Submitted:
09 September 2024
Posted:
09 September 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Clinical Sample Collection
DNA Extraction and PCV2 Detection
Complete Genome Sequencing of PCV2
Bioinformatics Analyses
Prevalence Data Analyses
Results
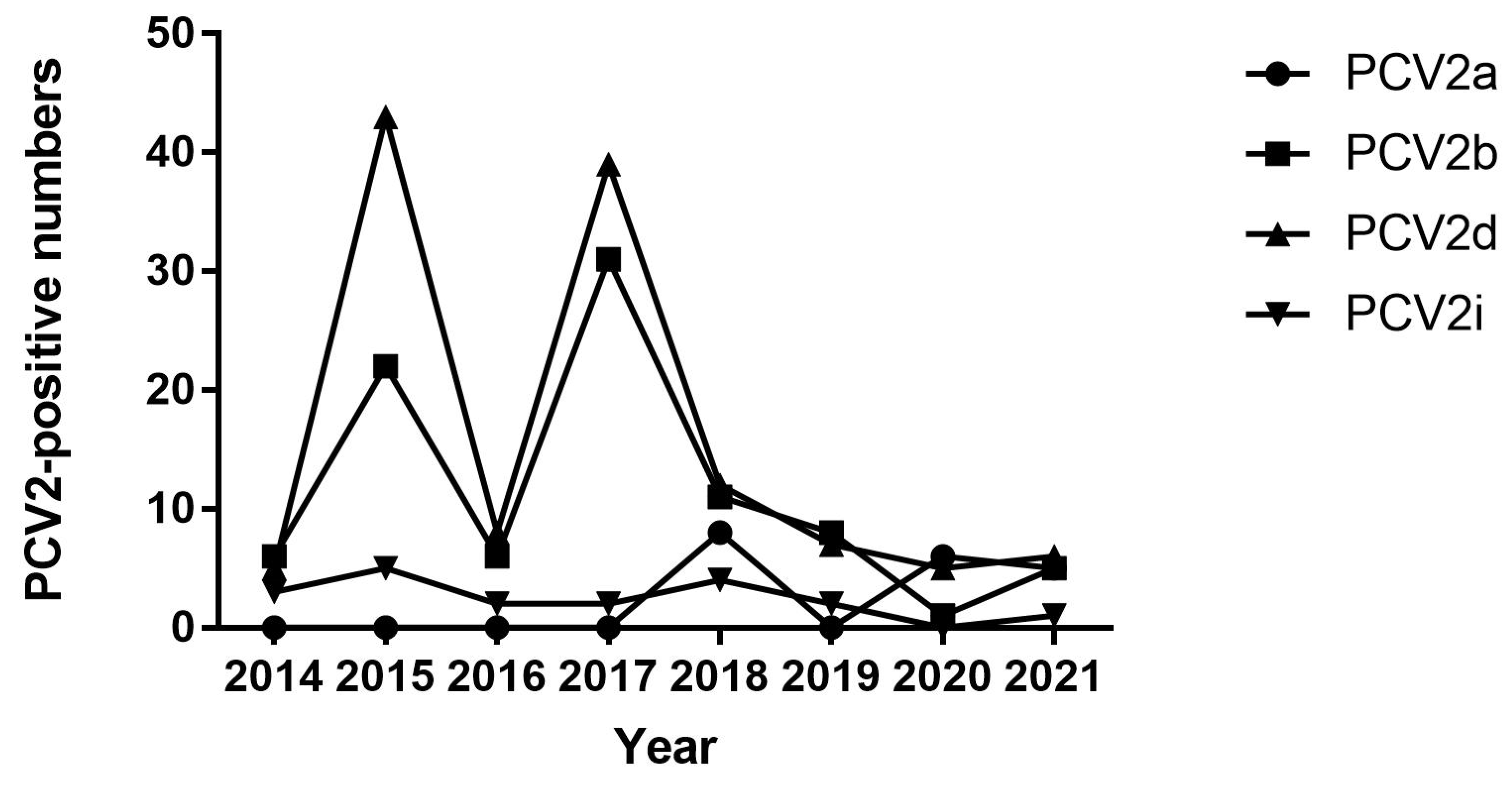
Prevalence of PCV2 in Jiangsu Province from 2014 to 2021
Genome Sequence Analysis
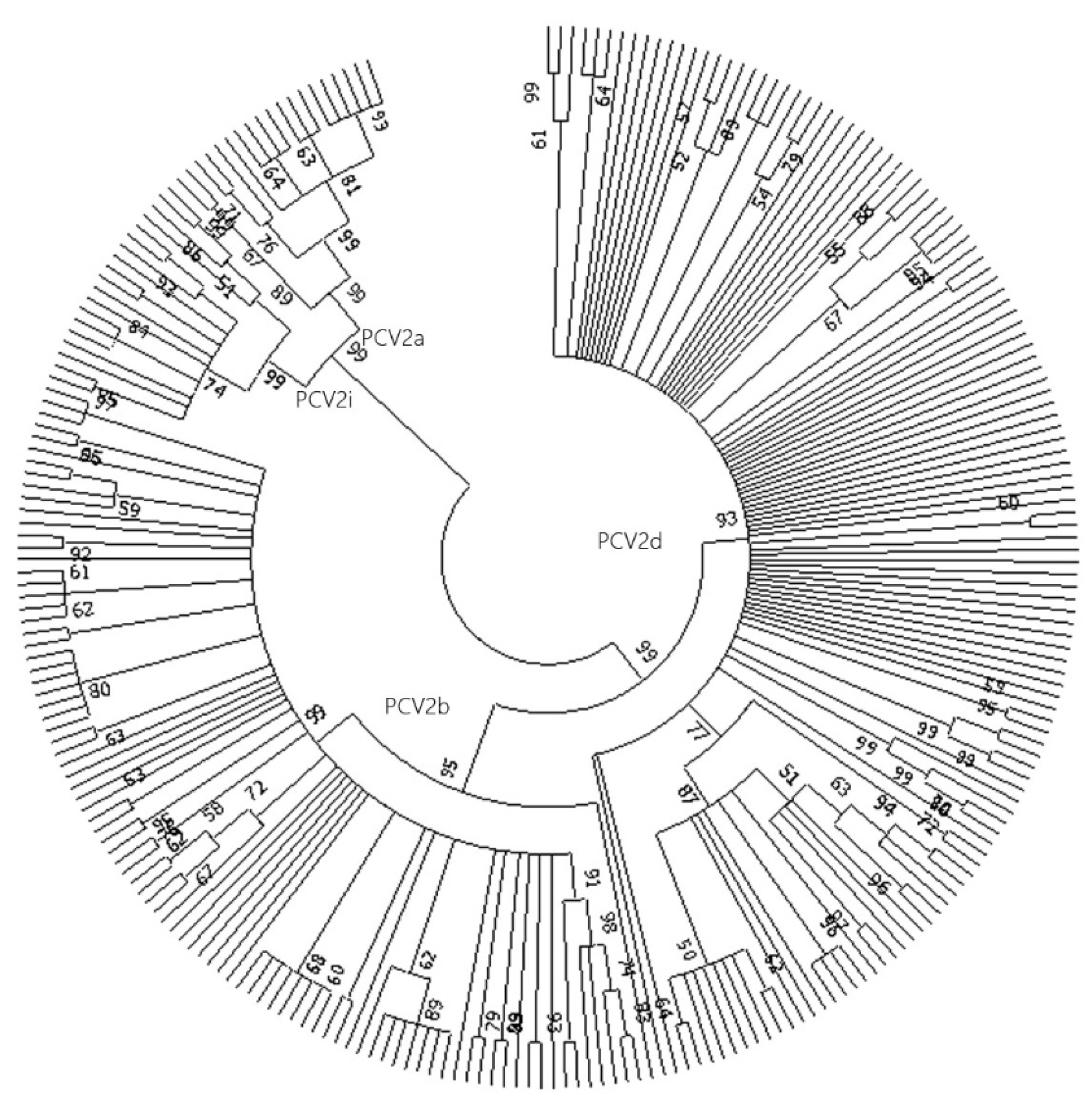
Phylogenetic Analysis
Analysis of Genomic Sequences of PCV2i Strains
Discussion
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tischer, I.; Gelderblom, H.; Vettermann, W.; Koch, M.A. A very small porcine virus with circular single-stranded DNA. Nature 1982, 295, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Meehan, B.M.; McNeilly, F.; Todd, D.; Kennedy, S.; Jewhurst, V.A.; Ellis, J.A.; Hassard, L.E; Clark, E.G.; Haines, D.M.; Allan, G.M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 1998, 79 Pt 9, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2016, 91, e01879–16. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.B.; He, K.W. Genomic rearrangement and recombination of porcine circovirus type 2 and porcine circovirus-like virus P1 in China. Front. Vet. Sci. 2021, 8, 736366. [Google Scholar] [CrossRef]
- Allan, G.M.; Ellis, J.A. Porcine circoviruses: a review. J. Vet. Diagn. Invest. 2000, 12, 3–14. [Google Scholar] [CrossRef]
- Chae, C. A review of porcine circovirus 2-associated syndromes and diseases. Vet. J. 2005, 169, 326–336. [Google Scholar] [CrossRef]
- Opriessnig, T.; Meng, X.J.; Halbur, P.G. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Invest. 2007, 19, 591–615. [Google Scholar] [CrossRef]
- Cheung, A.K. Transcriptional analysis of porcine circovirus type 2. Virology. 2003, 305, 168–180. [Google Scholar] [CrossRef]
- Mankertz, A.; Hillenbrand, B. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 2001, 279, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Grau-Roma, L.; Crisci, E.; Sibila, M.; López-Soria, S.; Nofrarias, M.; Cortey, M.; Fraile, L.; Olvera, A.; Segalés, J. A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence. Vet. Microbiol. 2008, 128, 23–35. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016-2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.J.; Lu, Y.H.; Wei, Y.W.; Huang, L.P.; Liu, C.M. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol. J. 2010, 7, 273. [Google Scholar] [CrossRef]
- Jantafong, T.; Boonsoongnern, A.; Poolperm, P.; Urairong, K.; Lekcharoensuk, C.; Lekcharoensuk, P. Genetic characterization of porcine circovirus type 2 in piglets from PMWS-affected and -negative farms in Thailand. Virol. J. 2011, 8, 88. [Google Scholar] [CrossRef]
- Bao, F.; Mi, S.; Luo, Q.; Guo, H.; Tu, C.; Zhu, G.; Gong, W. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound. Emerg. Dis. 2018, 65, 432–440. [Google Scholar] [CrossRef]
- Weissenbacher-Lang, C.; Kristen, T.; Mendel, V.; Brunthaler, R.; Schwarz, L.; Weissenböck, H. Porcine circovirus type 2 (PCV2) genotyping in Austrian pigs in the years 2002 to 2017. BMC Vet. Res. 2020, 16, 198. [Google Scholar] [CrossRef]
- Trible, B.R.; Rowland, R.R. Genetic variation of porcine circovirus type 2 (PCV2) and its relevance to vaccination, pathogenesis and diagnosis. Virus Res. 2012, 164, 68–77. [Google Scholar] [CrossRef]
- Kwon, T.; Lee, D.U.; Yoo, S.J.; Je, S.H.; Shin, J.Y.; Lyoo, Y.S. Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res. 2017, 228, 24–29. [Google Scholar] [CrossRef]
- Zou, J.; Liu, H.; Chen, J.; Zhang, J.; Li, X.; Long, Y.; Jiang, Y.; Li, W.; Zhou, B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in East China from 2020 to 2022. Vet. Sci. 2022, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; He, K.; Yang, H.; Yu, Z.; Mao, A.; Zhong, S.; Ni, Y.; Zhang, X.; Li, B.; Wang, X.; Zhou, J.; Guo, R.; Lv, L.; Jiang, J. Prevalence of porcine circovirus-like agent P1 in Jiangsu, China. Virol. J. 2011, 8, 543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; He, K.; Wen, L.; Fan, H. Genetic and phylogenetic analysis of a new porcine circovirus type 2 (PCV2) strain in China. Arch. Virol. 2015, 160, 3149–3151. [Google Scholar] [CrossRef]
- Allan, G.M.; Ellis, J.A. Porcine circoviruses: a review. J. Vet. Diagn. Invest. 2000, 12, 3–14. [Google Scholar] [CrossRef]
- Segalés, J.; Domingo, M. Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet. Q. 2002, 24, 109–124. [Google Scholar] [CrossRef]
- Chae, C. Postweaning multisystemic wasting syndrome: a review of aetiology, diagnosis and pathology. Vet. J. 2004, 168, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Fehér, E.; Jakab, F.; Bányai, K. Mechanisms of circovirus immunosuppression and pathogenesis with a focus on porcine circovirus 2: a review. Vet. Q. 2023, 43, 1–18. [Google Scholar] [CrossRef]
- Lang, H.W.; Zhang, G.C.; Wu, F.Q.; Zhang, C.G. Detection of serum antibody against postweaning multisystemic wasting syndrome in pigs. Chin. J. Vet. Sci. Technol. (in Chinese). 2000, 3, 3–5. [Google Scholar]
- Fan, M.; Bian, L.; Tian, X.; Hu, Z.; Wu, W.; Sun, L.; Yuan, G.; Li, S.; Yue, L.; Wang, Y.; Wu, L.; Wang, Y.; Yan, Z.; Ren, J.; Li, X. Infection characteristics of porcine circovirus type 2 in different herds from intensive farms in China, 2022. Front. Vet. Sci. 2023, 10, 1187753. [Google Scholar] [CrossRef]
- Ma, Z.C.; Liu, M.D.; Liu, Z.H.; Meng, F.L.; Wang, H.Y.; Cao, L.L.; Li, Y.; Jiao, Q.L.; Han, Z.F.; Liu, S.D. Epidemiological investigation of porcine circovirus type 2 and its coinfection rate in Shandong Province in China from 2015 to 2018. BMC Vet. Res. 2021, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.M.; Lu, Q.X.; Wang, F.Y.; Xing, G.X.; Feng, H.; Jin, Q.Y.; Guo, Z.H.; Teng, M.; Hao, H.F.; Li, D.L.; Wei, X.; Zhang, Y.H.; Deng, R.G.; Zhang, G.P. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Vet.Res. 2020, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef] [PubMed]
- Hamel, A.L.; Lin, L.L.; Sachvie, C.; Grudeski, E.; Nayar, G.P. PCR detection and characterization of type-2 porcine circovirus. Can. J. Vet. Res. 2000, 64, 44–52. [Google Scholar]
- Takahagi, Y.; Nishiyama, Y.; Toki, S.; Yonekita, T.; Morimatsu, F.; Murakami, H. Genotypic change of porcine circovirus type 2 on Japanese pig farms as revealed by restriction fragment length polymorphism analysis. J. Vet. Med. Sci. 2008, 70, 603–606. [Google Scholar] [CrossRef]
- Wen, L.; Guo, X.; Yang, H. Genotyping of porcine circovirus type 2 from a variety of clinical conditions in China. Vet. Microbiol. 2005, 110, 141–146. [Google Scholar] [CrossRef]
- Wang, H.J.; Gu, J.Y.; Xing, G.; Qiu, X.H.; An, S.T.; Wang, Y.X.; Zhang, C.; Liu, C.M.; Gong, W.J.; Tu, C.C.; Su, S.; Zhou, J.Y. Genetic diversity of porcine circovirus type 2 in China between 1999-2017. Transbound. Emerg. Dis. 2019, 66, 599–605. [Google Scholar] [CrossRef]
- Li, N.; Liu, J.; Qi, J.L.; Hao, F.; Xu, L.; Guo, K.K. Genetic Diversity and Prevalence of Porcine Circovirus Type 2 in China During 2000-2019. Front. Vet. Sci. 2021, 8, 788172. [Google Scholar]
- Guo, L.; Fu, Y.; Wang, Y.; Lu, Y.; Wei, Y.; Tang, Q.; Fan, P.; Liu, J.; Zhang, L.; Zhang, F.; Huang, L.; Liu, D.; Li, S.; Wu, H.; Liu, C. A porcine circovirus type 2 (PCV2) mutant with 234 amino acids in capsid protein showed more virulence in vivo, compared with classical PCV2a /b strain. PLoS ONE 2012, 7, e41463. [Google Scholar] [CrossRef]
- Suh, J.; Oh, T.; Park, K.; Yang, S.; Cho, H.; Chae, C. A comparison of virulence of three porcine circovirus type 2 (PCV2) genotypes (a, b, and d) in pigs singularly inoculated with PCV2 and dually inoculated with PCV2 and porcine reproductive and respiratory syndrome virus. Pathogens 2021, 10, 891. [Google Scholar] [CrossRef]
- Oh, T.; Suh, J.; Park, K.H.; Yang, S.; Cho, H.; Chae, C. A comparison of pathogenicity and virulence of three porcine circovirus type 2 (PCV2) genotypes (a, b, and d) in pigs singularly inoculated with PCV2 and dually inoculated with Mycoplasma hyopneumoniae and PCV2. Pathogens 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhan, Y.; Wang, D.D.; Xie, X.H.; Liu, T.B.; Liu, W.; Wang, N.D.; Deng, Z.B.; Lei, H.Y.; Yang, Y.; Wang, A.B. Evidence of natural co-infection with PCV2b subtypes in vivo. Arch. Virol. 2017, 162, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, T.; Zhang, X.W.; Liu, X.H.; Ren, L.Z. Co-infection of swine with porcine circovirus type 2 and other swine viruses. Viruses 2019, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Yin, L.; Zhu, J.; Li, H.; Zhang, F.; Hu, Q.; Xiao, Q.; Xie, J.; He, K. Nearly 20 Years of Genetic Diversity and Evolution of Porcine Circovirus-like Virus P1 from China. Viruses 2022, 14, 696. [Google Scholar] [CrossRef]
- Zhan, Y.; Yu, W.; Cai, X.; Lei, X.; Lei, H.; Wang, A.; Sun, Y.; Wang, N.; Deng, Z.; Yang, Y. The carboxyl terminus of the porcine circovirus type 2 capsid protein is critical to virus-like particle assembly, cell entry, and propagation. J. Virol. 2020, 94, e00042–e00020. [Google Scholar] [CrossRef]
- Liu, Q.; Tikoo, S.K.; Babiuk, L.A. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 2001, 285, 91–99. [Google Scholar] [CrossRef]
- Lekcharoensuk, P.; Morozov, I.; Paul, P.S.; Thangthumniyom, N.; Wajjawalku, W.; Meng, X.J. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 2004, 78, 8135–8145. [Google Scholar] [CrossRef]
- Shang, S.B.; Jin, Y.L.; Jiang, X.T.; Zhou, J.Y.; Zhang, X.; Xing, G.; He, J.L.; Yan, Y. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol. Immunol. 2009, 46, 327–334. [Google Scholar] [CrossRef]
| Collection Year | Sample type | Sample number | Clinical symptoms |
|---|---|---|---|
| 2014 | lung, serum, diarrheal stool, and lymph node | 408 | PMWS |
| 2015 | lung, diarrheal stool, and lymph node | 553 | PMWS |
| 2016 | lung, diarrheal stool, and lymph node | 459 | PMWS |
| 2017 | lung, serum, diarrheal stool, and lymph node | 463 | PMWS |
| 2018 | lung | 337 | PMWS |
| 2019 | lung, and lymph node | 170 | PMWS |
| 2020 | lung, diarrheal stool | 135 | PMWS |
| 2021 | lung | 150 | PMWS |
| 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 | |
|---|---|---|---|---|---|---|---|---|
| Changzhou | 0.00 | 24.39 | 47.37 | 0.00 | 42.86 | 44.44 | 0.00 | 50.00 |
| Huaian | 26.67 | 30.00 | 14.56 | 30.97 | 15.38 | 66.67 | 36.36 | 68.00 |
| Lianyungang | 58.33 | 65.85 | 32.73 | 48.89 | 22.58 | 71.43 | 47.37 | 62.50 |
| Nanjing | 62.50 | 30.77 | 47.06 | 30.00 | 75.00 | 60.00 | 57.14 | 66.67 |
| Nantong | 54.05 | 26.92 | 35.71 | 65.52 | 25.00 | 87.50 | 50.00 | 56.25 |
| Suzhou | 52.17 | 31.25 | 39.29 | 66.67 | 0.00 | 20.00 | 0.00 | 40.00 |
| Taizhou | 33.90 | 36.17 | 72.73 | 68.29 | 56.25 | 40.00 | 40.00 | 66.67 |
| Wuxi | 100.00 | 53.13 | 88.89 | 0.00 | 66.67 | 0.00 | 40.00 | 33.33 |
| Suqian | 72.22 | 40.28 | 18.95 | 45.45 | 38.89 | 91.67 | 62.50 | 80.00 |
| Xuzhou | 20.00 | 35.71 | 43.75 | 42.42 | 20.18 | 27.27 | 43.75 | 60.00 |
| Yancheng | 17.39 | 62.50 | 20.00 | 33.33 | 45.45 | 100.00 | 41.67 | 55.56 |
| Yangzhou | 76.19 | 66.67 | 56.52 | 20.00 | 33.33 | 100.00 | 85.71 | 80.00 |
| Zhenjiang | 63.16 | 47.62 | 100.00 | 100.00 | 27.89 | 0.00 | 50.00 | 50.00 |
| Nt position | PCV2a | PCV2b | PCV2c | PCV2d | PCV2e | PCV2f | PCV2g | PCV2h | PCV2i |
|---|---|---|---|---|---|---|---|---|---|
| 617 | C | C | C | C | C | C | C | C | T |
| 1164 | C | C | C | C | C | C | G | G | T |
| 1596 | G | G | G | G | G | G | G | G | C |
| 1604 | G | G or T | G | G | G | G | G | G | A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





