Submitted:
16 May 2023
Posted:
16 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample preparation
2.2. DNA extraction
2.3. Molecular Detection of viral nucleic acid
2.4. DNA sequencing and phylogenetic analysis
2.5. Statistical analysis
3. Results
3.1. Prevalence of PPV1 to 6 and PCV2 and 3 in pigs
| PPV1 | PPV2 | PPV3 | PPV4 | PPV5 | PPV6 | PCV2 | PCV3 | |
|---|---|---|---|---|---|---|---|---|
| Aborted pig fetus (n = 272) |
23.5% (64/272) |
16.9% (46/272) |
15.8% (43/272) |
7.0% (19/272) |
18.8% (51/272) |
18.0% (49/272) |
13.2% (36/272) |
9.9% (27/272) |
| Lung tissues from abattoirs (n = 654) |
59.6% (390/654) |
75.4% (493/654) |
71.6% (468/654) |
48.6% (318/654) |
62.5% (409/654) |
60.9% (398/654) |
67.4% (441/654) |
52.1% (341/654) |
| Total (n = 926) |
49.0% (454/926) |
58.2% (539/926) |
55.2% (511/926) |
36.4% (337/926) |
49.7% (460/926) |
48.3% (447/926) |
51.5% (477/926) |
39.7% (368/926) |
3.2. Prevalence of PPV1 co-infections with PPV2–6 or PCV3
| PPV2 | PPV3 | PPV4 | PPV5 | PPV6 | PCV3 | |
|---|---|---|---|---|---|---|
| Aborted pig fetus (n = 272) |
4.8% (13/272) |
5.1% (14/272) |
1.1% (3/272) |
4.8% (13/272) |
5.9% (16/272) |
1.5% (4/272) |
| Lung tissues from abattoirs (n = 654) |
47.2% (309/654) |
45.1% (295/654) |
32.0% (209/654) |
43.3% (283/654) |
41.0% (268/654) |
33.3% (218/654) |
| Total (n = 926) |
34.8% (322/926) |
33.4% (309/926) |
22.9% (212/926) |
32.0% (296/926) |
30.7% (284/926) |
24.0% (222/926) |
3.3. Prevalence of PCV2 co-infections with PPV1–6 or PCV3
| PPV1 | PPV2 | PPV3 | PPV4 | PPV5 | PPV6 | PCV3 | |
|---|---|---|---|---|---|---|---|
| Aborted pig fetus (n = 272) |
1.8% (5/272) |
5.1% (14/272) |
5.1% (14/272) |
2.9% (8/272) |
5.1% (14/272) |
5.9% (16/272) |
4.0% (11/272) |
| Lung tissues from abattoirs (n = 654) |
40.4% (264/654) |
52.9% (346/654) |
51.2% (335/654) |
35.3% (231/654) |
43.0% (281/654) |
44.5% (291/654) |
39.9% (261/654) |
| Total (n = 926) |
29.0% (269/926) |
38.9% (360/926) |
37.7% (349/926) |
25.8% (239/926) |
31.9% (295/926) |
33.2% (307/926) |
29.4% (272/926) |
3.4. Genetic analyses of sequenced genomes
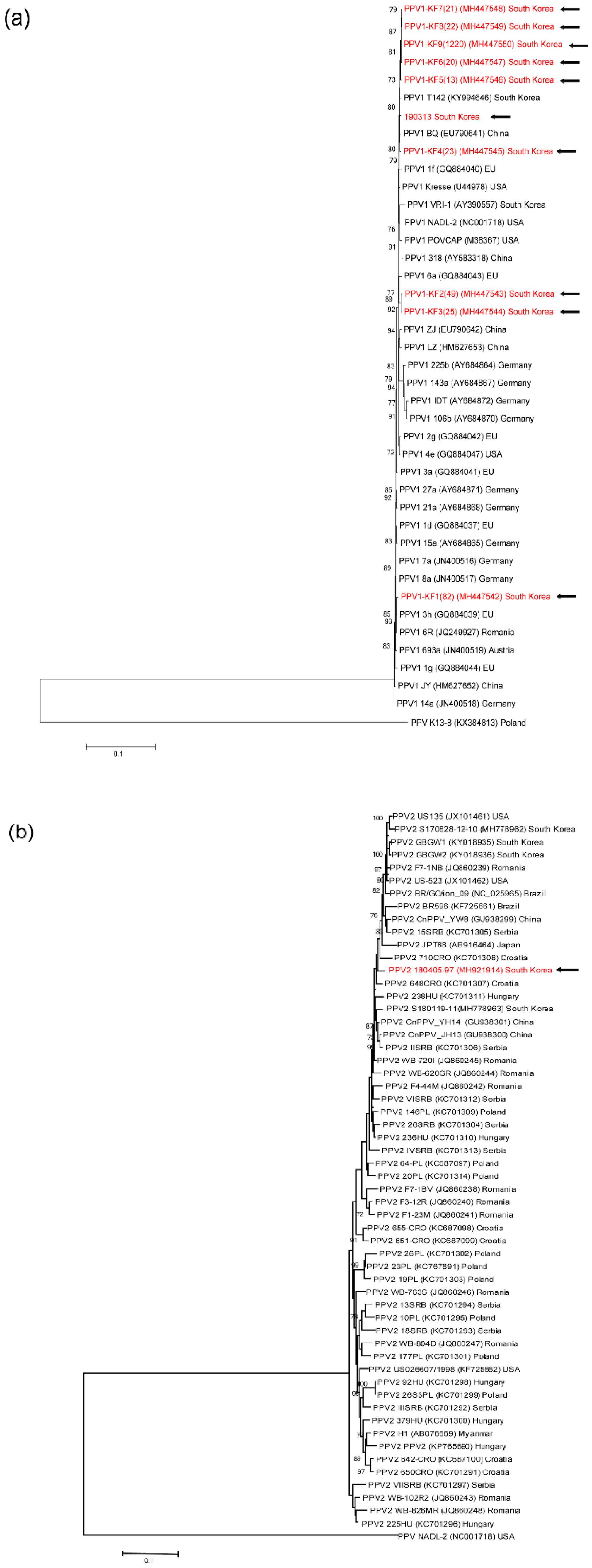
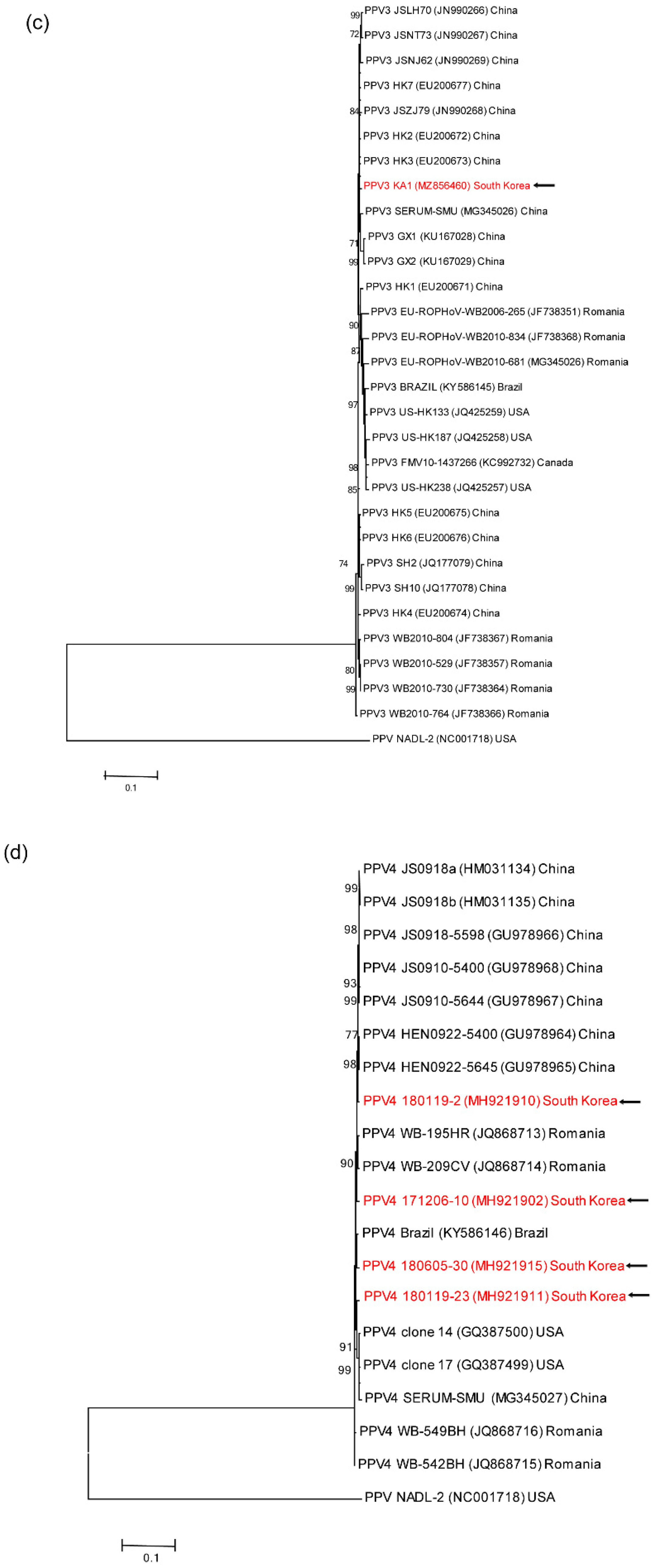

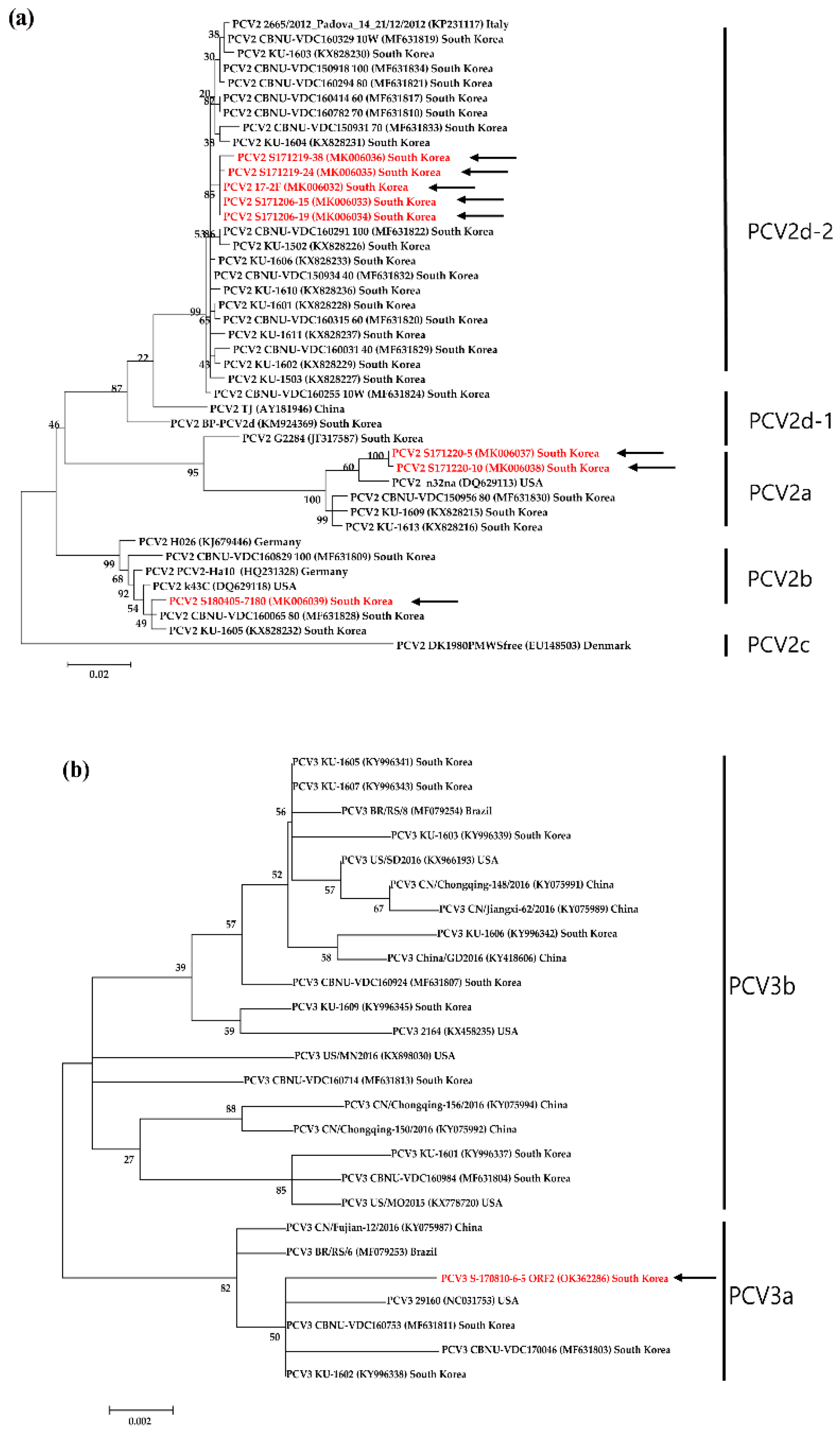
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernandes, S.; Boisvert, M.; Tijssen, P. Genetic Elements in the VP Region of Porcine Parvovirus Are Critical to Replication Efficiency in Cell Culture. J. Virol. 2011, 85 (6), 3025–3029; DOI: 10.1128/JVI.02215-10. [CrossRef]
- Hueffer, K.; Parrish, C. R. Parvovirus Host Range, Cell Tropism and Evolution. Curr. Opin. Microbiol. 2003, 6 (4), 392–398; DOI: 10.1016/s1369-5274(03)00083-3. [CrossRef]
- Xiao, C. T.; Giménez-Lirola, L. G.; Jiang, Y. H.; Halbur, P. G.; Opriessnig, T. Characterization of a Novel Porcine Parvovirus Tentatively Designated PPV5. PLOS ONE 2013, 8 (6), e65312; DOI: 10.1371/journal.pone.0065312. [CrossRef]
- Hijikata, M.; Abe, K.; Win, K. M.; Shimizu, Y. K.; Keicho, N.; Yoshikura, H. Identification of New Parvovirus DNA Sequence in Swine Sera from Myanmar. Jpn. J. Infect. Dis. 2001, 54 (6), 244–245.
- Wang, F.; Wei, Y.; Zhu, C.; Huang, X.; Xu, Y.; Yu, L.; Yu, X. Novel Parvovirus Sublineage in the Family of Parvoviridae. Virus Genes 2010, 41 (2), 305–308; DOI: 10.1007/s11262-010-0506-3. [CrossRef]
- Lau, S. K. P.; Woo, P. C. Y.; Tse, H.; Fu, C. T. Y.; Au, W. K.; Chen, X. C.; Tsoi, H. W.; Tsang, T. H. F.; Chan, J. S. Y.; Tsang, D. N. C.; Li, K. S. M.; Tse, C. W. S.; Ng, T. K.; Tsang, O. T. Y.; Zheng, B. J.; Tam, S.; Chan, K. H.; Zhou, B.; Yuen, K. Y. Identification of Novel Porcine and Bovine Parvoviruses Closely Related to Human Parvovirus 4. J. Gen. Virol. 2008, 89 (8), 1840–1848; DOI: 10.1099/vir.0.2008/000380-0. [CrossRef]
- Cheung, A. K.; Wu, G.; Wang, D.; Bayles, D. O.; Lager, K. M.; Vincent, A. L. Identification and Molecular Cloning of a Novel Porcine Parvovirus. Arch. Virol. 2010, 155 (5), 801–806; DOI: 10.1007/s00705-010-0646-8. [CrossRef]
- Ni, J.; Qiao, C.; Han, X.; Han, T.; Kang, W.; Zi, Z.; Cao, Z.; Zhai, X.; Cai, X. Identification and Genomic Characterization of a Novel Porcine Parvovirus (PPV6) in China. Virol. J. 2014, 11, 203; DOI: 10.1186/s12985-014-0203-2. [CrossRef]
- Palinski, R. M.; Mitra, N.; Hause, B. M. Discovery of a Novel Parvovirinae Virus, Porcine Parvovirus 7, by Metagenomic Sequencing of Porcine Rectal Swabs. Virus Genes 2016, 52 (4), 564–567; DOI: 10.1007/s11262-016-1322-1. [CrossRef]
- Streck, A. F.; Canal, C. W.; Truyen, U. Molecular Epidemiology and Evolution of Porcine Parvoviruses. Infect. Genet. Evol. 2015, 36, 300–306; DOI: 10.1016/j.meegid.2015.10.007. [CrossRef]
- Tuke, P. W.; Parry, R. P.; Appleton, H. Parvovirus PARV4 Visualization and Detection. J. Gen. Virol. 2010, 91 (2), 541–544; DOI: 10.1099/vir.0.014852-0. [CrossRef]
- Schirtzinger, E. E.; Suddith, A. W.; Hause, B. M.; Hesse, R. A. First Identification of Porcine Parvovirus 6 in North America by Viral Metagenomic Sequencing of Serum from Pigs Infected with Porcine Reproductive and Respiratory Syndrome Virus. Virol. J. 2015, 12, 170; DOI: 10.1186/s12985-015-0401-6. [CrossRef]
- Sun, J.; Huang, L.; Wei, Y.; Wang, Y.; Chen, D.; Du, W.; Wu, H.; Liu, C. Prevalence of Emerging Porcine Parvoviruses and Their Co-Infections with Porcine Circovirus type 2 in China. Arch. Virol. 2015, 160 (5), 1339–1344; DOI: 10.1007/s00705-015-2373-7. [CrossRef]
- Saekhow, P.; Kishizuka, S.; Sano, N.; Mitsui, H.; Akasaki, H.; Mawatari, T.; Ikeda, H. Coincidental Detection of Genomes of Porcine Parvoviruses and Porcine Circovirus type 2 Infecting Pigs in Japan. J. Vet. Med. Sci. 2016, 77 (12), 1581–1586; DOI: 10.1292/jvms.15-0167. [CrossRef]
- Sliz, I.; Vlasakova, M.; Jackova, A.; Vilcek, S. Characterization of Porcine Parvovirus type 3 and Porcine Circovirus type 2 in Wild Boars (sus Scrofa) in Slovakia. J. Wildl. Dis. 2015, 51 (3), 703–711; DOI: 10.7589/2015-01-005. [CrossRef]
- Sharma, R.; Saikumar, G. Porcine Parvovirus- and Porcine Circovirus 2-Associated Reproductive Failure and Neonatal Mortality in Crossbred Indian Pigs. Trop. Anim. Health Prod. 2010, 42 (3), 515–522; DOI: 10.1007/s11250-009-9454-0. [CrossRef]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of Papovavirus- and Picornavirus-Like Particles in Permanent Pig Kidney Cell Lines. Zentralbl. Bakteriol. Orig. A 1974, 226 (2), 153–167.
- Allan, G. M.; Kennedy, S.; McNeilly, F.; Foster, J. C.; Ellis, J. A.; Krakowka, S. J.; Meehan, B. M.; Adair, B. M. Experimental Reproduction of Severe Wasting Disease by Co-Infection of Pigs with Porcine Circovirus and Porcine Parvovirus. J. Comp. Pathol. 1999, 121 (1), 1–11; DOI: 10.1053/jcpa.1998.0295. [CrossRef]
- Phan, T. G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T. P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a Novel Circovirus PCV3 in Pigs with Cardiac and Multi-Systemic Inflammation. Virol. J. 2016, 13 (1), 184; DOI: 10.1186/s12985-016-0642-z. [CrossRef]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel Circovirus Species Identified in Farmed Pigs Designated as Porcine Circovirus 4, Hunan Province, China. Transbound. Emerg. Dis. 2020, 67 (3), 1057-1061; DOI: 10.1111/tbed.13446. [CrossRef]
- Meehan, B. M.; McNeilly, F.; Todd, D.; Kennedy, S.; Jewhurst, V. A.; Ellis, J. A.; Hassard, L. E.; Clark, E. G.; Haines, D. M.; Allan, G. M. Characterization of Novel Circovirus DNAs Associated with Wasting Syndromes in Pigs. J. Gen. Virol. 1998, 79 (9), 2171–2179; DOI: 10.1099/0022-1317-79-9-2171. [CrossRef]
- Dias, A. S.; Gerber, P. F.; Araújo, A. S.; Auler, P. A.; Gallinari, G. C.; Lobato, Z. I. Lack of Antibody Protection Against Porcine Circovirus 2 and Porcine Parvovirus in Naturally Infected Dams and Their Offspring. Res. Vet. Sci. 2013, 94 (2), 341–345; DOI: 10.1016/j.rvsc.2012.09.009. [CrossRef]
- Finsterbusch, T.; Mankertz, A. Porcine Circoviruses-Small but Powerful. Virus Res. 2009, 143 (2), 177–183; DOI: 10.1016/j.virusres.2009.02.009. [CrossRef]
- Ren, L.; Chen, X.; Ouyang, H. Interactions of Porcine Circovirus 2 with Its Hosts. Virus Genes 2016, 52 (4), 437–444; DOI: 10.1007/s11262-016-1326-x. [CrossRef]
- Segalés, J. Porcine Circovirus type 2 (pcv2) Infections: Clinical Signs, Pathology and Laboratory Diagnosis. Virus Res. 2012, 164 (1–2), 10–19; DOI: 10.1016/j.virusres.2011.10.007. [CrossRef]
- Liu, X.; Ouyang, T.; Ma, T.; Ouyang, H.; Pang, D.; Ren, L. Immunogenicity Evaluation of Inactivated Virus and Purified Proteins of Porcine Circovirus type 2 in Mice. B.M.C. Vet. Res. 2018, 14 (1), 137; DOI: 10.1186/s12917-018-1461-9. [CrossRef]
- Chae, C. Chae, C.A. Review of Porcine Circovirus 2-Associated Syndromes and Diseases. Vet. J. 2005, 169 (3), 326–336; DOI: 10.1016/j.tvjl.2004.01.012. [CrossRef]
- Chen, G. H.; Mai, K. J.; Zhou, L.; Wu, R. T.; Tang, X. Y.; Wu, J. L.; He, L. L.; Lan, T.; Xie, Q. M.; Sun, Y.; Ma, J. Y. Detection and Genome Sequencing of Porcine Circovirus 3 in Neonatal Pigs with Congenital Tremors in South China. Transbound. Emerg. Dis. 2017, 64 (6), 1650–1654; DOI: 10.1111/tbed.12702. [CrossRef]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B. M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91 (1), e01879-16; DOI: 10.1128/JVI.01879-16. [CrossRef]
- Stadejek, T.; Woźniak, A.; Miłek, D.; Biernacka, K. First Detection of Porcine Circovirus type 3 on Commercial Pig Farms in Poland. Transbound. Emerg. Dis. 2017, 64 (5), 1350–1353; DOI: 10.1111/tbed.12672. [CrossRef]
- Faccini, S.; Barbieri, I.; Gilioli, A.; Sala, G.; Gibelli, L. R.; Moreno, A.; Sacchi, C.; Rosignoli, C.; Franzini, G.; Nigrelli, A. Detection and Genetic Characterization of Porcine Circovirus type 3 in Italy. Transbound. Emerg. Dis. 2017, 64 (6), 1661–1664; DOI: 10.1111/tbed.12714. [CrossRef]
- Cadar, D.; Cságola, A.; Lorincz, M.; Tombácz, K.; Spînu, M.; Tuboly, T. Distribution and Genetic Diversity of Porcine Hokovirus in Wild Boars. Arch. Virol. 2011, 156 (12), 2233–2239; DOI: 10.1007/s00705-011-1125-6. [CrossRef]
- Gava, D.; Souza, C. K.; Schaefer, R.; Vincent, A. L.; Cantão, M. E.; Coldebella, A.; Ciacci-Zanella, J. R. A Taqman-Based Real-Time PCR for Detection and Quantification of Porcine Parvovirus 4. J. Virol. Methods 2015, 219, 14–17; DOI: 10.1016/j.jviromet.2015.03.011. [CrossRef]
- Schirtzinger, E. E.; Suddith, A. W.; Hause, B. M.; Hesse, R. A. First Identification of Porcine Parvovirus 6 in North America by Viral Metagenomic Sequencing of Serum from Pigs Infected with Porcine Reproductive and Respiratory Syndrome Virus. Virol. J. 2015, 12, 170; DOI: 10.1186/s12985-015-0401-6. [CrossRef]
- Ouardani, M.; Wilson, L.; Jetté, R.; Montpetit, C.; Dea, S. Multiplex PCR for Detection and Typing of Porcine Circoviruses. J. Clin. Microbiol. 1999, 37 (12), 3917–3924; DOI: 10.1128/JCM.37.12.3917-3924.1999. [CrossRef]
- Li, L.; Yuan, W.; Guo, H.; Ma, Z.; Song, Q.; Wang, X.; Li, H. Prevalence and Genetic Variation of Porcine Circovirus type 2 in Hebei, China from 2004 to 2014. Gene 2016, 586 (2), 222–227; DOI: 10.1016/j.gene.2016.04.014. [CrossRef]
- Ku, X.; Chen, F.; Li, P.; Wang, Y.; Yu, X.; Fan, S.; Qian, P.; Wu, M.; He, Q. Identification and Genetic Characterization of Porcine Circovirus type 3 in China. Transbound. Emerg. Dis. 2017, 64 (3), 703–708; DOI: 10.1111/tbed.12638. [CrossRef]
- Altschul, S. F.; Gish, W.; Miller, W.; Myers, E. W.; Lipman, D. J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215 (3), 403–410; DOI: 10.1016/S0022-2836(05)80360-2. [CrossRef]
- Cadar, D.; Dán, Á.; Tombácz, K.; Lőrincz, M.; Kiss, T.; Becskei, Z.; Spînu, M.; Tuboly, T.; Cságola, A. Phylogeny and Evolutionary Genetics of Porcine Parvovirus in Wild Boars. Infect. Genet. Evol. 2012, 12 (6), 1163–1171; DOI: 10.1016/j.meegid.2012.04.020. [CrossRef]
- Miłek, D.; Woźniak, A.; Podgórska, K.; Stadejek, T. Do Porcine Parvoviruses 1 Through 7 (PPV1-PPV7) have an Impact on Porcine Circovirus type 2 (PCV2) Viremia in Pigs?. Vet. Microbiol. 2020, 242, 108613; DOI: 10.1016/j.vetmic.2020.108613. [CrossRef]
- Lagan Tregaskis, P.; Staines, A.; Gordon, A.; Sheridan, P.; McMenamy, M.; Duffy, C.; Collins, P. J.; Mooney, M. H.; Lemon, K. Co-Infection Status of Novel Parvovirus’s (PPV2 to 4) with Porcine Circovirus 2 in Porcine Respiratory Disease Complex and Porcine Circovirus-Associated Disease from 1997 to 2012. Transbound. Emerg. Dis. 2021, 68 (4), 1979–1994; DOI: 10.1111/tbed.13846. [CrossRef]
- Miłek, D.; Woźniak, A.; Guzowska, M.; Stadejek, T. Detection Patterns of Porcine Parvovirus (PPV) and Novel Porcine Parvoviruses 2 Through 6 (PPV2-PPV6) in Polish Swine Farms. Viruses 2019, 11 (5), 474; DOI: 10.3390/v11050474. [CrossRef]
- Xiao, C. T.; Halbur, P. G.; Opriessnig, T. Complete Genome Sequence of a Novel Porcine Parvovirus (PPV) Provisionally Designated PPV5. Gnome Announc. Genome Announc. 2013, 1 (1), e00021-12; DOI: 10.1128/genomeA.00021-12. [CrossRef]
- Huang, L.; Zhai, S. L.; Cheung, A. K.; Zhang, H. B.; Long, J. X.; Yuan, S. S. Detection of a Novel Porcine Parvovirus, PPV4, in Chinese Swine Herds. Virol. J. 2010, 7, 333; DOI: 10.1186/1743-422X-7-333. [CrossRef]
- Oh, W. T.; Kim, R. Y.; Nguyen, V. G.; Chung, H. C.; Park, B. K. Perspectives on the Evolution of Porcine Parvovirus. Viruses 2017, 9 (8), 196; DOI: 10.3390/v9080196. [CrossRef]
- Cságola, A.; Lőrincz, M.; Cadar, D.; Tombácz, K.; Biksi, I.; Tuboly, T. Detection, Prevalence and Analysis of Emerging Porcine Parvovirus Infections. Arch. Virol. 2012, 157 (6), 1003–1010; DOI: 10.1007/s00705-012-1257-3. [CrossRef]
- Xiao, C. T.; Gerber, P. F.; Giménez-Lirola, L. G.; Halbur, P. G.; Opriessnig, T. Characterization of Porcine Parvovirus type 2 (PPV2) Which Is Highly Prevalent in the USA. Vet. Microbiol. 2013, 161 (3–4), 325–330; DOI: 10.1016/j.vetmic.2012.07.038. [CrossRef]
- Streck, A. F.; Homeier, T.; Foerster, T.; Fischer, S.; Truyen, U. Analysis of Porcine Parvoviruses in Tonsils and Hearts from Healthy Pigs Reveals High Prevalence and Genetic Diversity in Germany. Arch. Virol. 2013, 158 (6), 1173–1180; DOI: 10.1007/s00705-013-1603-0. [CrossRef]
- Saekhow, P.; Mawatari, T.; Ikeda, H. Coexistence of Multiple Strains of Porcine Parvovirus 2 in Pig Farms. Microbiol. Immunol. 2014, 58 (7), 382–387; DOI: 10.1111/1348-0421.12159. [CrossRef]
- Saekhow, P.; Ikeda, H. Prevalence and Genomic Characterization of Porcine Parvoviruses Detected in Chiangmai Area of Thailand in 2011. Microbiol. Immunol. 2015, 59 (2), 82–88; DOI: 10.1111/1348-0421.12218. [CrossRef]
- Lee, J. Y.; Kim, E. J.; Cho, I. S.; Lee, K. K.; Shin, Y. K. Complete Genome Sequences of Porcine Parvovirus 2 Isolated from Swine in the Republic of Korea. Genome Announc. 2017, 5 (15), e01738-16; DOI: 10.1128/genomeA.01738-16. [CrossRef]
- Ouh, I. O.; Park, S.; Lee, J. Y.; Song, J. Y.; Cho, I. S.; Kim, H. R.; Park, C. K. First Detection and Genetic Characterization of Porcine Parvovirus 7 from Korean Domestic Pig Farms. J. Vet. Sci. 2018, 19 (6), 855–857; DOI: 10.4142/jvs.2018.19.6.855. [CrossRef]
- Huang, L.; Zhai, S. L.; Cheung, A. K.; Zhang, H. B.; Long, J. X.; Yuan, S. S. Detection of a Novel Porcine Parvovirus, PPV4, in Chinese Swine Herds. Virol. J. 2010, 7, 333; DOI: 10.1186/1743-422X-7-333. [CrossRef]
- Saade, G.; Deblanc, C.; Bougon, J.; Marois-Créhan, C.; Fablet, C.; Auray, G.; Belloc, C.; Leblanc-Maridor, M.; Gagnon, C.A.; Zhu, J.; Gottschalk, M.; Summerfield, A.; Simon, G.; Bertho, N.; Meurens, F. Coinfections and Their Molecular Consequences in the Porcine Respiratory Tract. Vet. Res. 2020, 51 (1), 80; DOI: 10.1186/s13567-020-00807-8. [CrossRef]
- Opriessnig, T.; Fenaux, M.; Yu, S.; Evans, R.B.; Cavanaugh, D.; Gallup, J.M. Pallares, F.J.; Thacker, E.L.; Lager, K.M.; Meng, X.J.; Halbur, P.G. Effect of Porcine Parvovirus Vaccination on the Development of PMWS in Segregated Early Weaned Pigs Coinfected with Type 2 Porcine Circovirus and Porcine Parvovirus. Vet. Microbiol. 2004, 98 (3-4), 209–220; DOI: 10.1016/j.vetmic.2005.04.010. [CrossRef]
- Opriessnig, T.; Xiao, C. T.; Gerber, P. F.; Halbur, P. G. Identification of Recently Described Porcine Parvoviruses in Archived North American Samples from 1996 and Association with Porcine Circovirus Associated Disease. Vet. Microbiol. 2014, 173 (1–2), 9–16; DOI: 10.1016/j.vetmic.2014.06.024. [CrossRef]
- Li, X.; Qiao, M.; Sun, M.; Tian, K. A Duplex Real-Time PCR Assay for the Simultaneous Detection of Porcine Circovirus 2 and Circovirus 3. Virol. Sin. 2018, 33 (2), 181–186; DOI: 10.1007/s12250-018-0025-2. [CrossRef]
- Kim, H. R.; Park, Y. R.; Lim, D. R.; Park, M. J.; Park, J. Y.; Kim, S. H.; Lee, K. K.; Lyoo, Y. S.; Park, C. K. Multiple Real-Time Polymerase Chain Reaction for the Differential Detection of Porcine Circovirus 2 and 3. J. Virol. Methods 2017, 250, 11–16; DOI: 10.1016/j.jviromet.2017.09.021. [CrossRef]
- Wen, S.; Sun, W.; Li, Z.; Zhuang, X.; Zhao, G.; Xie, C.; Zheng, M.; Jing, J.; Xiao, P.; Wang, M.; Han, J.; Ren, J.; Liu, H.; Lu, H.; Jin, N. The Detection of Porcine Circovirus 3 in Guangxi, China. Transbound. Emerg. Dis. 2018, 65 (1), 27–31; DOI: 10.1111/tbed.12754. [CrossRef]
- Zheng, S.; Wu, X.; Zhang, L.; Xin, C.; Liu, Y.; Shi, J.; Peng, Z.; Xu, S.; Fu, F.; Yu, J.; Sun, W.; Xu, S.; Li, J.; Wang, J. The Occurrence of Porcine Circovirus 3 Without Clinical Infection Signs in Shandong Province. Transbound. Emerg. Dis. 2017, 64 (5), 1337–1341; DOI: 10.1111/tbed.12667. [CrossRef]
- Zhao, D.; Wang, X.; Gao, Q.; Huan, C.; Wang, W.; Gao, S.; Liu, X. Retrospective Survey and Phylogenetic Analysis of Porcine Circovirus type 3 in Jiangsu Province, China, 2008 to 2017. Arch. Virol. 2018, 163 (9), 2531–2538; DOI: 10.1007/s00705-018-3870-2. [CrossRef]
- Kim, S. C.; Nazki, S.; Kwon, S.; Juhng, J. H.; Mun, K. H.; Jeon, D. Y.; Jeong, C. G.; Khatun, A.; Kang, S. J.; Kim, W. I. The Prevalence and Genetic Characteristics of Porcine Circovirus type 2 and 3 in Korea. B.M.C. Vet. Res. 2018, 14 (1), 294; DOI: 10.1186/s12917-018-1614-x. [CrossRef]
- Jiang, H.; Wang, D.; Wang, J.; Zhu, S.; She, R.; Ren, X.; Tian, J.; Quan, R.; Hou, L.; Li, Z.; Chu, J.; Guo, Y.; Xi, Y.; Song, H.; Yuan, F.; Wei, L.; Liu, J. Induction of Porcine Dermatitis and Nephropathy Syndrome in Piglets by Infection with Porcine Circovirus type 3. J. Virol. 2019, 93 (4), e02045-18; DOI: 10.1128/JVI.02045-18. [CrossRef]
- Fu, X.; Fang, B.; Ma, J.; Liu, Y.; Bu, D.; Zhou, P.; Wang, H.; Jia, K.; Zhang, G. Insights into the Epidemic Characteristics and Evolutionary History of the Novel Porcine Circovirus type 3 in Southern China. Transbound. Emerg. Dis. 2018, 65 (2), e296–e303; DOI: 10.1111/tbed.12752. [CrossRef]
- Cibulski, S. P.; Teixeira, T. F.; Varela, A. P. M.; Scheffer, C. M.; Santos, H. F.; Lima, F. E. S.; Roehe, P. M. Ungulate Copiparvovirus 2 in Healthy and Postweaning Multisystemic Wasting Syndrome-Affected Pigs. Trop. Anim. Health Prod. 2017, 49 (5), 945–949; DOI: 10.1007/s11250-017-1279-7. [CrossRef]
- Woźniak, A.; Miłek, D.; Matyba, P.; Stadejek, T. Real-Time PCR Detection Patterns of Porcine Circovirus Type 2 (PCV2) in Polish Farms with Different Statuses of Vaccination against PCV2. Viruses 2019, 11 (12), 1135; DOI: 10.3390/v11121135. [CrossRef]
- Zeeuw, E. J. L.; Leinecker, N.; Herwig, V.; Selbitz, H. J.; Truyen, U. Study of the Virulence and Cross-Neutralization Capability of Recent Porcine Parvovirus Field Isolates and Vaccine Viruses in Experimentally Infected Pregnant Gilts. J. Gen. Virol. 2007, 88 (2), 420–427; DOI: 10.1099/vir.0.82302-0. [CrossRef]
- Jóźwik, A.; Manteufel, J.; Selbitz, H. J.; Truyen, U. Vaccination against Porcine Parvovirus Protects against Disease, but Does Not Prevent Infection and Virus Shedding After Challenge Infection with a Heterologous Virus Strain. J. Gen. Virol. 2009, 90 (10), 2437–2441; DOI: 10.1099/vir.0.012054-0. [CrossRef]
- Bergeron, J.; Hébert, B.; Tijssen, P. Genome Organization of the Kresse Strain of Porcine Parvovirus: Identification of the Allotropic Determinant and Comparison with Those of NADL-2 and Field Isolates. J. Virol. 1996, 70 (4), 2508–2515; DOI: 10.1128/JVI.70.4.2508-2515.1996. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





