Submitted:
08 December 2023
Posted:
11 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
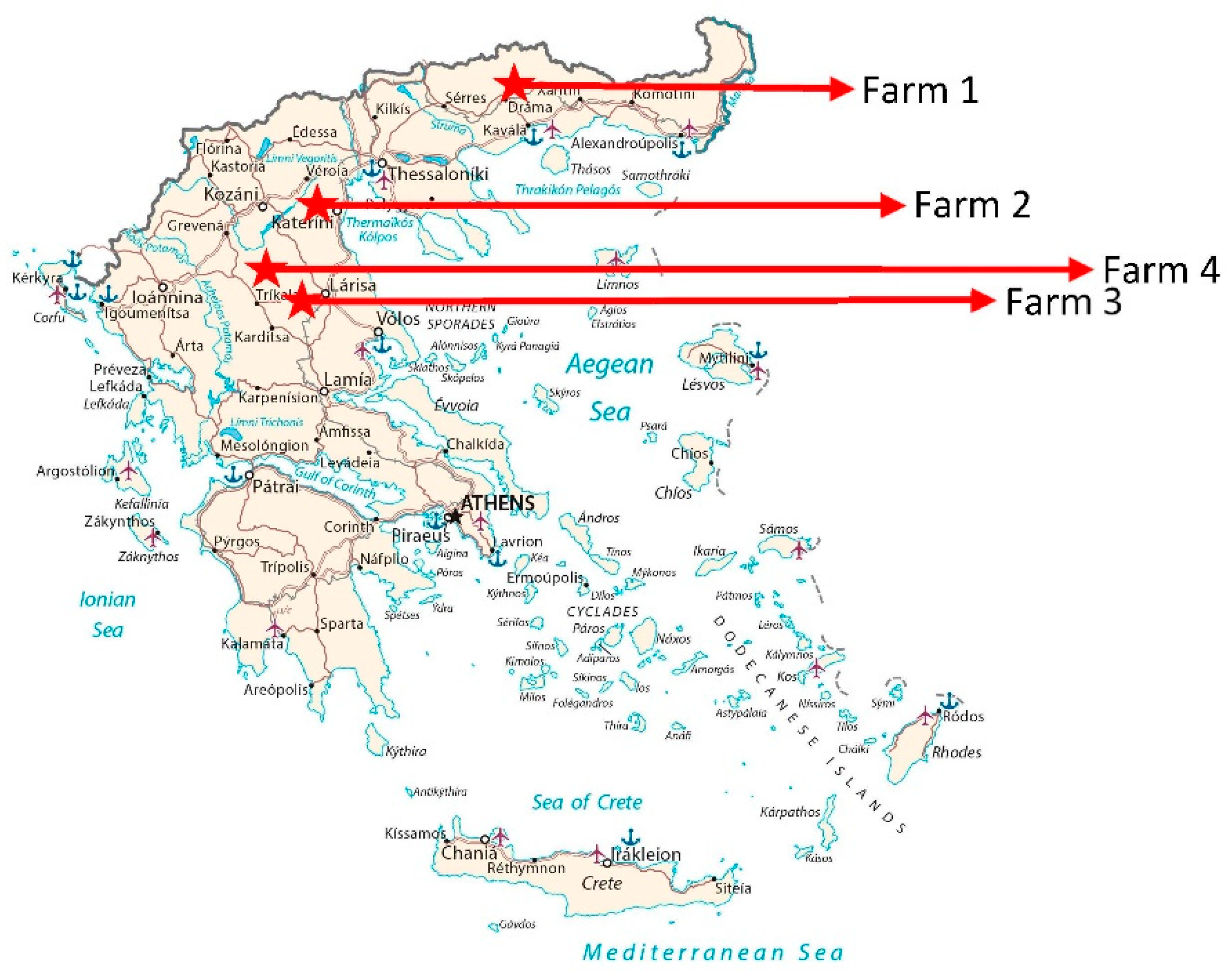
2.1. Animals and tissues
2.2. DNA and RNA isolation
2.3. Real-time PCR for the detection of DNA viruses
2.4. Real-time reverse transcriptase PCR for the detection of HEV
2.5. Conventional PCR for the detection of PERVs
2.6. Determination of the sensitivity
| Virus | Primer/Probe | Sequence 5′-3′ | Reference | |
|---|---|---|---|---|
| HEV | JVHEV-Fwd | GGT GGT TTC TGG GGT GAC | Jothikumar et al. 2006 [45] | |
| JVHEV-Rev | AGG GGT TGG TTG GAT GAA | |||
| JVHEV-Probe | 6FAM-TGA TTC TCA GCC CTT CGC-BHQ | |||
| PCMV/PRV | PCMV-Fwd | ACT TCG TCG CAG CTC ATC TGA | Mueller et al. 2002, [38] | |
| PCMV-Rev | GTT CTG GGA TTC CGA GGT TG | |||
| PCMV-Probe | 6FAM-CAG GGC GGC GGT CGA GCT C-BHQ | |||
| PLHV-1 | PLHV-1 (1125)-Fwd | CTC ACC TCC AAA TAC AGC GA | Chmielewicz et al. 2003 [39] | |
| PLHV-1 (1125)-Rev | GCT TGA ATC GTG TGT TCC ATA G | |||
| PLHV-1 (1125)-Probe | 6FAM-CTG GTC TAC TGA ATC GCC GCT AAC AG-TAMR | |||
| PLHV-2 | PLHV-2 (1155)-Fwd | GTC ACC TGC AAA TAC ACA GG | Chmielewicz et al. 2003 [39] | |
| PLHV-2 (1155)-Rev | GGC TTG AAT CGT ATG TTC CAT AT | |||
| PLHV-2 (1155)-Probe | 6FAM-CTG GTC TAC TGA AGC GCT GCC AAT AG-TAMRA | |||
| PLVH-3 | PLHV-3 (210s)-Fwd | AAC AGC GCC AGA AAA AAA GG | McMahon et al. 2006 [40] | |
| PLHV-3 (210as)-Rev | GGA AAG GTA GAA GGT GAA CCA TAA AA | |||
| PLHV-3 (210)-Probe | 6-FAM CCA AAG AGG AAA ATC-MGB | |||
| PCV2 | PCV2 (F2020)-Fwd | CTG AGT CTT TTT TAT CAC TTC GTA ATG GT | Chen et al. 2021, [41] | |
| PCV2 (F2020)-Rev | ACT GCG TTC GAA AAC AGT ATA TAC GA | |||
| PCV2 (F2020)-Probe | 6FAM-TTA AGT GGG GGG TCT TTA AGA TTA AAT TCT CTG AAT TGT-BHQ2 | |||
| PCV3 | PCV3-Fwd | AGT GCT CCC CAT TGA ACG | Palinski et al. 2017 [42] | |
| PCV3-Rev | ACA CAG CCG TTA CTT CAC | |||
| PCV3-Probe | 6FAM-ACC CCA TGG CTC AAC ACA TAT GAC C-BHQ1 | |||
| PCV4 | PCV4 (F2020)-Fwd | ATT ATT AAA CAG ACT TTA TTT GTG TCA TCA CTT | Chen et al. 2021 [41] | |
| PCV4 (F2020)-Rev | ACA GGG ATA ATG CGT AGT GAT CAC T | |||
| PCV4 (F2020)-Probe | 6FAM-ATA CTA CAC TTG ATC TTA GCC AAA AGG CTC GTT GA-BHQ1 | |||
| PPV1 | PPV1-Fwd | CAG AAT CAG CAA CCT CAC CA | Opriessnig et al. 2011 [43] | |
| PPV1-Rev | GCT GCT GGT GTG TAT GGA AG | |||
| PPV1-Probe | 6FAM-TGC AAG CTT/ZEN/AAT GGT CGC ACT AGA CA-BHQ1 | |||
| pGAPDH | pGAPDH-Fwd | ACA TGG CCT CCA AGG AGT AAG A | Duvigneau et al. 2005 [47] | |
| pGAPDH-Rev | GAT CGA GTT GGG GCT GTG ACT | |||
| pGAPDH-Probe | HEX-CCA CCA ACC CCA GCA AGA G-BHQ1 | |||
| PERV-C | PERV-envC-Fwd | GAT TAG AAC TGG AAG CCC CAA GTG CTC T | Kaulitz et al., 2013 [45] | |
| PERV-envC-Rev | TCT GAT CCA GAA GTT ATG TTA GAG GAT GGT | |||
| PERV-A/C | PERV-A env VRBF-Fwd | CCT ACC AGT TAT AAT CAA TTT AAT TAT GGC | Wood et al. 2004 [46] | |
| PERV-C env TMR-Rev | CTC AAA CCA CCC TTG AGT AGT TTC C | |||
2.7. Western blot to detect antibodies against PCMV/PRV
3. Results
3.1. Sensitivity of the assays
3.2. Screening for herpesviruses: PCMV/PRV, PLHV-1, PLHV-2, PLHV-3
3.3. Screening for circoviruses and PPV-1
3.4. Screening for RNA viruses. HEV3
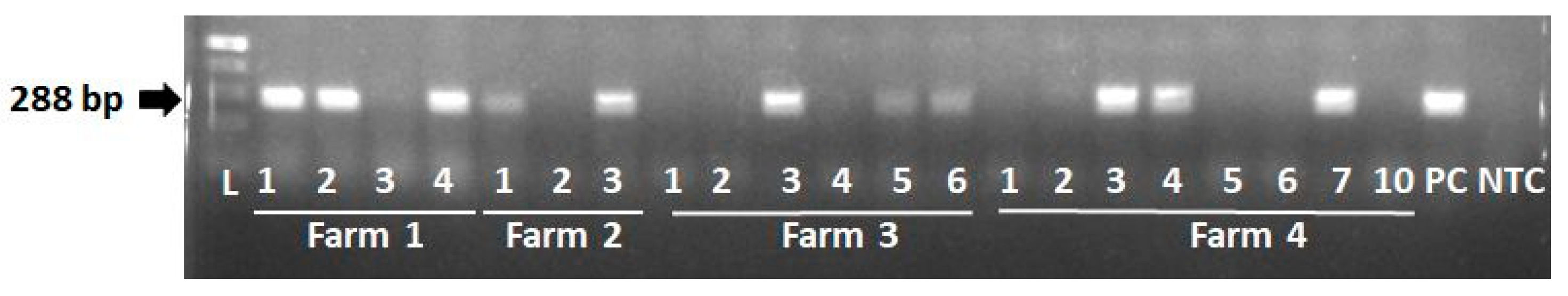
3.4. Screening for PERVs
| Virus | Method | Sensitivity (copy number per 100 ng DNA) |
Sensitivity R2 | Reference |
|---|---|---|---|---|
| PCMV/PRV | conventional PCR | 15 copies | Morozov et al., 2016 [51] | |
| nested PCR | 5 copies | |||
| real-time PCR | 2 copies | |||
| real-time PCR | 20 copies | Mueller et al., 2002 [38] | ||
| real-time PCR | 10 copy | 0.9964 | this manuscript | |
| HEV | real-time RT-PCR | 10 | Jothikumar et al., 2006 [44] | |
| real-time RT-PCR | 150-200 | Morozov et al., 2015 [17] | ||
| real-time RT-PCR | 10 copy | 0.9962 | this manuscript | |
| PCV2 | multiplex | 101/µl | Zhou et al., 2022 [52] | |
| real-time PCR | 1 copy | 0.9935 | this manuscript | |
| PCV3 | real-time PCR | 10 copies | 0.9906 | this manuscript |
| PCV4 | real-time PCR | 100 copies | 0.9906 | this manuscript |
| PLHV-1 | real-time PCR | 20 copies | Chmielewicz et al. 2003 [39] | |
| real-time PCR | 1 copy | 0.9964 | this manuscript | |
| PLHV2 | real-time PCR | 20 copies | Chmielewicz et al. 2003 [39] | |
| real-time PCR | 1 copy | 0.9953 | this manuscript | |
| PLHV3 | real-time PCR | 1 copies | 0.9983 | this manuscript |
| PPV1 | real-time PCR | 10 copy | 0.9961 | this manuscript |
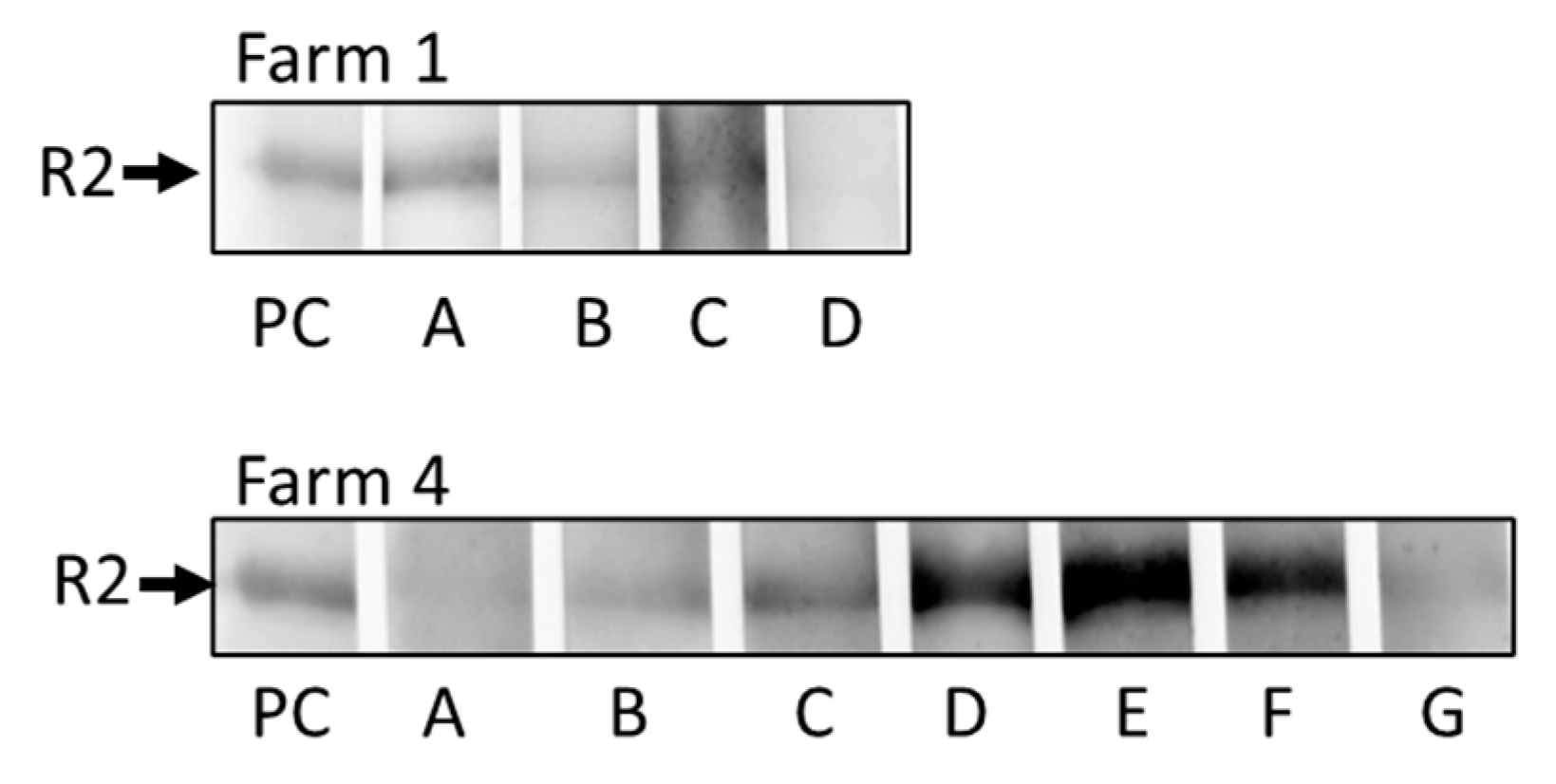
3.5. Western blot assay to detect antibodies against PCMV/PRV
4. Discussion
| Animal |
Age (months) |
PCMV/PRV | PLHV-1 |
PLHV -2 |
PLHV -3 |
PPV-1 | PCV2 | PCV3 | PCV4 | HEV | PERV-C | PERV-A/C | |
| Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time RT-PCR | PCR | PCR | |||
| Farm 1 | |||||||||||||
| 1 | 8-9 | n.d. | 33.75 | 33.31 | 28.49 | n.d. | 31.1 | n.d. | n.d. | n.d. | + | - | |
| 2 | 8-9 | 34.31 | n.d. | 28.74 | 28.09 | n.d. | 31.35 | 32.85 | n.d. | n.d. | + | - | |
| 3 | 8-9 | n.d. | n.d. | 27.33 | 34.24 | n.d. | 30.35 | n.d. | n.d. | n.d. | - | - | |
| 4 | 8-9 | 33.49 | n.d. | 27.55 | 26.72 | n.d. | 27.58 | 34.02 | n.d. | n.d. | + | - | |
| Farm 2 | |||||||||||||
| 1 | 11-12 | n.d. | 33.51 | n.d. | 29.91 | n.d. | 30.12 | n.d. | n.d. | n.d. | + | - | |
| 2 | 11-12 | 33.56 | 32.87 | n.d. | 22.74 | n.d. | 32.43 | n.d. | n.d. | n.d. | - | - | |
| 3 | 11-12 | 34.92 | 32.33 | 30.57 | 33.8 | n.d. | 34.52 | 25.17 | n.d. | n.d. | + | - | |
| Farm 3 | |||||||||||||
| 1 | 4 | 33.44 | n.d. | 31.15 | 22.86 | n.d. | 18.66 | 29.69 | n.d. | n.d. | - | - | |
| 2 | 36 | 35.32 | n.d. | 31.98 | 32.19 | n.d. | 33.37 | 29.45 | n.d. | n.d. | - | - | |
| 3 | 4 | 32.00 | n.d. | 32.34 | 27.27 | n.d. | 32.8 | 28.02 | n.d. | n.d. | + | - | |
| 4 | 4 | 31.94 | 28.99 | 27.04 | 36.44 | n.d. | 23.54 | n.d. | n.d. | n.d. | - | - | |
| 5 | 5 | n.d. | 33.46 | n.d. | 31.8 | n.d. | 34.51 | n.d. | n.d. | n.d. | + | - | |
| 6 | 5 | n.d. | 32.44 | n.d. | 34.33 | n.d. | 34.58 | n.d. | n.d. | n.d. | + | - | |
| Farm 4 | |||||||||||||
| 1 | 10-11 | 29.94 | 33.98 | 15.69 | 24.24 | n.d. | 28.99 | n.d. | n.d. | n.d. | - | - | |
| 2 | 10-11 | 29.8 | 28.23 | 25.58 | 24.54 | n.d. | 29.98 | n.d. | n.d. | n.d. | - | - | |
| 3 | 10-11 | 29.42 | 30.22 | n.d. | 31.52 | n.d. | 21.69 | n.d. | n.d. | n.d. | + | - | |
| 4 | 10-11 | 32.60 | n.d. | 29.88 | 27.26 | n.d. | 28.62 | n.d. | n.d. | n.d. | + | - | |
| 5 | 10-11 | 32.47 | n.d. | 28.43 | 20.98 | n.d. | 31.14 | n.d. | n.d. | n.d. | - | - | |
| 6 | 10-11 | 30.21 | 31.38 | 28.85 | 22.51 | n.d. | 26.1 | n.d. | n.d. | n.d. | - | - | |
| 7 | 10-11 | 31.41 | 29.74 | n.d. | 31.79 | n.d. | 23.02 | n.d. | n.d. | n.d. | + | - | |
| 8 | 10-11 | 31.70 | n.d. | 29.83 | 21.95 | n.d. | 26.03 | n.d. | n.d. | n.d. | - | - | |
| Animal | Organ | PCMV | pGAPDH |
|---|---|---|---|
| 1 | spleen | n.d. | 19.10 |
| liver | n.d. | 19.72 | |
| 2 | spleen | 31.34 | 18.58 |
| liver | 34.31 | 19.41 | |
| 3 | spleen | n.d. | 19.57 |
| liver | n.d. | 19.89 | |
| 4 | spleen | 32.32 | 20.00 |
| liver | 33.49 | 19.17 |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denner, J. The porcine virome and xenotransplantation. Virol. J. 2017, 14, 171. [CrossRef]
- Karlsson, O.E.; Larsson, J.; Hayer, J.; Berg, M.; Jacobson, M. The intestinal eukaryotic virome in healthy and diarrhoeic neonatal piglets. PLoS One. 2016, 11, e0151481. [CrossRef]
- Zhang, B.; Tang, C.; Yue, H.; Ren, Y.; Song, Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J. Gen. Virol. 2014, 95(Pt 7), 1603-1611. [CrossRef]
- Paim, W.P.; Maggioli, M.F.; Weber, M.N.; Rezabek, G.; Narayanan, S.; Ramachandran, A.; Canal, C.W.; Bauermann, F.V. Virome characterization in serum of healthy show pigs raised in Oklahoma demonstrated great diversity of ssDNA viruses. Virology. 2021, 556, 87-95. [CrossRef]
- Schuele, L.; Lizarazo-Forero, E.; Strutzberg-Minder, K.; Schütze, S.; Löbert, S.; Lambrecht, C.; Harlizius, J.; Friedrich, A.W.; Peter, S.; Rossen, J.W.A.; Couto, N. Application of shotgun metagenomics sequencing and targeted sequence capture to detect circulating porcine viruses in the Dutch-German border region. Transbound. Emerg. Dis. 2022, 69, 2306-2319. [CrossRef]
- Griffith, B.P.; Goerlich, C.E.; Singh, A.K.; Rothblatt, M.; Lau, C.L.; Shah, A.; Lorber, M.; Grazioli, A.; Saharia, K.K.; Hong, S.N.; Joseph, S.M.; Ayares, D.; Mohiuddin, M.M. Genetically Modified Porcine-to-Human Cardiac Xenotransplantation. N. Engl. J. Med. 2022, 387, 35-44. [CrossRef]
- https://www.statnews.com/2023/09/22/pig-heart-transplant-university-of-maryland/ (assessed November 13, 2023).
- Denner, J. Zoonosis and xenozoonosis in xenotransplantation: A proposal for a new classification. Zoonoses Public Health. 2023. [CrossRef]
- Denner, J. Reduction of the survival time of pig xenotransplants by porcine cytomegalovirus. Virol. J. 2018, 15, 171. [CrossRef]
- Denner, J.; Längin, M.; Reichart, B.; Krüger, L.; Fiebig, U.; Mokelke, M.; Radan, J.; Mayr, T.; Milusev, A.; Luther, F.; Sorvillo, N.; Rieben, R.; Brenner, P.; Walz, C.; Wolf, E.; Roshani, B.; Stahl-Hennig, C.; Abicht, J.M. Impact of porcine cytomegalovirus on long-term orthotopic cardiac xenotransplant survival. Sci. Rep. 2020, 10, 17531. [CrossRef]
- Mohiuddin, M.M.; Singh, A.K.; Scobie, L.; Goerlich, C.E.; Grazioli, A.; Saharia, K.; Crossan, C.; Burke, A.; Drachenberg, C.; Oguz, C.; Zhang, T.; Lewis, B.; Hershfeld, A.; Sentz, F.; Tatarov, I.; Mudd, S.; Braileanu, G.; Rice, K.; Paolini, J.F.; Bondensgaard, K.; Vaught, T.; Kuravi, K.; Sorrells, L.; Dandro, A.; Ayares, D.; Lau, C.; Griffith, B.P. Graft dysfunction in compassionate use of genetically engineered pig-to-human cardiac xenotransplantation: a case report. Lancet. 2023, 29,. [CrossRef]
- Denner, J. Virus Safety of Xenotransplantation. Viruses. 2022, 14, 1926. [CrossRef]
- Denner, J. Sensitive detection systems for infectious agents in xenotransplantation. Xenotransplantation. 2020, e12594. [CrossRef]
- Wynyard, S.; Nathu, D.; Garkavenko, O.; Denner, J.; Elliott, R. Microbiological safety of the first clinical pig islet xenotransplantation trial in New Zealand. Xenotransplantation. 2014, 21, 309-23.
- Garkavenko, O.; Dieckhoff, B.; Wynyard, S.; Denner, J.; Elliott, R.B.; Tan, P.L.; Croxson, M.C. Absence of transmission of potentially xenotic viruses in a prospective pig to primate islet xenotransplantation study. J. Med. Virol. 2008, 80, 2046-52. [CrossRef]
- Semaan, M.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Screening pigs for xenotransplantation: prevalence and expression of porcine endogenous retroviruses in Göttingen minipigs. Xenotransplantation. 2013, 20, 148-56.
- Morozov, V.A.; Morozov, A.V.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Extended Microbiological Characterization of Göttingen Minipigs in the Context of Xenotransplantation: Detection and Vertical Transmission of Hepatitis E Virus. PLoS One. 2015, 10, e0139893.
- Morozov, V.A.; Ludwig, S.; Ludwig, B.; Rotem, A.; Barkai, U.; Bornstein, S.R.; Denner, J. Islet cell transplantation from Göttingen minipigs to cynomolgus monkeys: analysis of virus safety. Xenotransplantation. 2016, 23, 320-7. [CrossRef]
- Morozov, V.A.; Plotzki, E.; Rotem, A.; Barkai, U.; Denner, J. Extended microbiological characterization of Göttingen minipigs: porcine cytomegalovirus and other viruses. Xenotransplantation. 2016, 23, 490-496.
- Fiebig, U.; Fischer, K.; Bähr, A.; Runge, C.; Schnieke, A.; Wolf, E.; Denner, J. Porcine endogenous retroviruses: Quantification of the copy number in cell lines, pig breeds, and organs. Xenotransplantation. 2018, 25, e12445.
- Krüger, L.; Kristiansen, Y.; Reuber, E.; Möller, L.; Laue, M.; Reimer, C.; Denner, J. A Comprehensive Strategy for Screening for Xenotransplantation-Relevant Viruses in a Second Isolated Population of Göttingen Minipigs. Viruses. 2019, 12, 38.
- Halecker, S.; Krabben, L.; Kristiansen, Y.; Krüger, L.; Möller, L.; Becher, D.; Laue, M.; Kaufer, B.; Reimer, C.; Denner, J. Rare isolation of human-tropic recombinant porcine endogenous retroviruses PERV-A/C from Göttingen minipigs. Virol. J. 2022, 19, 30.
- Jhelum, H.; Grand, N.; Jacobsen, K.R.; Halecker, S.; Salerno, M.; Prate, R.; Krüger, L.; Kristiansen, Y.; Krabben, L.; Möller, L.; Laue, M.; Kaufer, B.; Kaaber, K.; Denner, J. First virological and pathological study of Göttingen Minipigs with Dippity Pig Syndrome (DPS). PLoS One. 2023, 18, e0281521. [CrossRef]
- Plotzki, E.; Heinrichs, G.; Kubícková, B.; Ulrich, R.G.; Denner, J. Microbiological characterization of a newly established pig breed, Aachen Minipigs. Xenotransplantation. 2016, 23, 159-67.
- Halecker, S.; Metzger, J.; Strube, C.; Krabben, L.; Kaufer, B.; Denner, J. Virological and Parasitological Characterization of Mini-LEWE Minipigs Using Improved Screening Methods and an Overview of Data on Various Minipig Breeds. Microorganisms. 2021, 9, 2617.
- Abicht, J.M.; Mayr, T.A.; Reichart, B.; Plotzki, E.; Güthoff, S.; Falkenau, A.; Kind, A.; Denner, J. Hepatic Failure After Pig Heart Transplantation Into a Baboon: No Involvement of Porcine Hepatitis E Virus. Ann. Transplant. 2016, 21, 12-6.
- Morozov, V.A.; Abicht, J.M.; Reichart, B.; Mayr, T.; Guethoff, S.; Denner, J. Active Replication of Porcine Cytomegalovirus (PCMV) Following Transplantation of a Pig Heart into a Baboon despite Undetected Virus in the Donor Pig. Ann. Virol. Res. 2016, 2, 1018.
- Egerer, S.; Fiebig, U.; Kessler, B.; Zakhartchenko, V.; Kurome, M.; Reichart, B.; Kupatt, C.; Klymiuk, N.; Wolf, E.; Denner, J.; Bähr, A. Early weaning completely eliminates porcine cytomegalovirus from a newly established pig donor facility for xenotransplantation. Xenotransplantation. 2018, 25, e12449.
- Fiebig, U.; Abicht, J.M.; Mayr, T.; Längin, M.; Bähr, A.; Guethoff, S.; Falkenau, A.; Wolf, E.; Reichart, B.; Shibahara, T.; Denner, J. Distribution of Porcine Cytomegalovirus in Infected Donor Pigs and in Baboon Recipients of Pig Heart Transplantation. Viruses. 2018, 10, 66.
- Krüger, L.; Längin, M.; Reichart, B.; Fiebig, U.; Kristiansen, Y.; Prinz, C.; Kessler, B.; Egerer, S.; Wolf, E.; Abicht, J.M.; Denner, J. Transmission of Porcine Circovirus 3 (PCV3) by Xenotransplantation of Pig Hearts into Baboons. Viruses. 2019, 11, 650. [CrossRef]
- Halecker, S.; Hansen, S.; Krabben, L.; Ebner, F.; Kaufer, B.; Denner, J. How, where and when to screen for porcine cytomegalovirus (PCMV) in donor pigs for xenotransplantation. Sci. Rep. 2022, 12, 21545. [CrossRef]
- Halecker, S.; Papatsiros, V.; Psalla, D.; Krabben, L.; Kaufer, B.; Denner, J. Virological Characterization of Pigs with Erythema Multiforme. Microorganisms. 2022, 10, 652. [CrossRef]
- Krüger, L.; Stillfried, M.; Prinz, C.; Schröder, V.; Neubert, L.K.; Denner, J. Copy Number and Prevalence of Porcine Endogenous Retroviruses (PERVs) in German Wild Boars. Viruses. 2020, 12, 419.
- Hansen, S.; Menandro, M.L.; Franzo, G.; Krabben, L.; Marino, S.F.; Kaufer, B.; Denner, J. Presence of porcine cytomegalovirus, a porcine roseolovirus, in wild boars in Italy and Germany. Arch. Virol. 2023, 168, 55.
- Homer. The Odyssey. Translation by Ian Johnston. Book Fourteen. Odysseus Meets Eumaeus. Available online: https://www.hellenicaworld.com/Greece/Literature/Homer/en/Odyssey14.html (assessed on July, 27, 2023).
- Michailidou, S.; Kalivas, A.; Ganopoulos, I.; Stea, E.; Michailidis, G.; Tsaftaris, A.; Argiriou, A. A multi-farm assessment of Greek black pig genetic diversity using microsatellite molecular markers. Genet. Mol. Res. 2014, 13, 2752-2765.
- Papakonstantinou, G.I.; Arsenakis, I.; Pourlis, A.; Papatsiros, V.G. Animal Health and Productivity of Organic Greek Pig Farms: The Current Situation and Prospects for Sustainability. Animals. 2023, 13, 2834.
- Mueller, N.J.; Barth, R.N.; Yamamoto, S.; Kitamura, H.; Patience, C.; Yamada, K.; Cooper, D.K.; Sachs, D.H.; Kaur, A.; Fishman, J.A. Activation of cytomegalovirus in pig-to-primate organ xenotransplantation. J. Virol. 2002, 76, 4734-40. [CrossRef]
- Chmielewicz, B.; Goltz, M.; Franz, T.; Bauer, C.; Brema, S.; Ellerbrok, H.; Beckmann, S.; Rziha, H.-J.; Lahrmann, K.-H.; Romero, C.; et al. A novel porcine gammaherpesvirus. Virol. 2003, 308, 317–329. [CrossRef]
- McMahon, K.J.; Minihan, D.; Campion, E.M.; Loughran, S.T.; Allan, G.; McNeilly, F.; Walls, D. Infection of pigs in Ireland with lymphotropic gamma-herpesviruses and relationship to postweaning multisystemic wasting syndrome. Vet. Microbiol. 2006, 116, 60–6.
- Chen, N.; Xiao, Y.; Li, X.; Li, S.; Xie, N.; Yan, X.; Li, X.; Zhu, J. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 2021, 68, 1615–1624. [CrossRef]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879. [CrossRef]
- Opriessnig, T.; Shen, H.G.; Pal, N.; Ramamoorthy, S.; Huang, Y.W.; Lager, K.M.; Beach, N.M.; Halbur, P.G.; Meng, X. A Live-Attenuated Chimeric Porcine Circovirus Type 2 (PCV2) Vaccine Is Transmitted to Contact Pigs but Is Not Upregulated by Concurrent Infection with Porcine Parvovirus (PPV) and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and Is Efficacious in a PCV2b-PRRSV-PPV Challenge Model. Clin. Vaccine Immunol. 2011, 18, 1261–1268.
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods. 2006, 131, 65–71.
- Kaulitz, D.; Mihica, D.; Adlhoch, C.; Semaan, M.; Denner, J. Improved pig donor screening including newly identified variants of porcine endogenous retrovirus-C (PERV-C). Arch. Virol. 2013, 158, 341-8.
- Wood, J.C.; Quinn, G.; Suling, K.M.; Oldmixon, B.A.; Van Tine, B.A.; Cina, R.; Arn, S.; Huang, C.A.; Scobie, L.; Onions, D.E.; et al. Identification of Exogenous Forms of Human-Tropic Porcine Endogenous Retrovirus in Miniature Swine. J. Virol. 2004, 78, 2494–2501.
- Duvigneau, J.; Hartl, R.; Groiss, S.; Gemeiner, M. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J. Immunol. Methods. 2005, 306, 16–27.
- Plotzki E, Keller M, Ivanusic D, Denner J. A new Western blot assay for the detection of porcine cytomegalovirus (PCMV). J Immunol Methods. 2016, 437, 37-42. [CrossRef]
- Hansen S, Fischer K, Krabben L, Rinke Carrapeiro A, Klinger B, Schnieke A, Kaufer B, Denner J. Detection of porcine cytomegalovirus, a roseolovirus, in pig ovaries and follicular fluid: implications for somatic cells nuclear transfer, cloning and xenotransplantation. Virol J. 2023, 20, 15. [CrossRef]
- Denner J, Mankertz A. Porcine Circoviruses and Xenotransplantation. Viruses. 2017, 9, 83. [CrossRef]
- Morozov, V.A.; Morozov, A.V.; Denner, J. New PCR diagnostic systems for the detection and quantification of the porcine cytomegalovirus (PCMV). Arch. Virol. 2016, 161, 1159–1168. [CrossRef]
- Zou, J.; Liu, H.; Chen, J.; Zhang, J.; Li, X.; Long, Y.; Jiang, Y.; Li, W.; Zhou, B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in East China from 2020 to 2022. Vet. Sci. 2022, 10, 29.
- Fischer N, Gulich B, Keßler B, Längin M, Fishman JA, Wolf E, Boller K, Tönjes RR, Godehardt AW. PCR and peptide based PCMV detection in pig - development and application of a combined testing procedure differentiating newly from latent infected pigs. Xenotransplantation. 2023, 30, e12803. [CrossRef]
- Denner J, Jhelum H, Hansen S, Kaufer BB, Comparison of methods for the detection of porcine cytomegalovirus/roseolovirus in relation to biosafety monitoring of xenotransplantation products. Xenotransplantation, in press.
- Denner J. Porcine Lymphotropic Herpesviruses (PLHVs) and Xenotranplantation. Viruses. 2021;13:1072.
- Hartline, C.B.; Conner, R.L.; James, S.H.; Potter, J.; Gray, E.; Estrada, J.; Tector, M.; Tector, A.J.; Prichard, M.N. Xenotransplantation panel for the detection of infectious agents in pigs. Xenotransplantation 2018, 25, e12427. [CrossRef]
- Mueller, N.J.; Livingston, C.; Knosalla, C.; Barth, R.N.; Yamamoto, S.; Gollackner, B.; Dor, F.J.; Buhler, L.; Sachs, D.H.; Yamada, K.; et al. Activation of porcine cytomegalovirus, but not porcine lymphotropic herpesvirus, in pig-to-baboon xenotransplantation. J. Infect.Dis. 2004, 189, 1628–1633.
- Segalés J, Sibila M. Revisiting Porcine Circovirus Disease Diagnostic Criteria in the Current Porcine Circovirus 2 Epidemiological Context. Vet Sci. 2022, 9, 110.
- Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Investig. 2007, 19, 591–615.
- Faccini S, Barbieri I, Gilioli A, Sala G, Gibelli LR, Moreno A, Sacchi C, Rosignoli C, Franzini G, Nigrelli A. Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound. Emerging Dis. 2017, 64, 1661. [CrossRef]
- Jiang, H., Wang, D., Wang, J., Zhu, S., She, R., Ren, X., Tian, J., Quan, R., Hou, L., Li, Z., Chu, J., Guo, Y., Xi, Y., Song, H., Yuan, F., Wei, L., Liu, J., 2019. Induction of Porcine Dermatitis and Nephropathy Syndrome in Piglets by Infection with Porcine Circovirus Type 3. J. Virol. 2019, 93, e02045-18.93. [CrossRef]
- Franzo, G., He, W., Correa-Fiz, F., Li, G., Legnardi, M., Su, S., Segalés, J., 2019b. A Shift in Porcine Circovirus 3 (PCV-3) History Paradigm: Phylodynamic Analyses Reveal an Ancient Origin and Prolonged Undetected Circulation in the Worldwide Swine Population. Adv Sci (Weinh) 2019, 6, 1901004.
- Zhang HH, Hu WQ, Li JY, et al. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transboundary and emerging diseases. 2020, 67, 1057-1061.
- Holgado-Martín R, Arnal JL, Sibila M, Franzo G, Martín-Jurado D, Risco D, Segalés J, Gómez L. First detection of porcine circovirus 4 (PCV-4) in Europe. Virol J. 2023, 20, 230.
- Mengeling W, Lager K, Vorwald A. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim Reprod Sci. 2000, 60, 199–210. [CrossRef]
- Wang B, Meng XJ. Hepatitis E virus: host tropism and zoonotic infection. Curr Opin Microbiol. 2021, 59, 8-15. [CrossRef]
- Denner J. Xenotransplantation and Hepatitis E virus. Xenotransplantation. 2015, 22, 167-73. [CrossRef]
- Krüger L, Stillfried M, Prinz C, Schröder V, Neubert LK, Denner J. Copy Number and Prevalence of Porcine Endogenous Retroviruses (PERVs) in German Wild Boars. Viruses. 2020, 12, 419. [CrossRef]
- Pal, N.; Baker, R.; Schalk, S.; Scobie, L.; Tucker, A.W.; Opriessnig, T. Detection of porcine endogenous retrovirus (PERV) viremia in diseased versus healthy US pigs by qualitative and quantitative real-time RT-PCR. Transbound. Emerg. Dis. 2011, 58, 344–351.
- Liu, G.; Li, Z.; Pan, M.; Ge, M.; Wang, Y.; Gao, Y. Genetic prevalence of porcine endogenous retrovirus in chinese experimental miniature pigs. Transplant. Proc. 2011, 43, 2762–2769. [CrossRef]
- Wu, J.; Ma, Y.; Lv, M.; Yang, Y.; Guo, Y.; Yu, X.; Tian, K.; Zhang, J. Large-scale survey of porcine endogenous retrovirus in Chinese miniature pigs. Comp. Immunol. Microbiol. Infect. Dis. 2008, 31, 367–371. [CrossRef]
- Harrison I, Takeuchi Y, Bartosch B, Stoye JP. Determinants of high titer in recombinant porcine endogenous retroviruses. J Virol. 2004, 78, 13871-1389. [CrossRef]
- Scobie L, Taylor S, Wood JC, Suling KM, Quinn G, Meikle S, Patience C, Schuurman HJ, Onions DE. Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature Swine. J Virol. 2004, 78, 2502-2509.
- Hayward JA, Tachedjian M, Johnson A, Irving AT, Gordon TB, Cui J, Nicolas A, Smith I, Boyd V, Marsh GA, Baker ML, Wang LF, Tachedjian G. Unique Evolution of Antiviral Tetherin in Bats. J Virol. 2022, 96, e0115222.
- Fernandes AP, Águeda-Pinto A, Pinheiro A, Rebelo H, Esteves PJ. Evolution of TRIM5 and TRIM22 in Bats Reveals a Complex Duplication Process. Viruses. 2022, 14, 345.
- Hayward JA, Tachedjian M, Cui J, Cheng AZ, Johnson A, Baker ML, Harris RS, Wang LF, Tachedjian G. Differential Evolution of Antiretroviral Restriction Factors in Pteropid Bats as Revealed by APOBEC3 Gene Complexity. Mol Biol Evol. 2018, 35, 1626-1637. [CrossRef]
- Weinberg M, Yovel Y. Revising the paradigm: Are bats really pathogen reservoirs or do they possess an efficient immune system? iScience. 2022, 25, 104782.
- Calisher, C.H., Childs, J.E., Field, H.E., Holmes, K.V., and Schountz, T. Bats: important reservoir hosts of emerging viruses. Clin.Microbiol. Rev. 2006, 19, 531–545. [CrossRef]
- Geraci C, et al., Genetic markers associated with resistance to infectious diseases have no effects on production traits and haematological parameters in Italian Large White pigs, Livestock Science 2019, 223, 32-38. [CrossRef]
- Ribani A. et al. Signatures of Admixture and Genetic Uniqueness in the Autochthonous Greek Black Pig Breed Deduced from Gene Polymorphisms Affecting Domestication-Derived Traits. Animals 2023, 13, 1763. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





