1. Introduction
Rare neurodevelopmental disorders caused by single-gene mutations often disrupt complex signaling networks rather than isolated molecular targets. IQSEC2-related encephalopathy is a paradigmatic example of this challenge [
1,
2,
3,
4,
5]. IQSEC2 (IQ motif and SEC7 domain-containing protein 2) encodes a guanine nucleotide exchange factor (GEF) that activates ARF small GTPases, controlling synaptic vesicle trafficking, actin cytoskeleton remodeling, and excitatory synapse development. Pathogenic variants in IQSEC2 lead to intellectual disability, epilepsy, autistic traits, and disrupted synaptic plasticity. Despite its clinical importance, there are no mechanism-based therapies targeting IQSEC2 [
1,
2,
3,
4,
5] IQSEC2 is an X-linked gene encoding a guanine nucleotide exchange factor (GEF) containing an IQ motif and a SEC7 catalytic domain, which specifically activates ARF small GTPases. The name IQSEC2 literally highlights its key functional domains: it has an IQ motif and a Sec7 domain, and it is family member 2 [
1,
2,
3,
4,
5].
IQSEC2 is a GEF (guanine nucleotide exchange factor) for ARF GTPases [
1,
2,
3,
4,
5]. Pathogenic variants in IQSEC2 lead to severe intellectual disability, epilepsy, autistic features, and impaired synaptic plasticity [
5,
6,
7]. IQSEC2 activates ARF GTPases, which then control key neuronal processes. If IQSEC2 is mutated, this “switching” fails, which can lead to intellectual disability and epilepsy [
5,
6,
7] (
https://www.uniprot.org/uniprotkb/Q5JU85/entry).
At the cellular level, IQSEC2 regulates excitatory synapse development, actin cytoskeleton remodeling, and membrane trafficking through ARF activation [
8,
9,
10]. ARF proteins (ARF1–ARF6) coordinate vesicular trafficking and interact with the broader Ras superfamily, particularly Rab GTPases, which regulate cargo transport, membrane identity, and vesicle fusion. Dysregulation of ARF–Rab–Ras signaling is implicated in neurodevelopmental disorders and other diseases. Natural compounds, especially polyphenols, represent attractive candidates for probing these pathways due to their multitarget activity and favorable safety profiles [
11,
12,
13]. ARF proteins, in turn, function in close coordination with other members of the Ras superfamily, particularly Rab GTPases, which orchestrate vesicle transport, endosomal recycling, and synaptic vesicle dynamics. Disruption of this tightly connected ARF–Rab–Ras network is increasingly recognized as a convergent mechanism underlying several neurodevelopmental disorders [
11,
12,
13].
Despite this clear biological relevance, IQSEC2 remains an underexplored pharmacological target. No small-molecule modulators of IQSEC2 or its downstream signaling cascade are currently available [
1,
2,
3,
4,
5].
Natural polyphenols and flavonoids represent attractive candidates for exploratory studies due to their structural diversity, multitarget behavior, and favorable safety profiles in humans [
23].
The aim of this study was therefore to systematically investigate, using molecular docking [
14,
15], whether selected natural compounds could interact with IQSEC2 and associated small GTPases, providing a computational foundation for future experimental research. Polydatin, a glycosylated derivative of resveratrol, has demonstrated neuroprotective, anti-inflammatory, and antioxidant properties [
23,
24,
25,
26]. Here, we integrate new in silico docking results for IQSEC2/ARFs with previously reported Polydatin-Rab/Ras interactions to identify promising network modulators of small GTPase signaling.
2. Computational Methods
2.1. Protein Targets
Fourteen natural compounds were selected, including flavonoids (quercetin, baicalein), glycosylated flavonoids (rutin, hesperidin), stilbenes (resveratrol, polydatin), curcuminoids (curcumin), anthraquinones (hypericin), and biflavonoids (amentoflavone, bilobetin, ginkgetin). Ligands were geometry-optimized using the MMFF94 force field prior to docking.
Proteins were prepared by removing water molecules, adding hydrogens, and assigning Gasteiger charges through Chimera software [
15,
16,
17,
18].
2.2. Docking Protocol
AutoDock Vina was employed for docking. Exhaustiveness was set to 8, and grid boxes were centered on the active or binding sites of each protein. Binding energies (kcal/mol) were recorded, and top poses were analyzed for hydrogen bonds, π-π stacking, and hydrophobic interactions [
14,
15,
16,
17]. Docking simulations were performed using a rigid-receptor/flexible-ligand approach. Binding energies (kcal/mol) were used as comparative indicators of interaction strength. Results were analyzed comparatively across protein families to identify multitarget interaction patterns [
14,
15,
16,
17]. Docking was performed with AutoDock Vina [
14,
15,
16,
17] using rigid receptors and flexible ligands. Grid boxes were defined according to ligand-binding sites reported in PDB. Binding energies were evaluated in kcal/mol and analyzed comparatively across protein families.
2.3. Protein Structures
Protein structures were obtained from the Protein Data Bank (PDB). The following targets were included:
ARF1 (ADP-ribosylation factor 3) (PDB ID: 1HUR)
ARF2 (ADP-ribosylation factor 2) (PDB ID: 1KU1)
ARF3 (ADP-ribosylation factor 3) (PDB ID: 6II6)
ARF4 (ADP-ribosylation factor 4) (PDB ID: 1Z6X)
ARF5 (ADP-ribosylation factor 5) (PDB IDs: 2B6H,)
ARF6 (ADP-ribosylation factor 6) (PDB IDs: 2A5D)
IQSEC2 (PDB ID: 6FAE)
IQSEC1 (PDB ID: 7VMB)
ADP-ribosylation factor GTPase-activating protein 1 (PDB IDs: 3DWD)
Crystal structure of the Sec7 domain of human ARNO (PDB IDs: 1PBV)
Rab/Ras proteins: Rab-4A, Rab-5A, Rab-6A/B, Rab-18, Rab-31, HRas, R-Ras, Rap-2A, Rab-3D, Rab-11B (PDB codes as in Ferrari et al., 2023 preprint) [
31]
3. Results
3.1. Docking to ARF GTPases
Docking analysis revealed that several natural compounds bind strongly to ARF ( ADP-Ribosylation Factors) proteins [
19,
20,
21,
22]. Biflavonoids such as amentoflavone, bilobetin, and ginkgetin consistently showed high binding affinities across ARF1–ARF6, with binding energies frequently below −9.0 kcal/mol. ARF3 and ARF6 appeared particularly susceptible to ligand binding, suggesting potential hot spots for modulation of trafficking-related signaling [
19,
20,
21,
22]. All docking results are presented in
Table 4.
Polydatin ( also known as piceid, a natural stilbenoid, a glucosylated form of resveratrol, it exhibits multiple biological activities [
23,
24,
25,
26]) and it displayed moderate but consistent binding across all ARF isoforms, indicating a stable interaction profile rather than isoform-specific selectivity.
3.2. Docking to IQSEC2 and IQSEC1
Docking against IQSEC2 (6FAE) yielded strong interactions for several compounds. Rutin (−9.9 kcal/mol), hesperidin (−9.8 kcal/mol), amentoflavone (−9.5 kcal/mol), and ginkgetin (−9.4 kcal/mol) were among the top binders. Polydatin also showed reproducible affinity toward IQSEC2.
Comparable binding trends were observed for IQSEC1, suggesting a conserved ligand-recognition pattern within the IQSEC family and supporting the biological relevance of the docking poses.
3.3. Integration with Rab and Ras GTPase Data
Previously reported docking data (doi:10.20944/preprints202312.0367.v1) [
31] demonstrated that polydatin strongly binds multiple Rab proteins (Rab-31, Rab-3D, Rab-6A/B, Rab-5A, Rab-4A), as well as HRas and Rap proteins, with binding energies reaching −10.5 kcal/mol. When integrated with the present ARF and IQSEC2 results, these data highlight polydatin as a unique molecule capable of interacting with several interconnected branches of the Ras superfamily ( See below
Table 1,
Table 2 and
Table 3).
Table 4.
Comparative docking results (Binding energies in kcal/mol) performed by Autodock Vina with Pyrx software [
14].
Table 4.
Comparative docking results (Binding energies in kcal/mol) performed by Autodock Vina with Pyrx software [
14].
| Ligand |
ARF1 (1HUR) |
ARF2 (1KU1) |
ARF3 (6II6) |
ARF4 (1Z6X) |
ARF5 (2B6H) |
ARF6 (2A5D) |
IQSEC2 (6FAE) |
IQSEC1 (7VMB) |
ARF GAP1 (3DWD) |
Sec7 (1PBV) |
| Amentoflavone |
-8.0 |
-7.9 |
-9.7 |
-7.6 |
-8.2 |
-9.9 |
-9.5 |
-9.2 |
-8.4 |
-8.4 |
| Baicalein |
-6.6 |
-7.2 |
-8.3 |
-6.5 |
-6.7 |
-8.2 |
-7.0 |
-8.1 |
-6.9 |
-6.4 |
| Baicalin |
-7.0 |
-7.5 |
-8.0 |
-6.6 |
-7.5 |
-8.5 |
-8.3 |
-8.0 |
-8.1 |
-7.4 |
| Bilobetin |
-7.9 |
-7.9 |
-9.5 |
-8.1 |
-8.1 |
-9.2 |
-9.1 |
-9.2 |
-8.0 |
-7.9 |
| Curcumin |
-6.3 |
-7.4 |
-8.2 |
-6.0 |
-6.3 |
-8.3 |
-5.9 |
-7.0 |
-6.7 |
-6.1 |
| Ginkgetin |
-7.9 |
-7.9 |
-9.5 |
-7.4 |
-8.0 |
-9.1 |
-9.4 |
-8.9 |
-7.8 |
-7.9 |
| Hesperetin |
-6.7 |
-6.8 |
-7.3 |
-6.8 |
-6.9 |
-7.5 |
-7.1 |
-7.3 |
-6.8 |
-6.6 |
| Hesperidin |
-6.2 |
-8.4 |
-8.3 |
-5.5 |
-7.6 |
-8.7 |
-9.8 |
-8.1 |
-8.1 |
-7.7 |
| Hypericin |
-7.2 |
-8.5 |
-9.6 |
-5.3 |
-5.9 |
-9.8 |
-8.6 |
-8.7 |
-8.4 |
-7.9 |
| Polydatin |
-7.0 |
-7.2 |
-8.4 |
-6.3 |
-7.5 |
-9.1 |
-7.6 |
-7.6 |
-7.0 |
-6.6 |
| Quercetin |
-6.7 |
-7.2 |
-8.1 |
-6.2 |
-6.6 |
-8.0 |
-7.9 |
-7.9 |
-6.7 |
-6.7 |
| Resveratrol |
-5.6 |
-6.3 |
-6.3 |
-5.5 |
-5.9 |
-6.6 |
-7.1 |
-6.6 |
-6.5 |
-6.1 |
| Rutin |
-7.1 |
-8.3 |
-8.2 |
-5.6 |
-7.4 |
-7.9 |
-9.9 |
-8.6 |
-7.5 |
-7.5 |
The docking results focused on Polydatin shows the strongest interaction with ARF6, which is a critical downstream effector of IQSEC2. This suggests that Polydatin could modulate the IQSEC2–ARF6 signaling pathway, making it a biologically plausible candidate for experimental exploration in IQSEC2-related disorders.
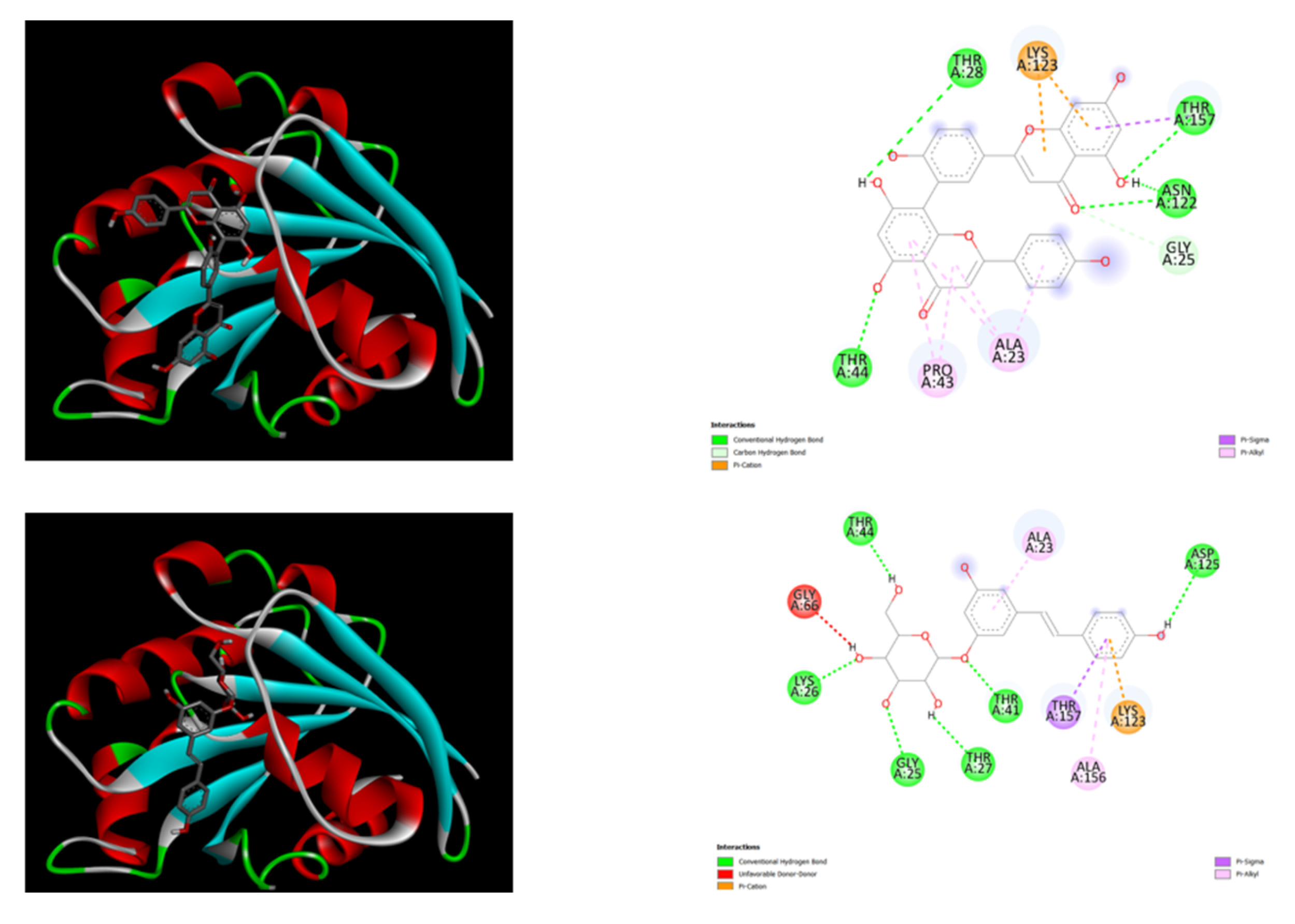
Figure 1.
displays the docking outcomes of ARF6 (ADP-ribosylation factor 6) (PDB IDs: 2A5D) in conjunction with Polydatin and Amentoflavone respectively within the Ligand Binding Site, as analyzed by Autodock Vina through Pyrx software. On the right side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, theleft side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin and Amentoflavone, respectively. The figure was reproduced by Pymol [
32] and Discovery Studio software [33].
Figure 1.
displays the docking outcomes of ARF6 (ADP-ribosylation factor 6) (PDB IDs: 2A5D) in conjunction with Polydatin and Amentoflavone respectively within the Ligand Binding Site, as analyzed by Autodock Vina through Pyrx software. On the right side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, theleft side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin and Amentoflavone, respectively. The figure was reproduced by Pymol [
32] and Discovery Studio software [33].
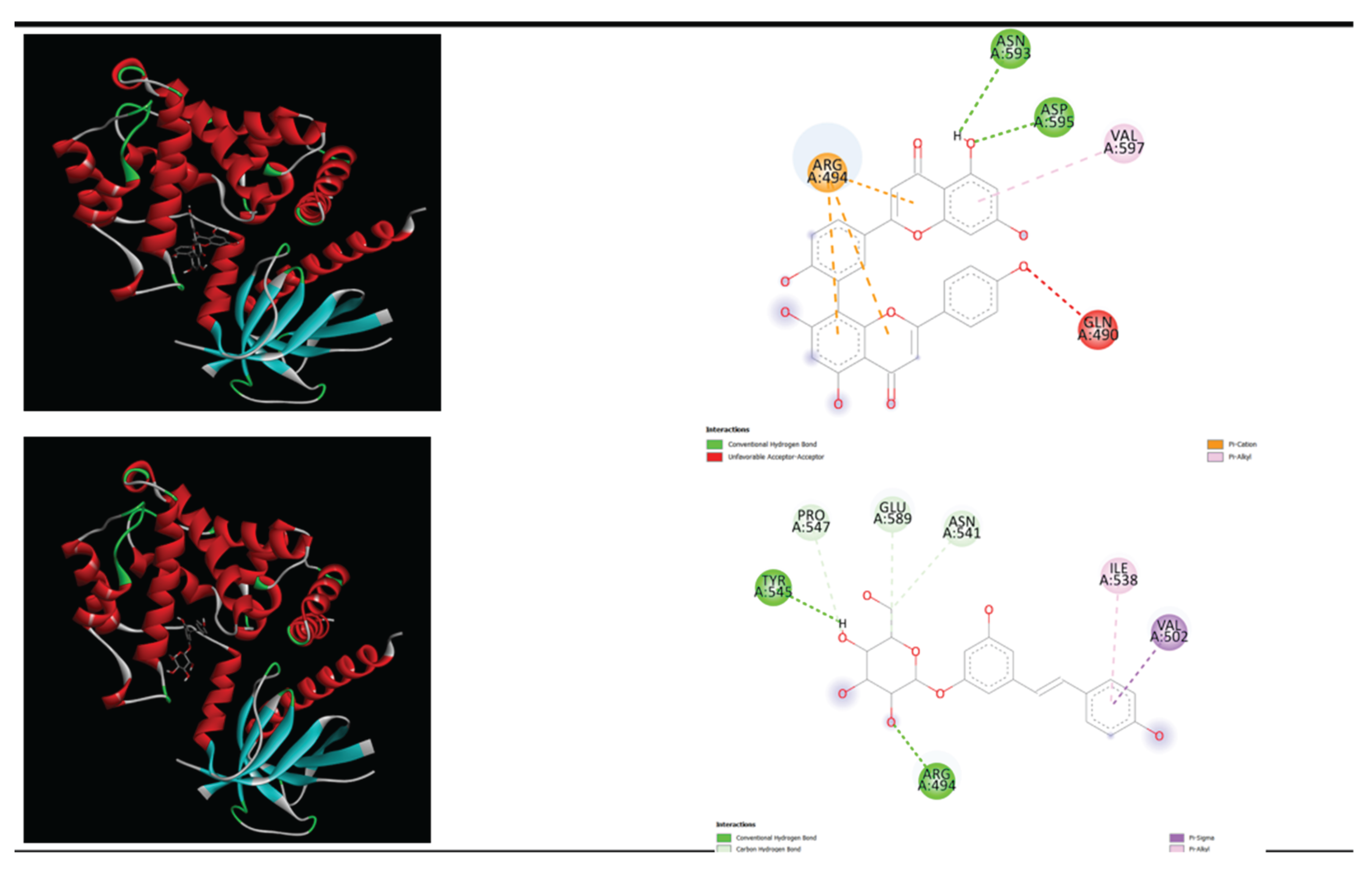
Figure 2.
displays the docking outcomes of IQSEC2 (PDB ID: 6FAE)in conjunction with Polydatin and Amentoflavone respectively within the Ligand Binding Site, as analyzed by Autodock Vina through Pyrx software. On the right side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, theleft side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin and Amentoflavone, respectively. The figure was reproduced by Pymol [
32] and Discovery Studio software [33].
Figure 2.
displays the docking outcomes of IQSEC2 (PDB ID: 6FAE)in conjunction with Polydatin and Amentoflavone respectively within the Ligand Binding Site, as analyzed by Autodock Vina through Pyrx software. On the right side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, theleft side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin and Amentoflavone, respectively. The figure was reproduced by Pymol [
32] and Discovery Studio software [33].
4. Discussion
This study provides a network-oriented interpretation of molecular docking results focused on IQSEC2-related neurodevelopmental disorders. Rather than identifying a single high-affinity inhibitor, our findings suggest that modulation of the ARF–Rab–Ras signaling network may represent a more realistic strategy for conditions driven by synaptic trafficking dysfunction [
11,
12,
13]. Based on the corrected docking table, All docking results are presented in
Table 1) we can now identify and discuss the best natural compounds in terms of binding affinity across ARFs, IQSEC1/2, ARF GAP1, and Sec7 domains.
From the docking results ( See
Table 4) several compounds consistently show strong binding affinities (more negative binding energies) across multiple protein targets. The top candidates include:
-Amentoflavone
Binding energies: Ranged from -7.6 to -9.9 kcal/mol across all ARFs and IQSEC proteins.
-Bilobetin
Binding energies: -7.9 to -9.5 kcal/mol.
-Ginkgetin
Binding energies: -7.4 to -9.5 kcal/mol.
-Rutin
Binding energies: Moderate to strong (-7.1 to -9.9 kcal/mol).
-Hypericin
Binding energies: -5.3 to -9.8 kcal/mol.
From all docking results ( See
Table 4), Amentoflavone emerges as a leading natural modulator of ARF/IQSEC2 signaling, demonstrating high and consistent binding across ARF3, ARF5, IQSEC2, and the Sec7 domain. Its biflavonoid structure, rich in hydroxyl groups, enables robust hydrogen bonding and hydrophobic interactions, suggesting potential to stabilize or inhibit ARF/IQSEC activity in neuronal contexts. Polydatin, while less potent, offers enhanced solubility and bioavailability, highlighting its relevance as a complementary in vivo modulator. These findings position Amentoflavone as a promising candidate for therapeutic exploration in ARF/IQSEC-related pathways.
Biflavonoids exhibited the strongest binding energies but are limited by poor bioavailability [
27,
28] and lack of clinical safety data, particularly in pediatric populations. In contrast, Polydatin emerged as a biologically and translationally plausible candidate [
29,
30].
Although its binding energies are slightly lower than those of biflavonoids, Polydatin shows consistent interactions across IQSEC2, ARF, Rab, and Ras proteins, aligning well with the systems-level nature of IQSEC2 pathology.
Importantly, polydatin is a glycosylated form of resveratrol with improved stability and a favorable safety profile, widely used as a nutraceutical. This does not imply therapeutic efficacy, but it supports its potential role as an experimental probe for studying small GTPase network modulation.
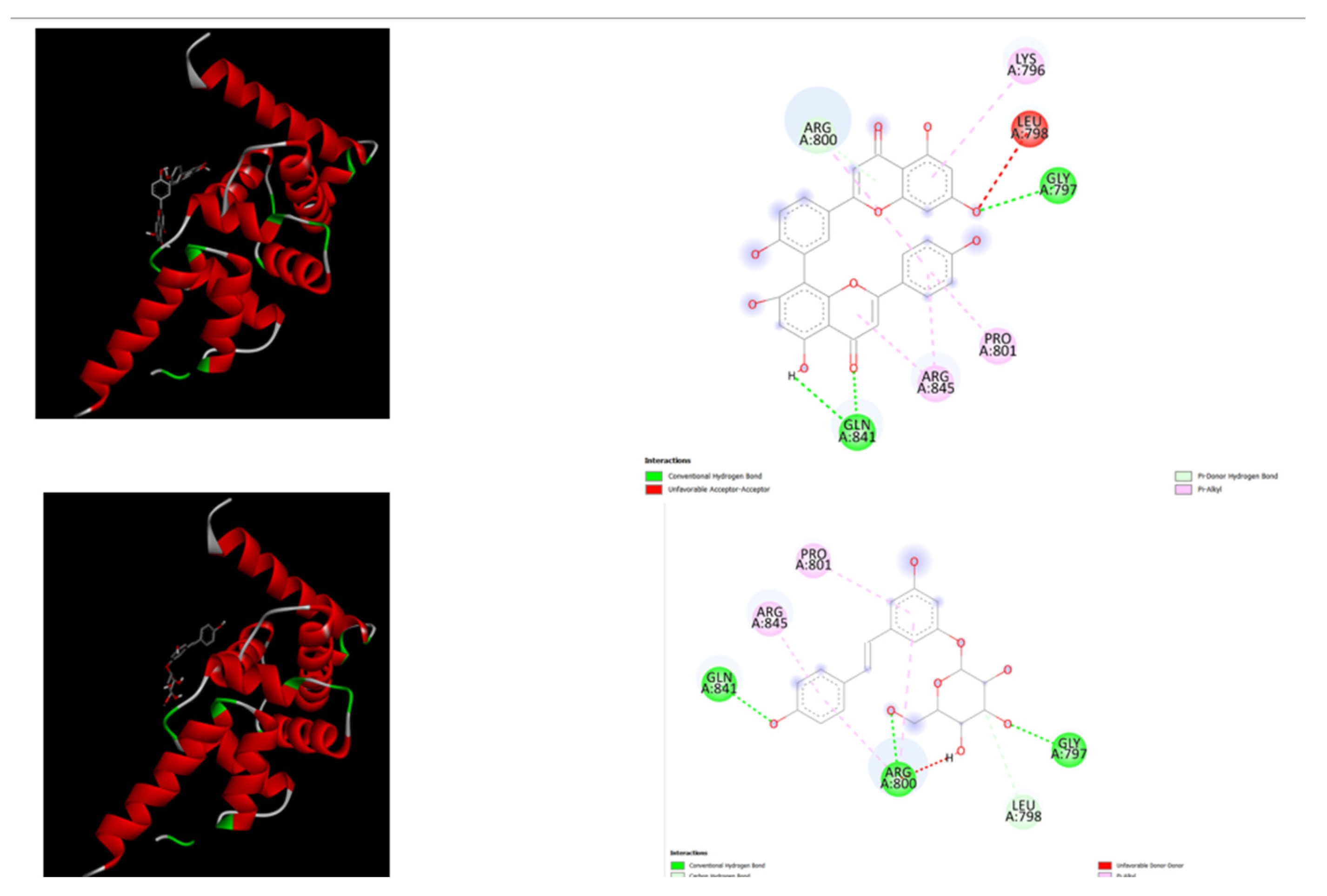
Another aspect of this computational docking study focused on a critical region of IQSEC2 (residues 740–900) to evaluate the binding potential of two natural compounds, Amentoflavone and Polydatin, before and after disease-associated point mutations. Four missense mutations—R758Q, A789V, Q801P, and R863W—were introduced to model clinically relevant variants (
https://www.uniprot.org/uniprotkb/Q5JU85/entry)
In the wild-type IQSEC2 sequence, Amentoflavone and Polydatin displayed strong binding affinities of −9.5 kcal/mol and −7.6 kcal/mol, respectively. Following the introduction of the mutations, binding energies decreased markedly, with Amentoflavone at −6.7 kcal/mol and Polydatin at −5.6 kcal/mol. These results indicate that pathogenic mutations can significantly impair the interaction of potential natural modulators with IQSEC2, potentially limiting their therapeutic efficacy.
Comparison of the two ligands indicates that Amentoflavone maintains relatively stronger binding than Polydatin in both wild-type and mutant contexts, suggesting that biflavonoids may provide more robust interactions with IQSEC2. Polydatin, while exhibiting weaker binding, remains a promising candidate for experimental investigation, especially in wild-type or partially functional protein contexts.
These findings underscore the necessity of mutation-specific analyses in drug discovery for rare genetic disorders. Screening of natural compounds without consideration of patient-specific mutations may overestimate potential efficacy. Computational docking provides a valuable predictive tool to prioritize promising compounds, but experimental validation in vitro and in vivo is essential to confirm biological relevance.
From a network perspective, Polydatin has previously shown favorable interactions with ARF6 and Ras/Rab GTPases, yet the presence of IQSEC2 mutations may reduce its modulatory capacity. This observation highlights the importance of precision medicine approaches, where therapeutic strategies are tailored to the patient’s specific genetic variant.
5. Conclusions
This study presents a comprehensive computational evaluation of natural compounds Polydatin and Amentoflavone as potential modulators of IQSEC2-related signaling pathways. Polydatin exhibited strong binding to ARF6 (−9.1 kcal/mol), IQSEC2 (−7.6 kcal/mol), and IQSEC1 (−7.6 kcal/mol), as well as significant interactions with multiple Ras-related Rab and Ras GTPases, highlighting its potential to modulate synaptic vesicle trafficking and GTPase signaling at a network level. Amentoflavone also demonstrated high binding affinities across ARF isoforms and IQSEC proteins, further supporting its candidacy as a biologically relevant probe.
Figure 3.
displays the docking outcomes of mutated IQSEC2 (AlphaFold,
https://www.uniprot.org/uniprotkb/Q5JU85/entry) in conjunction with Polydatin and Amentoflavone respectively within the Ligand Binding Site, as analyzed by Autodock Vina through Pyrx software. On the right side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, theleft side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin and Amentoflavone, respectively. The figure was reproduced by Pymol [
32] and Discovery Studio software [33].
Figure 3.
displays the docking outcomes of mutated IQSEC2 (AlphaFold,
https://www.uniprot.org/uniprotkb/Q5JU85/entry) in conjunction with Polydatin and Amentoflavone respectively within the Ligand Binding Site, as analyzed by Autodock Vina through Pyrx software. On the right side, 2D diagrams illustrate the residue interactions between the protein and Polydatin. Meanwhile, theleft side exhibits the Ligand Binding Site of the protein, highlighting the specific location of Polydatin and Amentoflavone, respectively. The figure was reproduced by Pymol [
32] and Discovery Studio software [33].
These findings suggest that both Polydatin and Amentoflavone are computationally plausible compounds for experimental studies targeting IQSEC2 dysfunction. By potentially modulating the IQSEC2–ARF6 axis and associated GTPase networks, they may offer a rational starting point for therapeutic exploration in rare IQSEC2-related disorders. However, it is important to note that these results are predictive, and in vitro and in vivo validation is essential to confirm their biological activity and safety. Overall, this work provides a strong rationale for prioritizing Polydatin and Amentoflavone in future experimental investigations aimed at ameliorating the molecular defects underlying IQSEC2 pathology.
Author Contributions
Ivan Vito Ferrari conceived the idea, designed the studies, carried out the research, interpreted the results, and wrote the manuscript.
References
- [1] Mignot, C., McMahon, A. C., Bar, C., Campeau, P. M., Davidson, C., Buratti, J., ... & Depienne, C. (2019). IQSEC2-related encephalopathy in males and females: a comparative study including 37 novel patients. Genetics in Medicine, 21(4), 837-849. [CrossRef]
- [2] Liu, X., Zhang, S., Wan, L., Zhang, X., Wang, H., Zhang, H., ... & Yang, G. (2022). IQSEC2-related encephalopathy in male children: Novel mutations and phenotypes. Frontiers in Molecular Neuroscience, 15, 984776. [CrossRef]
- [3] Leoncini, S., Boasiako, L., Lopergolo, D., Altamura, M., Fazzi, C., Canitano, R., ... & De Felice, C. (2023). Natural course of IQSEC2-related encephalopathy: an Italian national structured survey. Children, 10(9), 1442. [CrossRef]
- [4] Zerem, A., Haginoya, K., Lev, D., Blumkin, L., Kivity, S., Linder, I., ... & Lerman-Sagie, T. (2016). The molecular and phenotypic spectrum of IQSEC 2-related epilepsy. Epilepsia, 57(11), 1858-1869. [CrossRef]
- [5] Shoubridge, C., Dudding-Byth, T., Pasquier, L., Goel, H., Yap, P., & McConnell, V. (2022). IQSEC2-related encephalopathy in males due to missense variants in the pleckstrin homology domain. Clinical Genetics, 102(1), 72-77. [CrossRef]
- [6] Wayhelova, M., Ryzí, M., Oppelt, J., Hladilkova, E., Vallova, V., Krskova, L., ... & Kuglik, P. (2020). Novel familial IQSEC2 pathogenic sequence variant associated with neurodevelopmental disorders and epilepsy. neurogenetics, 21(4), 269-278. [CrossRef]
- [7] Ren, Y., Luo, X., Tong, H., Wang, S., Yan, J., Lin, L., & Chen, Y. (2024). Preliminary Study on Clinical Characteristics and Pathogenesis of IQSEC2 Mutations Patients. Pharmacogenomics and Personalized Medicine, 289-318.
- [8] Nakashima, M., Shiroshima, T., Fukaya, M., Sugawara, T., Sakagami, H., & Yamazawa, K. (2024). C-terminal truncations in IQSEC2: implications for synaptic localization, guanine nucleotide exchange factor activity, and neurological manifestations. Journal of Human Genetics, 69(3), 119-123. [CrossRef]
- [9] Levy, N. S., Borisov, V., Lache, O., & Levy, A. P. (2023). Molecular insights into IQSEC2 disease. International Journal of Molecular Sciences, 24(5), 4984. [CrossRef]
- [10] Jackson, M. R., Loring, K. E., Homan, C. C., Thai, M. H., Määttänen, L., Arvio, M., ... & Shoubridge, C. (2019). Heterozygous loss of function of IQSEC2/Iqsec2 leads to increased activated Arf6 and severe neurocognitive seizure phenotype in females. Life science alliance, 2(4).
- [11] Yang, W. (2021). A General Mechanism Of Ca2+ Regulation Occurs in Both Excitatory and Inhibitory Synapses (Doctoral dissertation, Hong Kong University of Science and Technology (Hong Kong)). [CrossRef]
- [12] Arrazola Sastre, A., Luque Montoro, M., Lacerda, H. M., Llavero, F., & Zugaza, J. L. (2021). Small GTPases of the Rab and Arf families: key regulators of intracellular trafficking in neurodegeneration. International journal of molecular sciences, 22(9), 4425. [CrossRef]
- [13] Balestrini, S., & Sisodiya, S. M. (2017). Treatment of epileptic encephalopathies. Current pharmaceutical design, 23(37), 5667-5690. [CrossRef]
- [14] Pawar, R. P., & Rohane, S. H. (2021). Role of autodock vina in PyRx molecular docking. Asian Journal of Research in Chemistry, 14(2), 132-134.
- [15] Butt, S. S., Badshah, Y., Shabbir, M., & Rafiq, M. (2020). Molecular docking using chimera and autodock vina software for nonbioinformaticians. JMIR Bioinformatics and Biotechnology, 1(1), e14232.
- [17] Ferrari, I. V., & Patrizio, P. (2021). Development and validation molecular docking analysis of human serum albumin (HSA). BioRxiv, 2021-07.
- [18] Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry, 25(13), 1605-1612. [CrossRef]
- [19] D’Souza-Schorey, C., & Chavrier, P. (2006). ARF proteins: roles in membrane traffic and beyond. Nature reviews Molecular cell biology, 7(5), 347-358. [CrossRef]
- [20] Gillingham, A. K., & Munro, S. (2007). The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol., 23(1), 579-611.
- [21] D’Souza, R. S., & Casanova, J. E. (2016). The BRAG/IQSec family of arf GEFs. Small GTPases, 7(4), 257-264.
- [22] Um, J. W. (2017). Synaptic functions of the IQSEC family of ADP-ribosylation factor guanine nucleotide exchange factors. Neuroscience Research, 116, 54-59.
- [23] Du, Q. H., Peng, C., & Zhang, H. (2013). Polydatin: a review of pharmacology and pharmacokinetics. Pharmaceutical biology, 51(11), 1347-1354.
- [24] Luo, J., Chen, S., Wang, L., Zhao, X., & Piao, C. (2022). Pharmacological effects of polydatin in the treatment of metabolic diseases: A review. Phytomedicine, 102, 154161. [CrossRef]
- [25] Karami, A., Fakhri, S., Kooshki, L., & Khan, H. (2022). Polydatin: pharmacological mechanisms, therapeutic targets, biological activities, and health benefits. Molecules, 27(19), 6474.
- [26] Lanzilli, G., Cottarelli, A., Nicotera, G., Guida, S., Ravagnan, G., & Fuggetta, M. P. (2012). Anti-inflammatory effect of resveratrol and polydatin by in vitro IL-17 modulation. Inflammation, 35(1), 240-248.
- [27] Chen, B., Wang, X., Zhang, Y., Huang, K., Liu, H., Xu, D., ... & Lin, X. (2020). Improved solubility, dissolution rate, and oral bioavailability of main biflavonoids from Selaginella doederleinii extract by amorphous solid dispersion. Drug Delivery, 27(1), 309-322. [CrossRef]
- [28] Carrillo-Hormaza, L., López-Parra, S., Stahr, P. L., Keck, C. M., & Osorio, E. (2023). Natural dispersion of biflavonoids from Garcinia madruno extracts: A green and sustainable processing to improve the solubility and dissolution rate. Food and Bioproducts Processing, 141, 199-209. [CrossRef]
- [29] Ding, X., Hou, X., Gao, S., Sun, M., Lin, F., Cai, G., & Xiao, K. (2014). Pharmacokinetics and bioavailability study of polydatin in rat plasma by using a LC-MS/MS method. Pak. J. Pharm. Sci, 27(6), 1931-1937.
- [30] Paczkowska-Walendowska, M., Miklaszewski, A., & Cielecka-Piontek, J. (2022). Is It Possible to Improve the Bioavailability of Resveratrol and Polydatin Derived from Polygoni cuspidati Radix as a Result of Preparing Electrospun Nanofibers Based on Polyvinylpyrrolidone/Cyclodextrin?. Nutrients, 14(19), 3897. [CrossRef]
- [31] FERRARI, I. V., & Ravagnan, G. (2023). Computational Insights into Polydatin: Unraveling Therapeutic Opportunities through Interactions with Rab Proteins.
- [32] Yuan, S., Chan, H. S., & Hu, Z. (2017). Using PyMOL as a platform for computational drug design. Wiley Interdisciplinary Reviews: Computational Molecular Science, 7(2), e1298.
- [33] Baroroh, U., Biotek, M., Muscifa, Z. S., Destiarani, W., Rohmatullah, F. G., & Yusuf, M. (2023). Molecular interaction analysis and visualization of protein-ligand docking using Biovia Discovery Studio Visualizer. Indonesian Journal of Computational Biology (IJCB), 2(1), 22-30. [CrossRef]
Table 1.
Binding energies Scores of Polydatin with ARF and IQSEC2 proteins.
Table 1.
Binding energies Scores of Polydatin with ARF and IQSEC2 proteins.
| Ligand |
Target |
Binding Energy (kcal/mol) |
| Polydatin |
ARF1 |
-7.0 |
| Polydatin |
ARF2 |
-7.2 |
| Polydatin |
ARF3 |
-8.4 |
| Polydatin |
ARF4 |
-6.3 |
| Polydatin |
ARF5 |
-7.5 |
| Polydatin |
ARF6 |
-9.1 |
| Polydatin |
IQSEC2 |
-7.6 |
| Polydatin |
IQSEC1 |
-7.6 |
Table 2.
Binding energies of Polydatin with Rab/Ras proteins [
31].
Table 2.
Binding energies of Polydatin with Rab/Ras proteins [
31].
| Protein |
Binding Energy (kcal/mol) |
| Rab-4A |
-9.6 |
| Rab-5A |
-10.0 |
| HRas |
-10.4 |
| C3 botulinum toxin substrate 3 |
-10.1 |
| Rab-6B |
-9.6 |
| Rab-31 |
-9.6 |
| Rab-18 |
-10.5 |
Table 3.
Comparison all Binding Energies of Ras-related protein Rab-proteins [
31].
Table 3.
Comparison all Binding Energies of Ras-related protein Rab-proteins [
31].
| Ras-related protein Rab-proteins |
Binding Energy (kcal/mol) |
| Ras-related protein Rab-28 |
-7.0 |
| Ras-related GTP-binding protein C |
-9.0 |
| Ras-related protein Rab-9A |
-8.1 |
| Ras-related protein Rab-18 |
-10.5 |
| Ras-related protein Rab-31 |
-9.6 |
| Ras-related protein Rap-2a |
-8.1 |
| Ras-related protein Rab-11B |
-9.1 |
| Ras-related protein Rab-5A |
-10 |
| Ras-related protein Rab-3D |
-9.2 |
| Ras-related protein Rab-4A |
-9.6 |
| GTPase HRas |
-10.4 |
| Ras-related protein R-Ras |
-9.1 |
| Ras-related protein Rap-2A |
-8.4 |
| Ras-related protein Rab-1B |
-8.9 |
| Ras-related C3 botulinum toxin substrate 3 |
-10.1 |
| Ras-related protein Rab-6B |
-9.6 |
| Ras-related protein Rab-6A |
-9.3 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).