2016, 39, 1049–57. [Google Scholar] [CrossRef]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol J 2020, 18, 1507–25. [Google Scholar] [CrossRef]
- Torres, N.V.; Santos, G. The (Mathematical) Modeling Process in Biosciences. Front Genet 2015, 6. [Google Scholar] [CrossRef]
- Vittadello, S.T.; Stumpf, M.P.H. Open problems in mathematical biology. Math Biosci 2022, 354, 108926. [Google Scholar] [CrossRef]
- Lischer, H.E.L.; Shimizu, K.K. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinformatics 2017, 18, 474. [Google Scholar] [CrossRef]
- Bao, E.; Jiang, T.; Girke, T. AlignGraph: algorithm for secondary de novo genome assembly guided by closely related references. Bioinformatics 2014, 30, i319–28. [Google Scholar] [CrossRef]
- Tang, H.; Sezen, U.; Paterson, A.H. Domestication and plant genomes. Curr Opin Plant Biol 2010, 13, 160–6. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A. Genetics and Consequences of Crop Domestication. J Agric Food Chem 2013, 61, 8267–76. [Google Scholar] [CrossRef]
- Scossa, F.; Brotman, Y.; de Abreu e Lima, F.; Willmitzer, L.; Nikoloski, Z.; Tohge, T.; et al. Genomics-based strategies for the use of natural variation in the improvement of crop metabolism. Plant Science 2016, 242, 47–64. [Google Scholar] [CrossRef]
- Wang, X.; Wang, S.; Lin, Q.; Lu, J.; Lv, S.; Zhang, Y.; et al. The wild allotetraploid sesame genome provides novel insights into evolution and lignan biosynthesis. J Adv Res 2022. [CrossRef]
- Li, Z.; Wang, J.; Fu, Y.; Jing, Y.; Huang, B.; Chen, Y.; et al. The Musa troglodytarum L. genome provides insights into the mechanism of non-climacteric behaviour and enrichment of carotenoids. BMC Biol 2022, 20, 186. [Google Scholar] [CrossRef]
- Gnerre, S.; Lander, E.S.; Lindblad-Toh, K.; Jaffe, D.B. Assisted assembly: how to improve a de novo genome assembly by using related species. Genome Biol 2009, 10, R88. [Google Scholar] [CrossRef]
- Sohn, J.; Nam, J.-W. The present and future of de novo whole-genome assembly. Brief Bioinform 2016, bbw096. [Google Scholar] [CrossRef]
- Schatz, M.C.; Witkowski, J.; McCombie, W.R. Current challenges in de novo plant genome sequencing and assembly. Genome Biol 2012, 13, 243. [Google Scholar] [CrossRef]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Brief Bioinform 2016, 17, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Deng, Z.-N.; Wu, K.-C.; Malviya, M.K.; Solanki, M.K.; Verma, K.K.; et al. Transcriptome analysis reveals a gene expression pattern that contributes to sugarcane bud propagation induced by indole-3-butyric acid. Front Plant Sci 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Han, S.; Yuan, M.; Ma, X.; Hagan, A.; He, G. Transcriptomic analyses reveal the expression and regulation of genes associated with resistance to early leaf spot in peanut. BMC Res Notes 2020, 13, 381. [Google Scholar] [CrossRef]
- Ji, X.; Liu, T.; Xu, S.; Wang, Z.; Han, H.; Zhou, S.; et al. Comparative transcriptome analysis reveals the gene expression and regulatory characteristics of broad-spectrum immunity to leaf rust in a wheat–Agropyron cristatum 2P addition line. Int J Mol Sci 2022, 23, 7370. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, C.; Yang, H.; Kuang, R.; Liu, K.; Huang, B.; et al. Comparative transcriptomics and genomic analyses reveal differential gene expression related to Colletotrichum brevisporum resistance in papaya (Carica papaya L.). Front Plant Sci 2022, 13. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, F.; Yu, Y.; Feng, L.; Jia, J.; Liu, B.; et al. A Comprehensive Online Database for Exploring ∼20,000 Public Arabidopsis RNA-Seq Libraries. Mol Plant 2020, 13, 1231–3. [Google Scholar] [CrossRef]
- Ejigu, G.F.; Jung, J. Review on the computational genome annotation of sequences obtained by Next-Generation Sequencing. Biology (Basel) 2020, 9, 295. [Google Scholar] [CrossRef]
- Salzberg, S.L. Next-generation genome annotation: we still struggle to get it right. Genome Biol 2019, 20, 92. [Google Scholar] [CrossRef]
- Petrey, D.; Fischer, M.; Honig, B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proceedings of the National Academy of Sciences 2009, 106, 17377–82. [Google Scholar] [CrossRef]
- Anthoney, N.; Foldi, I.; Hidalgo, A. Toll and Toll-like receptor signalling in development. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.-X.; Chen, D.; Zhao, Y.; Zhang, X.-Y.; Zhao, M.; Peng, R.; et al. RNA-seq analysis reveals key genes associated with seed germination of Fritillaria taipaiensis P.Y.Li by cold stratification. Front Plant Sci 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Lyv, Y.; Zheng, W.; Yang, C.; Li, Y.; Wang, X.; et al. Comparative metabolomics reveals two metabolic modules affecting seed germination in rice (Oryza sativa). Metabolites 2021, 11, 880. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.V.; Sanjinez-Argandoña, E.J.; Linzmeier, A.M.; Cardoso, C.A.L.; Macedo, M.L.R. Food value of mealworm grown on Acrocomia aculeata pulp flour. PLoS One 2016, 11, e0151275. [Google Scholar] [CrossRef]
- Jensen, L.M.; Halkier, B.A.; Burow, M. How to discover a metabolic pathway? An update on gene identification in aliphatic glucosinolate biosynthesis, regulation and transport. Biol Chem 2014, 395, 529–43. [Google Scholar] [CrossRef]
- Toubiana, D.; Puzis, R.; Wen, L.; Sikron, N.; Kurmanbayeva, A.; Soltabayeva, A.; et al. Combined network analysis and machine learning allows the prediction of metabolic pathways from tomato metabolomics data. Commun Biol 2019, 2, 214. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.S.; Shah, F.F.M.; Mohamad, M.S.; Hamran, N.L.; Salleh, A.H.M.; Deris, S.; et al. Database and tools for metabolic network analysis. Biotechnology and Bioprocess Engineering 2014, 19, 568–85. [Google Scholar] [CrossRef]
- Meeta Mistry, Mary Piper, Jihe Liu, Radhika Khetani. Differential gene expression workshop lessons from HCBC (first release). Zenodo 2021.
- Wang, L.; Dash, S.; Ng, C.Y.; Maranas, C.D. A review of computational tools for design and reconstruction of metabolic pathways. Synth Syst Biotechnol 2017, 2, 243–52. [Google Scholar] [CrossRef]
- Wang, P.; Schumacher, A.M.; Shiu, S.-H. Computational prediction of plant metabolic pathways. Curr Opin Plant Biol 2022, 66, 102171. [Google Scholar] [CrossRef]
- Mulvihill, M.M.; Nomura, D.K. Metabolomic strategies to map functions of metabolic pathways. American Journal of Physiology-Endocrinology and Metabolism 2014, 307, E237–44. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




