Introduction
Resistance-strain infections are a major source of morbidity and mortality among hospitalised patients, particularly among critically sick patients in the intensive care unit (ICU) (Hanberger et al., 1999; Vincent et al., 1995; Singh et al., 2002; Alam et al., 2023). According to Vandijck et al. (2008), Blot (2008), Blot (2007), Rice (2003), Bari et al. (2023), greater morbidity, less mobility, and increased usage of invasive equipment enhance the susceptibility of patients admitted to intensive care units to infection. Moreover, a number of medications that increase the risk of infection are routinely taken. For instance, muscle relaxants, sedatives, and stress ulcer prophylaxis can cause pneumonia by impairing cough and swallow reflexes, or they might alter the natural nonpathogenic bacterial flora, which increases the risk of infection (Marwick and Davey, 2009; Vincent et al, 2009, Faruk et al: 2023).
Many of the medical advancements of the last century are in risk of being undermined by the growing global health concern of antibiotic resistance. Globally, since the 1940s, when antibiotics were first used in medicine, people’s health and well-being have improved dramatically (Ferdous et al., 2023, Mithun et al., 2023; Tufael et al., 2023). The globe is now confronted with a severe threat of bacterial infections and antibiotic resistance, which is prevalent in every nation on the planet and adds to the worldwide concern of a "post-antimicrobial era," despite many decades of success with antibiotics. Developing nations also have significant prevalences of resistance in E. Coli and other Klebsiella spp., in addition to resistance to malaria. The ICU patients are at a 5-to7-fold increased risk of nosocomial infection in comparison to other patients because of underlying medical conditions, weakened immune systems, frequent invasive device use, exposure to wide spectrum antibiotics, and the colonisation of resistant microbes. According to Picard et al. (2006), Chakma et al. (2022), Hasan et al. (2023), Hossain et al. (2023), pneumonia, meningitis, intra-abdominal infections, and urinary tract infections, among others, may be suspected diseases. The continuous rise of resistance in the community and hospital is regarded as a serious threat to public health, in addition to the issue of nosocomial infections. Because of the unique risk profile of its patients, the intensive care unit (ICU) is considered the centre of resistance development. It has even been likened to a factory for the production, dissemination, and intensification of antimicrobial resistance (Carlet et al., 2007; Islam et al., 2018; Islam et al., 2023). Clinical and financial burdens are significant due to both infection and multidrug resistance (MDR). Thus, the detrimental effects of nosocomial infection are increased when MDR is present (Salgado et al, 2005; Cosgrove, 2006; Kuddus et al., 2020; Kuddus et al., 2021). However, the greater incidence of improper empirical antibiotic therapy linked to illnesses caused by multidrug-resistant bacteria is likely more responsible for this burden of resistance than the virulence of specific multidrug-resistant strains (Figueiredo Costa, 2008). Pseudomonas aeruginosa, Klebsiella spp., Escherichia coli, Acinetobacter spp., Staphylococcus aureus, and Streptococcus pyogenes were the most frequently found pathogens in an Indian ICU investigation (Patwardhan et al, 2008; Kuddus et al., 2022). However in an ICU in Europe, Staphylococcus aureus was discovered to be the most often isolated bacterium (30.1%), with Pseudomonas aeruginosa (28.7%), Coagulase-negative Staphylococcus (19.1%), and yeast (17.1%) following closely after (Spencer, 1996). In almost all cases, there is a need to initiate empirical antimicrobial treatment before obtaining the microbial culture results, but the situation is further complicated during the past decades, a shift in the MDR dilemma has been noted from gram-positive to gram-negative bacteria, especially due to the scarceness of new antimicrobial agent’s active against resistant gramme negative microorganisms (Boucher et al, 2009; Sazzad et al., 2023;). Gram-negative bacteria Klebsiella pneumonia, Escherichia coli, Proteus mirabilis, Enterobacter spp, Citrobacter spp, Pseudomonas aeruginosa, Acinetobacter spp., and Stenotrophomonas maltophilia are of great concern (Jones, 2001; Kaul et al, 2007; Sunny et al., 2022). Gram-positive organisms such as Staphylococcus aureus and vancomycin-resistant enterococci are of great concern (Boucher et al, 2009; Jones, 2001; Sunny et al., 2020). These important studies all point to the necessity of gathering information on antibiotic resistance in the intensive care unit (ICU) as well as the susceptibility pattern in order to inform future revisions to antibiotic policies and to assist physicians treat patients more effectively. This also eliminates unnecessary usage of broad spectrum antibiotics and prevents emergence of drug resistant bacterial strains. The current study was therefore conducted to ascertain the pattern of antimicrobial resistance of bacterial isolates among ICU patients, which would assist doctors in organizing antibiotic recommendations and antibiotic cycling in ICU environments.
Materials and Procedures
Location and Time of Study
This investigation was carried out in Bangladesh at the IBN Sina Specialised Hospital’s Microbiology Laboratory in Dhanmondi, Dhaka-1209. This investigation collected samples from patients hospitalised to this ICU between January and December of 2021 who had a clinical suspicion of infection.
Data Collection
Patients who met the inclusion criteria were asked a series of questions about their demographics and clinical history. Age, gender, major cause for admission, medical history, vital signs, and Glasgow coma score were ascertained in addition to specifics about the ICU hospitalisation. Standard biochemistry tests included blood urea nitrogen, blood glucose, serum creatinine, complete blood count (CBC), and blood electrolytes. Further testing, including an ECG, chest X-ray, arterial blood gases, and specialised diagnostics, were performed in the event that a symptom was detected. For patients undergoing mechanical ventilation, the initial configurations included of closed suctioning, assist control mode, 100% inspired oxygen fraction (FIO2), 10% tidal volume per kg, 14 repetitions per minute, and 1.2 seconds of inhalation time. After being on mechanical ventilation for 48 hours, they were assessed again for temperature, sputum category, oxygen demand, and antibiotic use.
Examine the Sample and the Microbial Isolates
A clinical suspicion regarding the source of infection led to the collection of patient samples, including blood, urine, sputum/tracheal aspirate (respiratory secretions), pus, and wound swabs. These were forwarded to the department of Microbiology, IBN Sina for routine procedures such as microbial culture, isolation, identification, and antibiotic susceptibility testing (Murray et al, 1999). To sum up, each specimen was put on a plate that contained either MacConkey agar (MCA), chocolate agar (CA), or blood agar (BA), and it was then incubated at 37°C for 18 to 24 hours. Blood cultures that show favourable growth were processed in an automated blood culture system (Bact Alert 3D, bioMeriux, France) and then subcultured onto the previously described bacterial culture substrate. Standard microbiological procedures, such as colony morphology, Gramme stain, biochemical reaction, serologic testing, and antibiotic susceptibility testing, were applied to further define and identify the positively growing suspected pathogenic bacteria (Murray et al, 1999). All Intensive Care Unit patients who gave informed consent to participate in the study were involved. We requested the patient’s next of kin for substituted consent if the patient was unable to grant informed consent. Patients who were pregnant or younger than eighteen years of age were excluded from the study.
Antimicrobial Resistance Assessment
The Antimicrobial Susceptibility Test was performed using a panel of antibiotic discs for both gramme positive and gramme negative microorganisms using the Kirby-Bauer method. In summary, Muller-Hinton broth was used to prepare test organism suspensions, turbidity was adjusted to meet McFarland 0.5 standards, and the mixture was then incubated for two hours. The Mueller Hinton Agar (MHA) plates were then covered with antibiotic discs after a bacterial lawn had been established on them. After the plates were incubated for 24 hours at 370C, the results were assessed for susceptible, intermediate, or resistant status using criteria supplied by the Clinical Laboratory Standard Institute (CLSI). The diameter of the zone of inhibition was also measured.
Data Analysis
All patient’s information was electronically saved in a relational database system created especially for the research. After that, a spreadsheet containing the data was exported for statistical analysis. The percentage of positive results throughout the whole study sample was used to determine the prevalence of antibiotic resistance. SPSS 17 has performed statistical analysis on Windows. Z-test of proportion has been used to compare quantitative data that has been reported as a percentage. P values are considered significant if they are less than 0.05.
Results
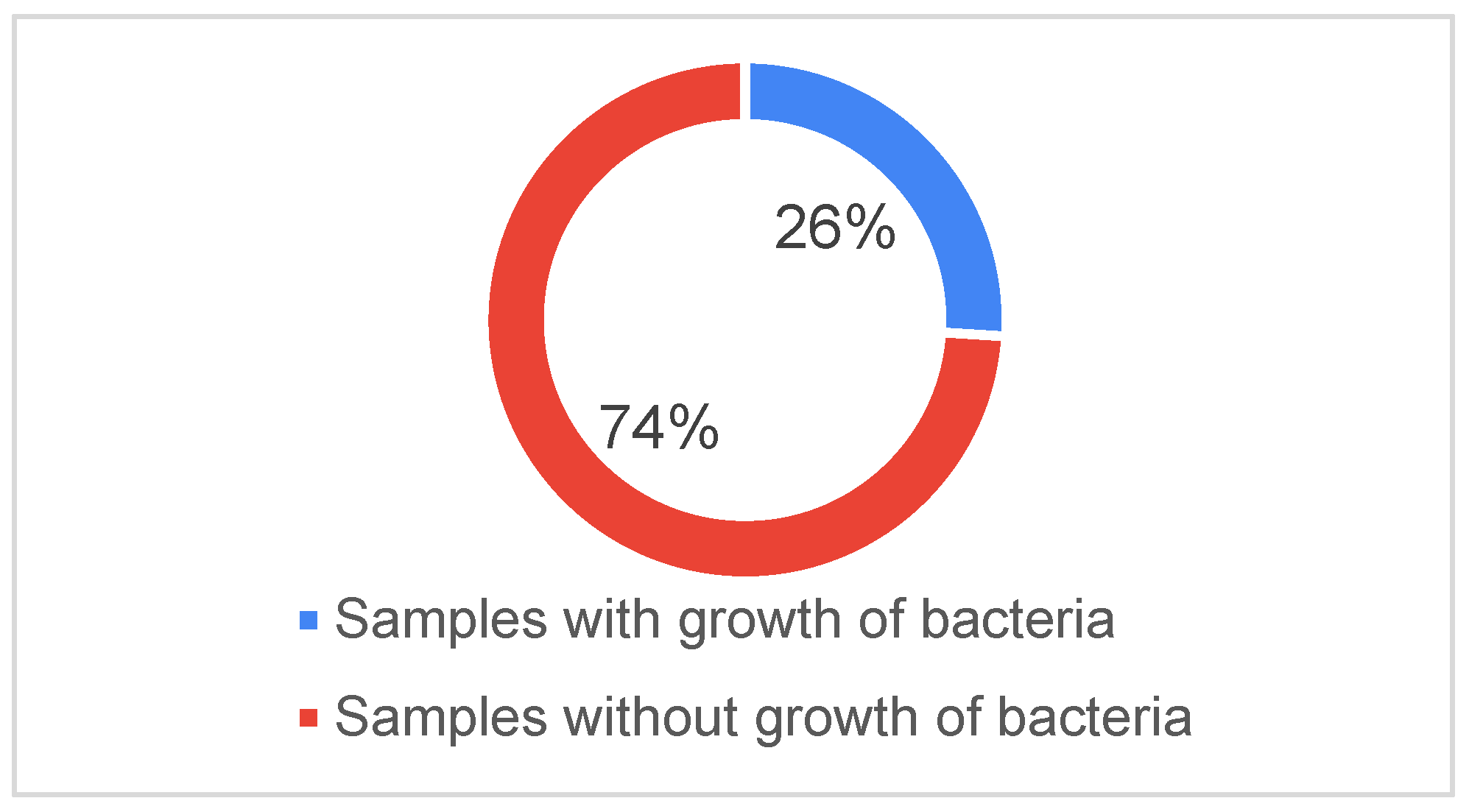
In the ICU, 200 patients were diagnosed between January and December of 2021. Of these patients, 250 blood samples were examined; of these, 65 samples from 55 patients produced the growth of 85 microorganisms (
Figure 1).
Table 2 lists the initial parameters for the patients whose blood cultures revealed the proliferation of microorganisms. Ninety-three percent of the patients had an age beyond thirty. Nonetheless, 58.2% of patients in this age category were over 60, followed by 41.8% of those between 46 and 60. 56.3% of all isolates recovered from patients, regardless of age group, were from male patients, and 43.7% came from female patients with male patients.
Aspiration pneumonia (29%) and diabetes mellitus (24%), in that order, were the most prevalent primary diagnoses (
Table 2). Urinary tract infections (12%), chronic kidney disease (12%), cerebral-vascular disease (10%), and COPD with respiratory failure (8%). The most common sites for infection were tracheal aspirate (64.71%) and urine (14.12%) (
Table 3).
Acinetobacter spp. (20%),
Klebsiella spp. (14.12%),
Escherichia coli (24.71%), and
pseudomonas spp. (30.59%) were the most frequently isolated bacteria and were primarily detected in the tracheal aspirate sample (
Table 4).
Table 5 shows the sensitivity and resistance of various microorganisms to familiar antibiotics. In
Table 6, out of 85 samples, 71.76% showed resistance to ceftriaxone and 75.29% found resistance to ceftazidime which were mostly for
Pseudomonas spp., Escherichia coli, Acinetobacter spp. and
Klebsiella spp. Klebsiella spp. showed high resistance to ceftriaxone (82.21%) followed by ciprofloxacin (81.33%), levofloxacin (81.21%), gentamicin (71.22%), amikacin (70.18%), ceftazidime (42.39%), netilmicin (37.89%), cotrimoxazole (36.89%), Piperacillin+tazobactam (36.89%), Meropenem (32.63%) and colistin (0.00%) (
Table 5).
Acinetobacter spp. showed high resistant to ciprofloxacin (89.67%) followed by ceftazidime (78.17%), levofloxacin (77.17%), Meropenem (74.44%), gentamicin (72.33%), amikacin (64.67%), ceftriaxone (57.44%), colistin (41.67%), netilmicin (49.21%), cotrimoxazole (32.33%) and Piperacillin+tazobactam (31.50%) (
Table 5).
Pseudomonas spp. showed high resistant to ceftazidime (92.29%) followed by levofloxacin (77.17%), ceftriaxone (76.24%), ciprofloxacin (75.15%), Meropenem (73.29%), gentamicin (72.29%), cotrimoxazole (72.29%), amikacin (67.57%), netilmicin (43.75%) colistin (41.86%), and Piperacillin+tazobactam (24.72%) (
Table 5).
E. coli showed high resistant to ceftriaxone (84.71%) followed by gentamicin (83.71%), ceftazidime (81.1%), ciprofloxacin (80.14%), levofloxacin (76.57%), amikacin (69.43%), cotrimoxazole (55.14%), Meropenem (48.98%), netilmicin (26.57%), Piperacillin+tazobactam (20.43%) and colistin (0.00%) (
Table 5). Meropenem was the most sensitive antibiotic against
Klebsiella spp. (64.61%) and
Acinetobacter spp. was found highly sensitive to cotrimoxazole (64.67%) and Piperacillin + tazobactam (60.50%) (
Table 5).
Escherichia coli was found greatly sensitive to netilmicin (70.48%) and meropenem (49.32%) where Pseudomonas spp. was mostly sensitive to colistin (55.14%) and netilmicin (52.25%).
Discussion
Since drug resistance, notably antimicrobial resistance, affects people worldwide, especially in impoverished nations, it is a global concern. The microbiology and epidemiology of infections in the ICU patients at IBN Sina Specialized Hospital are analyzed in this study. Aspiration pneumonia and diabetes mellitus are the two most prevalent disorders in this study; these findings are consistent with those of Kumari et al. (2007). Infections with gram-negative organisms and patterns of resistance have increased noticeably in recent years (Carlet et al, 2007). According to Bayram and Balci (2006), Acinetobacter spp., Klebsiella spp., Pseudomonas spp., and Escherichia coli are the organisms that have shown to be the most harmful for patients in the intensive care unit. Gram-negative organisms including Pseudomonas spp. (30.59%), Escherichia coli (24.71%), Acinetobacter spp. (20%), and Klebsiella spp. (14.2%) were the most often isolated microorganisms from samples in our investigation; these results are consistent with a study conducted in a private hospital in Dhaka (Islam et al, 2014). Acinetobacter spp. have become significant ICU pathogens in recent years; the majority of these bacteria are resistant to gentamicin, ampicillin, carbenicillin, cefotaxime, and chloramphenicol (Kumari et al., 2007; Islam et al., 2014). Based on samples obtained from the tracheal aspirate, Acinetobacter spp. was found to be the primary cause of pneumonia in our study. It also demonstrates resistance to high concentrations of ciprofloxacin (89.67%). Other resistance markers that were found to be associated with this pathogen include ceftazidime (78.17%), levofloxacin (77.17%), Meropenem (74.44%), gentamicin (72.33%), amikacin (64.67%), ceftriaxone (57.44%), colistin (41.67%), netilmicin (49.21%), cotrimoxazole (32.33%), Piperacillin+tazobactam (31.50%). These results align with those of related research carried out in Bangladesh and India (Kumari et al, 2007; Islam et al, 2014; Jamshdi et al, 2009). Acinetobacter spp. in our study were sensitive to cotrimoxazole (64.67%) but resistant to meropenem (74.44%); Similar results were found in another Bangladeshi investigation (60 % sensitivity to cotrimoxazole and 79.3% resistance to meropenem). (Islam et al, 2014).
Nosocomial infections in intensive care units are frequently associated with gram-negative bacteria. Intensive Care Unit (ICU) samples are frequently used to identify Pseudomonas species, and data from a multicenter ISS in the United States revealed that these organisms are especially resistant to fluoroquinolones. (Friedland et al, 2004). In this investigation, 30.59% of the isolates of Pseudomonas spp. were from tracheal aspirate. Pseudomonas spp. exhibited high resistance to ceftazidime (92.29%) in this study, which is in close agreement with other studies conducted by Bayram and Balci (2006) and Islam et al. (2014). Following Pseudomonas spp., there was high resistance to levofloxacin (77.17%), ceftriaxone (76.24%), ciprofloxacin (75.15%), Meropenem (73.29%), gentamicin (72.29%), cotrimoxazole (72.29%), amikacin (67.57%), netilmicin (43.75%), colistin (41.86%), and Piperacillin+tazobactam (24.72%).
Extended spectrum beta lactamase-producing Klebsiella spp. are another commonly observed resistant infection in intensive care unit (ICU) patients. (Jamshdi et al, 2009). A significant increase in ESBLs has resulted in multidrug-resistant Escherichia coli and Klebsiella pneumonia, making the choice of the best course of treatment challenging. Ceftriaxone (82.21%) was the most resistant antibiotic to which our isolates of Klebsiella spp. showed high resistance, followed by ciprofloxacin (81.33%), levofloxacin (81.21%), gentamicin (71.22%), amikacin (70.18%), ceftazidime (42.39%), netilmicin (37.89%), cotrimoxazole (36.89%), Piperacillin+tazobactam (36.89%), Meropenem (32.63%), and colistin (0.00%), but meropenem (64.61%) showed higher sensitivity. Once more, our results are strikingly comparable to those of a recent Dhaka study (Islam et al, 2014).
According to our observations, the most common pathogen found in UTI patients’ samples was Escherichia coli. The results of earlier research (Islam et al., 2014; Islam, 2012) are comparable to this. Escherichia coli in the study by Islam et al. was resistant to ceftriaxone but completely sensitive to meropenem. Escherichia coli in our study exhibited strong resistance to ceftriaxone (84.71%), followed by gentamicin (83.71%), ceftazidime (81.1%), ciprofloxacin (80.14%), and levofloxacin (76.57%). However, Escherichia coli was largely responsive to piperacillin + tazobactam (75.57%) and netilmicin (70.48%). Acinetobacter, E. coli, Klebsiella spp, and Pseudomonas spp that are resistant to many drugs have added additional facets to the issue of infections linked to hospitals. It is hoped that the combination of piperacillin and tazobactam shown less than 40% resistance against these four species. The concerning problem is the Acinetobacter spp. infection, for which there was no effective antibiotic sensitivity.
Our findings have important therapeutic implications for the management of patients in intensive care units, particularly those with ventilator-associated pneumonia. First and foremost, doctors should be aware that patients with ventilator-associated pneumonia are likely to be infected with one of the three common bacteria, and that treatment resistance to numerous medications is a real risk. Second, the high rate of multidrug resistance shown in this study raises serious concerns about the management of patients in intensive care units. It recommends lowering antibiotic resistance rates more systematically and minimizing the use of broad-spectrum antibiotics. Third, when multidrug resistance is present, developing rapid diagnostic tools is essential for prompt targeted therapy. To optimize drug distribution and enable a more customized treatment plan, a drug monitoring system also needs to be implemented. Nonetheless, the study’s benefits and drawbacks should be considered when interpreting the findings. The experiment included a well-characterized group of patients under close observation. Our capacity to look into a broad range of diseases and antibiotic treatments has allowed us to fairly fully document the prevalence of antimicrobial resistance in ICU patients.
However, our study is subject to certain important limitations. The study is a one-center investigation with a small sample size. As such, the results may not be generalizable. Anaerobic cultures or cultures suitable for isolating the finicky microorganisms were not worked on. It is conceivable that certain antibiotics that are used less frequently yet are becoming important for treating patients concurrently were not included in the sensitivity testing. Moreover, the disc diffusion method was utilized to determine the antibiotic sensitivity instead of the broth dilution approach, which leaves out information on the lowest inhibitory concentration of antibiotics. Because the study was carried out at a tertiary hospital and the data only covered ICU patients, the findings might not correctly reflect community-acquired illnesses in Bangladesh. The study’s tiny sample size prevented it from having the power to detect rare events or smaller effect sizes. It was unknown what specifically caused the infections and comorbidities. Furthermore, we did not thoroughly look into every aspect of our patients’ care that might have resulted in the prescription of needless antibiotics.
Conclusion
The rapid emergence and spread of antibiotic-resistant bacteria is a global concern for intensive care units. These days, the number of organisms and their resistance to the medications that are currently available is gradually increasing. Gramme negative bacteria make up the bulk of antibiotic-resistant organisms. Commercially available antibiotics might not always be effective against frequently identified microorganisms. The results of culture and sensitivity testing must, if possible, be used to guide antibiotic selection. When using empirically, third-generation cephalosporins or carbapenems (such as meropenem) may be the best choice to begin with. Piperacillin + tazobactam and colomycin (colistin) should be stored for later use. Antibiotic stewardship programs and infection control policies are crucial parts of the overall strategy to reduce antibiotic resistance and improve the care of critically ill patients, since inappropriate antibiotic usage is known to be a major driver of resistance and there aren’t many new drugs in the works. If not, we would quickly return to the pre-antibiotic era, when people would die from very minor ailments and simple infections would not be treated.
References
- Alam K, Chowdhury MZ, Jahan N, Rahman K, Chowdhury R, Mia MT, Mithun MH. Relationship between Brand Awareness and Customer Loyalty in Bangladesh: A Case Study of Fish Feed Company. J. Knowl. Learn. Sci. Technol. 2023, 2, 212–222. [Google Scholar] [CrossRef]
- Bari KF, Salam MT, Hasan SE, Sunny AR. Serum zinc and calcium level in patients with psoriasis. J. Knowl. Learn. Sci. Technol. 2023, 2, 7–14. [Google Scholar]
- Bayram A, Balci I. Patterns of antimicrobial resistance in a surgical intensive care unit of a university hospital in Turkey. BMC Infect Dis. 2006, 6, 155. [CrossRef]
- Blot S, Depuydt P, Vandewoude K, De Bacquer D. Measuring the impact of multidrug resistance in nosocomial infection. CurrOpin Infect Dis. 2007, 20, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Blot, S. Limiting the attributable mortality of nosocomial infection and multidrug resistance in intensive care units. Clin Microbiol Infect. 2008, 14, 5–13. [Google Scholar] [CrossRef]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J: Bad bugs, no drugs: no ESKAPE! Anupdate from the Infectious Diseases Society of America. Clin Infect Dis 2009, 48, 1–12. [CrossRef] [PubMed]
- Carlet J, Ben Ali A, Tabah A, Willems V, Philippart F, Chafine A, Garrouste- Orgeas M, Misset B: Multidrug resistant infections in the ICU: mechanisms, prevention and treatment. In 25 Years of Progress and Innovation in Intensive Care Medicine. Edited by: Kuhlen R, Moreno R, Ranieri VM, Rhodes A. Berlin, Germany: Medizinisch Wissenschaftliche Verlagsgesellschaft; 2007, 199-211.
- et al. Climate Change Impacts and Ongoing Adaptation Measures in the Bangladesh Sundarbans. Egyptian Journal of Aquatic Biology and Fisheries. 2022, 26, 329–348. [Google Scholar] [CrossRef]
- Cosgrove, SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006, 42 (Suppl. S2), S82–S89. [Google Scholar] [CrossRef]
- Faruk O, Hasan SE, Jubayer A, Akter K, Al Shiam SA, Rahman K, Ali MY. Microbial Isolates from Urinary Tract Infection and their Antibiotic Resistance Pattern in Dhaka city of Bangladesh. J. Knowl. Learn. Sci. Technol. 2023, 2, 76–87. [Google Scholar] [CrossRef]
- Ferdous J, Sunny AR, Khan RS, Rahman K, Chowdhury R, Mia MT, Al Shiam A, Mithun MH. Impact of Varying Synthetic Hormone on Mystus cavasius (Hamilton):: Fertilization, Hatching, and Survival Rates. Journal of Knowledge Learning and Science Technology 2023, 2, 88–105. [Google Scholar] [CrossRef]
- Figueiredo Costa, S. Impact of antimicrobial resistance on the treatment and outcome of patients with sepsis. Shock. 2008, 30 (Suppl. S1), 23–29. [Google Scholar] [CrossRef] [PubMed]
- Friedland I, Gallagher G, King T, Woods GL. Antimicrobial susceptibility patterns in Pseudomonas aeruginosa: data from a multicenter Intensive Care Unit Surveillance Study (ISS) in the United States. J Chemother. 2004, 16, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Hanberger H, Garcia-Rodriguez JA, Gobernado M, Goossens H, Nilsson LE, Struelens MJ. Antibiotic susceptibility among aerobic gram-negative bacilli in intensive care units in 5 European countries. French and Portuguese ICU Study Groups. JAMA. 1999, 281, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Hasan MR, Hossain MM, Islam MS, Sunny AR, Ferdous J, Chowdhury MZ, Maria AM, Sarder AA, Sultana A. Seasonal variation of quality and the total viable count of lean and fatty fish. Egyptian Journal of Aquatic Biology & Fisheries. 2023, 27.
- Hossain M, Kuddus MA, Khan RS, Mia MT, Rahman K, Chowdhury R, Al Shiam SA. Climate Change and Current Adaptation Strategies in the Haor Areas. J. Knowl. Learn. Sci. Technol. 2023, 2, 230–241. [Google Scholar] [CrossRef]
- Islam MR, Cansse T, Islam MS, Sunny AR. Climate Change and Its Impacts: The Case of Coastal Fishing Communities of the Meghna River in South-Central Bangladesh. International Journal of Marine and Environmental Sciences. 2018, 12, 368–376.
- Islam MR, Sunny AR, Sazzad SA, Dutta A, Hasan N, Miah MF, Ashrafuzzaman M, Prodhan SH. Environmental Jeopardy and Coping Strategies of the Small-Scale Fishers in the Bangladesh Sundarbans: The Precedent of the World’s Largest Mangrove. Egyptian Journal of Aquatic Biology & Fisheries. 2023, 27.
- Islam QT, Siddiqui MMR, Raz F, Asrafuzzaman M, Amin MR.Patterns of antimicrobial resistance among intensive care unit patients of a private medical college hospital in Dhaka. Bangladesh Journal of Medicine 2014, 25, 47–51. [CrossRef]
- Islam, QT. Antimicrobial resistance: A man made crisis. Journal of Bangladesh College of Physicians and Surgeons 2012, 29, 120–125. [Google Scholar] [CrossRef]
- Jamshdi M, Javadpour S, Eftekhari TE, Moradi N, Jomehpour F. Antimicrobial resistance pattern among Intensive Care Unit patients. Afr. J. Microbiol. Res. 2009, 3, 590–594. [Google Scholar]
- Jones RN: Resistance patterns among nosocomial pathogens: trends over the past few years. Chest 2001, 119, 397S–404S. [CrossRef]
- Kaul S, Brahmadathan K N, Jagannati M, Sudarsanam T D, Pitchamuthu K, Abraham O C, John G. One year Antimicrobial Resistance among Intensive Care Unit Patients in A Tertiary Care Hospital BJM Vol. 32 No. 1 trends in the gram-negative bacterial antibiotic susceptibility patterns in a medical intensive care unit in South India. Indian J Med Microbiol 2007, 25, 230–5. [Google Scholar] [CrossRef]
- Kuddus MA, Alam MJ, Datta GC, Miah MA, Sarker AK, Sunny MA. Climate resilience technology for year round vegetable production in northeastern Bangladesh. International Journal of Agricultural Research, Innovation and Technology (IJARIT). 2021, 11, 29–36.
- Kuddus MA, Datta GC, Miah MA, Sarker AK, Hamid SM, Sunny AR. Performance study of selected orange fleshed sweet potato varieties in north eastern bangladesh. Int. J. Environ. Agric. Biotechnol. 2020, 5, 673–682. [Google Scholar]
- Kuddus MA, Sunny AR, Sazzad SA, Hossain M, Rahman M, Mithun MH, Hasan SE, Ahmed KJ, Zandonadi RP, Han H, Ariza-Montes A. Sense and Manner of WASH and Their Coalition With Disease and Nutritional Status of Under-five Children in Rural Bangladesh: A Cross-Sectional Study. Frontiers in Public Health. 2022, 10, 890293. [Google Scholar] [CrossRef]
- Kumari HB, Nagarathna S, Chandramuki A. Antimicrobial resistance pattern among aerobic gram negative bacilli of lower respiratory tract specimens of intensive care unit patients in a neurocentre. Indian J Chest Dis Allied Sci. 2007, 49, 19–22. [Google Scholar]
- Marwick C, Davey P. Care bundles: the holy grail of infectious risk management in hospital? CurrOpin Infect Dis. 2009, 22, 364–369. [Google Scholar] [CrossRef]
- Mithun MH, Sunny AR, Billah M, Sazzad SA, Salehin S, Jahan N, Rahman K, Al Shiam A, Chowdhury R, Arafat J, Baten A. Assessing Impact of Microplastics on Aquatic Food System and Human Health.
- Murray,P. R., Baron, E.J., Pfaller, M.A., et al. (ed.).Manual of Clinical Microbiology, 7th ed. ASM press.,(1999); Washington, D.C.
- Patwardhan RB, Dhakephalkar PK, Niphadkar KB, Chopade BA. A study on nosocomial pathogens in ICU with special reference to multiresistant Acinetobacter baumanniiharbouring multiple plasmids. Indian J Med Res. 2008, 128, 178–187. [Google Scholar]
- Picard KM, O’Donoghue SC, Young-Kershaw DA, Russell KJ. Development and implementation of a multidisciplinary sepsis protocol. Crit Care Nurse. 2006, 26, 43–54. [Google Scholar] [CrossRef]
- Rice, LB. Controlling antibiotic resistance in the IC: Different bacteria, different strategies. Clevel Clin. J. Med. 2003, 70, 793–800. [Google Scholar] [CrossRef]
- Salgado CD, O’Grady N, Farr BM. Prevention and control of antimicrobial-resistant infections in intensive care patients. Crit Care Med. 2005, 33, 2373–2382. [Google Scholar] [CrossRef]
- Sazzad SA, Billah M, Sunny AR, Anowar S, Pavel JH, Rakhi MS, Rahman GM, Ahmed KT, Haider KM, Rahman MZ, Al-Mamun MA. Sketching Livelihoods and Coping Strategies of Climate Vulnerable Fishers. Egyptian Journal of Aquatic Biology & Fisheries. 2023, 27.
- Seifert H, Baginski R, Schulze A, Pulverer G. Antimicrobial susceptibility of Acinetobacter species. Antimicrob Agents Chemother. 1993, 37, 750–753. [CrossRef]
- Singh AK, Sen MR, Anupurba S, Bhattacharya P. Antibiotic sensitivity pattern of the bacteria isolated from nosocomial infection in ICU. J. Commun. Dis. 2002, 34, 257–263. [Google Scholar]
- Spencer RC. Predominant pathogens found in the Europian prevalence of infection in Intensive care study. Eur J Clin Microb. Infect Dis 1996, 15, 281–285.
- Sunny AR, Mithun MH, Prodhan SH, Ashrafuzzaman M, Rahman SM, Billah MM, Hussain M, Ahmed KJ, Sazzad SA, Alam MT, Rashid A. Fisheries in the context of attaining Sustainable Development Goals (SDGs) in Bangladesh: COVID-19 impacts and future prospects. Sustainability. 2021, 13, 9912. [Google Scholar] [CrossRef]
- Sunny AR, Reza MJ, Chowdhury MA, Hassan MN, Baten MA, Hasan MR, Monwar MM, Hossain MS, Hossain MM. Biodiversity assemblages and conservation necessities of ecologically sensitive natural wetlands of north-eastern Bangladesh. Indian J. Geo-Mar. Sci. (IJMS). 2022, 49, 135–148. [Google Scholar]
- Tufael, Hasan SE, Jubayer A, Akter K, Akter A, Akter F, Al Shiam SA, Sunny AR. Effects of Nigella Sativa and Syzygium Cumini Seed Extracts on Blood Glucose Levels in Swiss Albino Mice. J. Knowl. Learn. Sci. Technol. 2023, 2, 53–62. [Google Scholar] [CrossRef]
- Vandijck DM, Depaemelaere M, Labeau SO, Depuydt PO, Annemans L, Buyle FM, Oeyen S, Colpaert KE, Peleman RP, Blot SI, Decruyenaere JM. Daily cost of antimicrobial therapy in patients with Intensive Care Unit-acquired, laboratory-confirmed bloodstream infection. Int J Antimicrob Agents. 2008, 31, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-Chanoin MH, Wolff M, Spencer RC, Hemmer M. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA 1995, 274, 639–644. [Google Scholar] [CrossRef]
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K; EPIC II Group of Investigators. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009, 302, 2323–2329. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).