Submitted:
27 November 2025
Posted:
28 November 2025
You are already at the latest version
Abstract
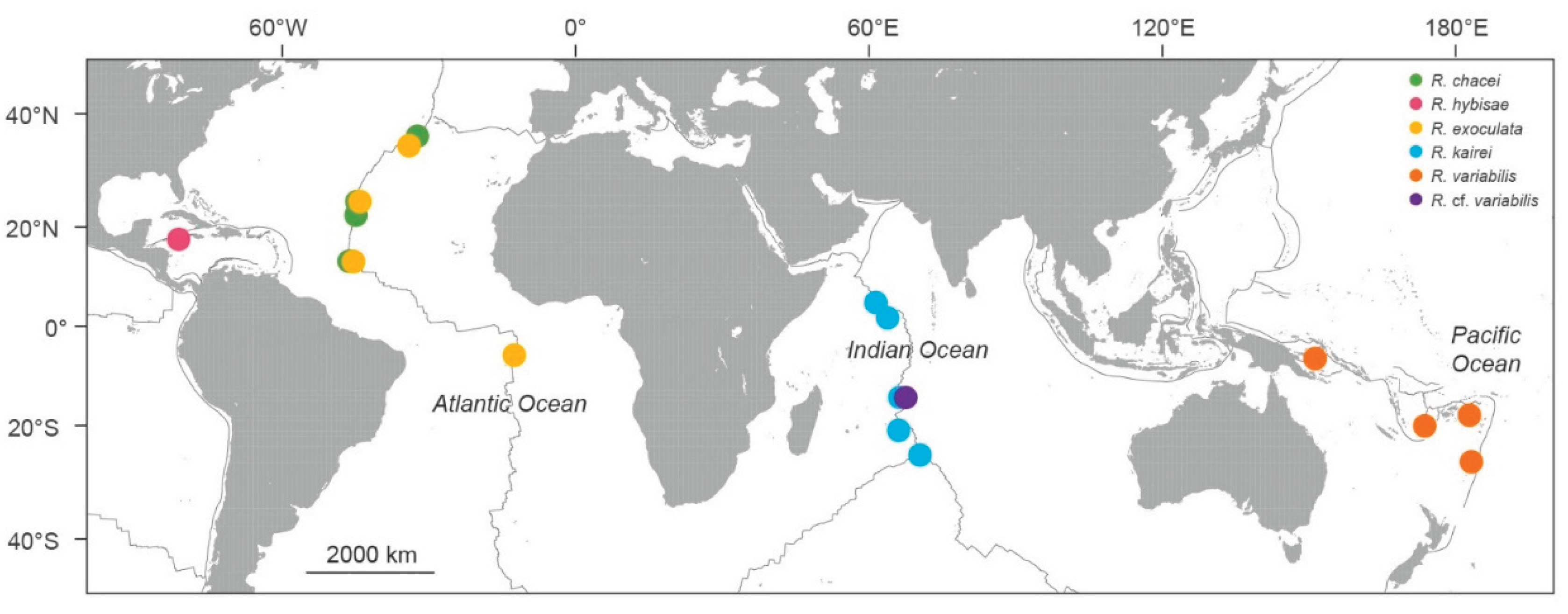
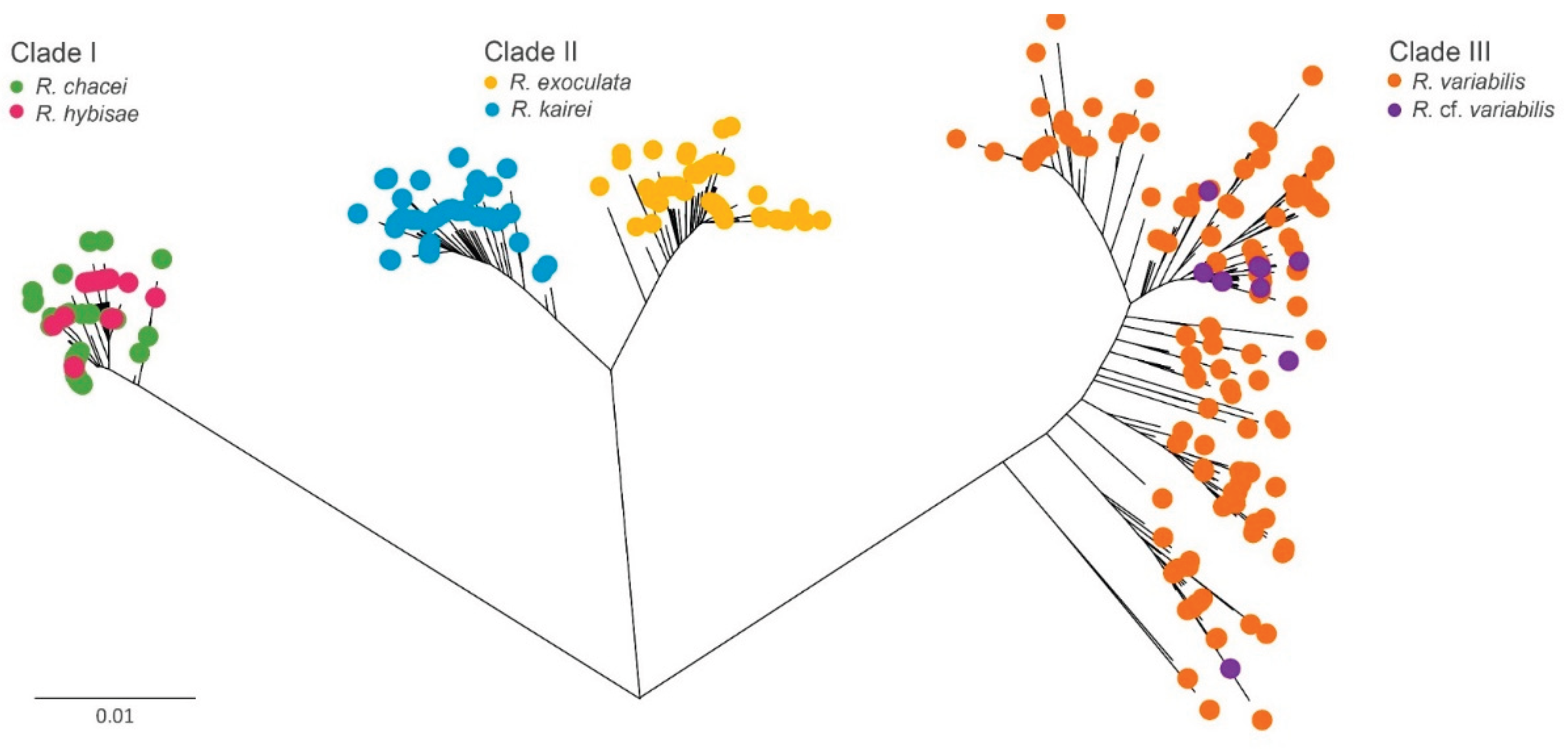
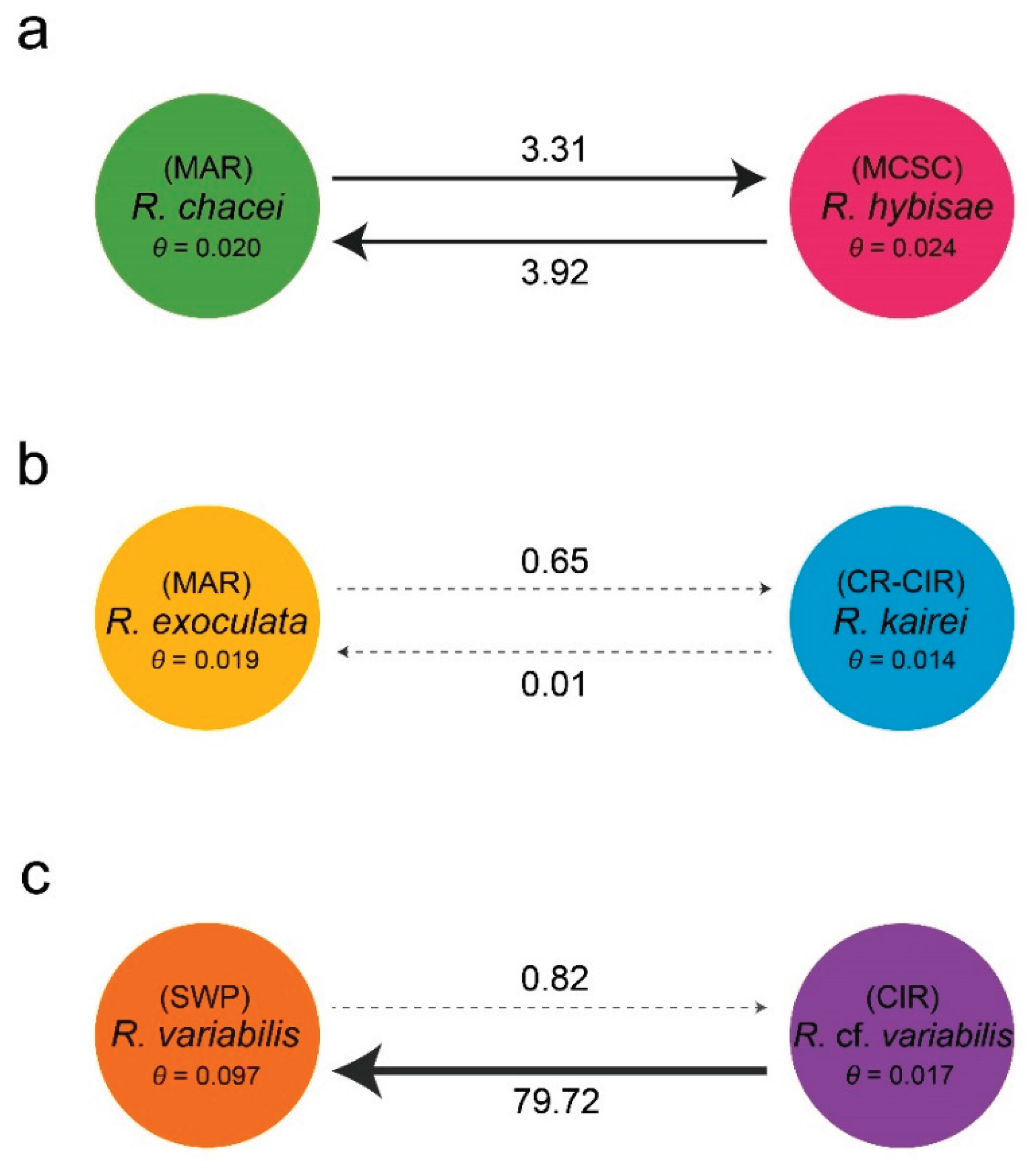
Deep-sea hydrothermal vent fauna is often regarded as highly endemic, although exceptions have been reported. We examined genetic connectivity across broad spatial scales within the alvinocaridid genus Rimicaris, which has undergone substantial adaptive radiation worldwide. We analyzed six Rimicaris species using three genetic markers (COI, 16S, and H3) and complete mitogenomes, using newly generated sequences combined with publicly available sequence data. Genetic tree and haplotype networks were constructed, and divergence analyses were performed. As a result, three clades of paired Rimicaris species were identified, each comprising taxa from different oceanic regions, but showing relatively low COI divergence (0.35–1.90%). In Clade I, Rimicaris chacei and Rimicaris hybisae are morphologically similar and exhibit bidirectional gene flow, suggesting a dispersal route between the Mid-Atlantic Ridge and Mid-Cayman Spreading Center. In Clade II, Rimicaris exoculata and Rimicaris kairei are morphologically, genetically, and ecologically distinct, reflecting restricted connectivity between the Mid-Atlantic Ridge and Carlsberg Ridge–Central Indian Ridge. In Clade III, Rimicaris variabilis and Rimicaris cf. variabilis differ in nutritional strategies, showing a unidirectional dispersal route from the northern Central Indian Ridge to the southwestern Pacific, but morphological data to distinguish them are currently lacking. Some Rimicaris lineages maintain connectivity across distinct oceanic regions while others still form unique regional populations. This finding highlights the need for conservation strategies that incorporate both global-scale connectivity and regional endemism, rather than treating vent ecosystems as a single homogeneous management unit.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Vent Shrimp Sampling and Identification
2.3. DNA Extraction, Partial Gene and Mitogenome Sequencing, and Sequence Data Preprocessing
2.4. Tree Construction, Nucleotide Divergence, Haplotype Network, and Gene Flow
2.5. Mitogenome Sequence Comparison
3. Results
3.1. Datasets Prepared from Multi-Gene Sequences
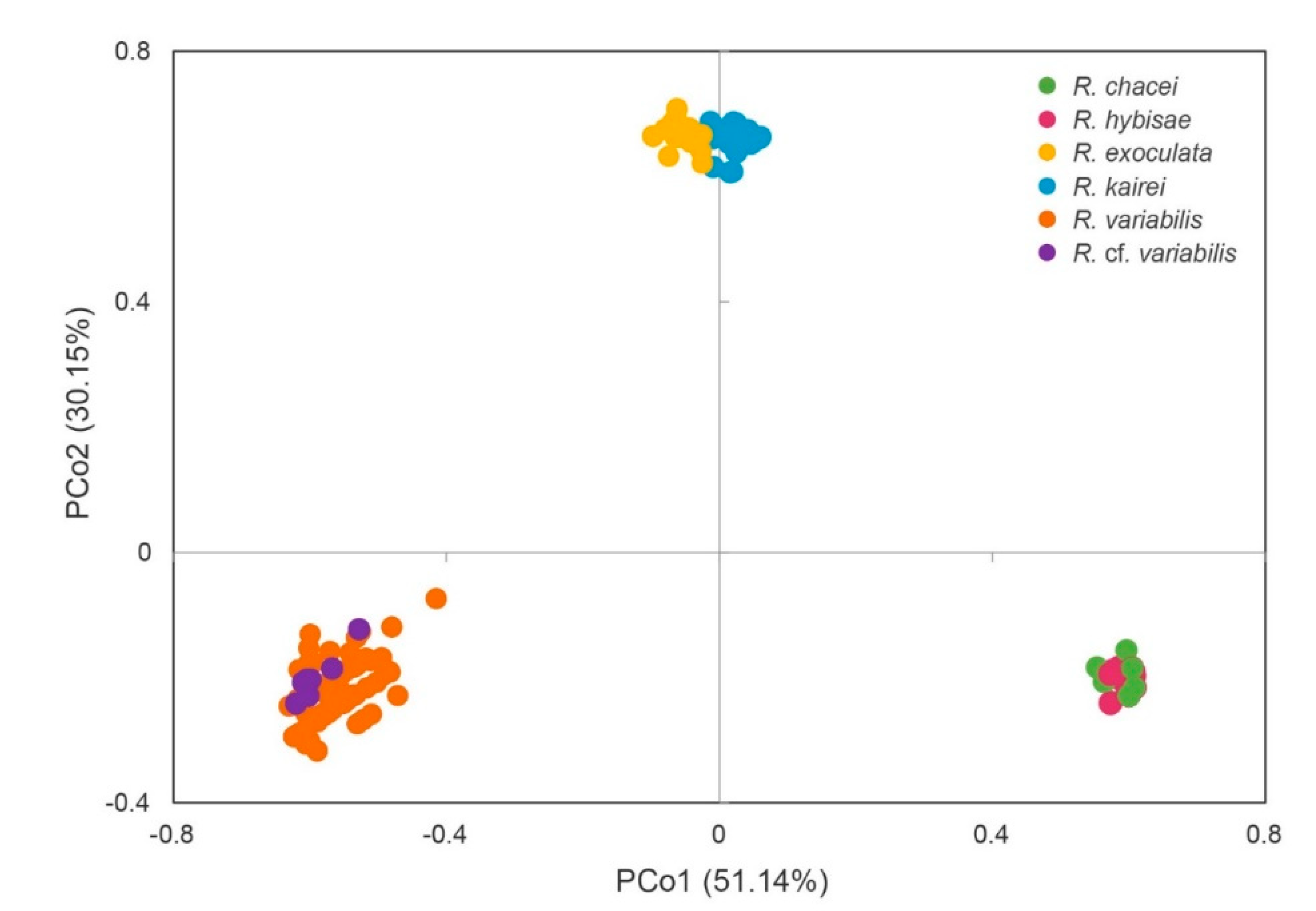
3.2. Genetic Clusters of Rimicaris Species
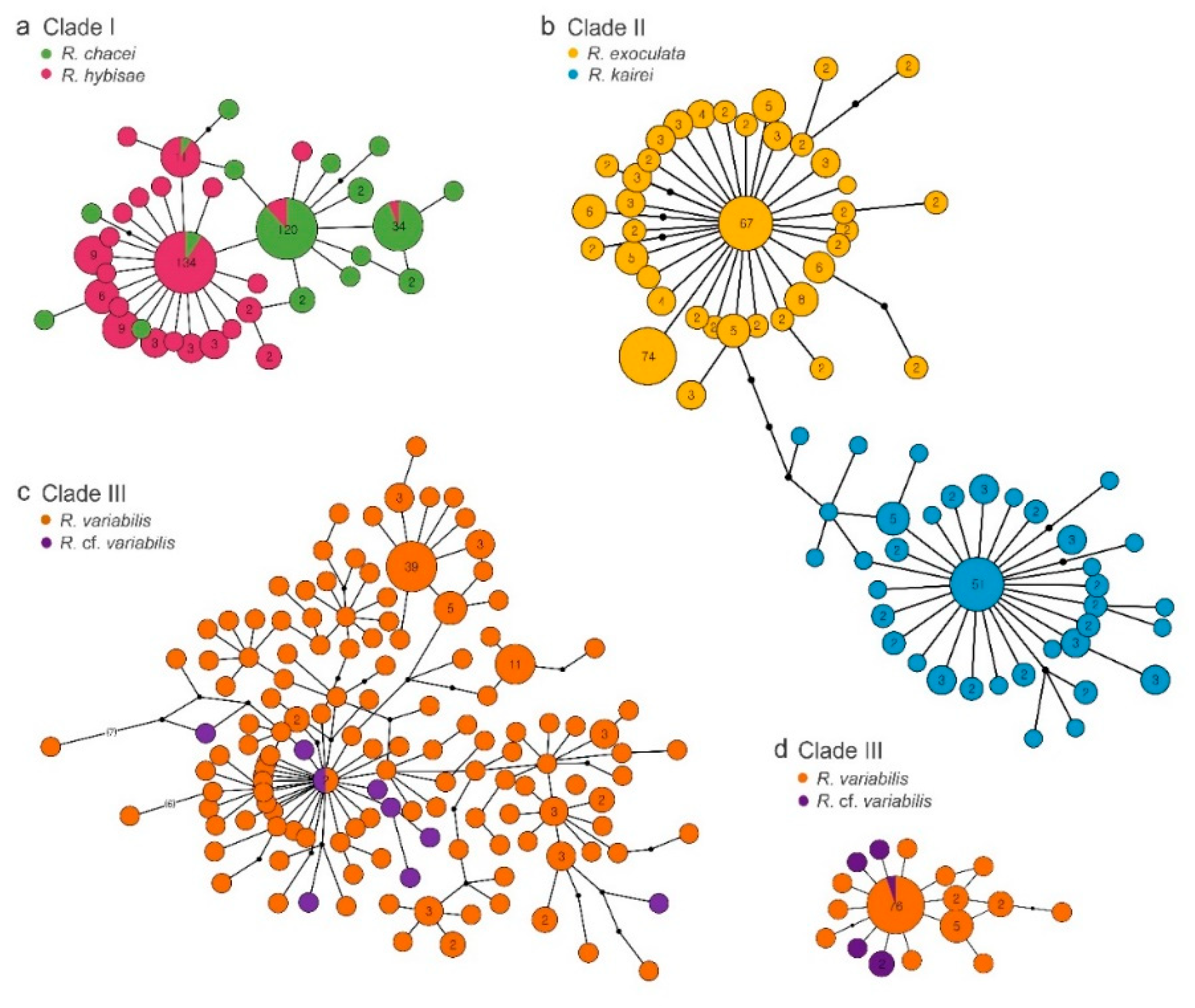
3.3. Genetic Connectivity Between Paired Rimicaris Species Within Each Clade
3.4. Mitogenomic Similarity Between Paired Rimicaris Species
4. Discussion
4.1. Clade-Specific Patterns of Genetic Similarity in Rimicaris
4.2. Adaptive Divergence, Eastward Dispersal, and Regional Barriers in Clade III
4.3. New Perspective on Vent Organism Conservation
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agiadi, K., N. Hohmann, E. Gliozzi, D. Thivaiou, F. R. Bosellini, M. Taviani, G. Bianucci, et al. 2024. Late Miocene transformation of Mediterranean Sea biodiversity. Science Advances 10: eadp1134.
- Apremont, V., M.-A. Cambon-Bonavita, V. Cueff-Gauchard, D. François, F. Pradillon, L. Corbari, and M. Zbinden. 2018. Gill chamber and gut microbial communities of the hydrothermal shrimp Rimicaris chacei Williams and Rona 1986: A possible symbiosis C.-H. Kuo [ed.],. PLOS ONE 13: e0206084.
- Assié, A. 2016. Deep Se(a)quencing: A study of deep sea ectosymbioses using next generation sequencing. [Doctoral dissertion, Universität Bremen].
- Bachraty, C., P. Legendre, and D. Desbruyères. 2009. Biogeographic relationships among deep-sea hydrothermal vent faunas at global scale. Deep Sea Research Part I: Oceanographic Research Papers 56: 1371–1378.
- Beerli, P. Beerli, P. 2006. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22: 341–345.
- Beerli, P., S. Mashayekhi, M. Sadeghi, M. Khodaei, and K. Shaw. 2019. Population Genetic Inference With MIGRATE. Current Protocols in Bioinformatics 68: e87.
- Bernt, M. Bernt, M., A. Donath, F. Jühling, F. Externbrink, C. Florentz, G. Fritzsch, J. Pütz, et al. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution 69: 313–319.
- Bialik, O. M., M. Frank, C. Betzler, R. Zammit, and N. D. Waldmann. 2019. Two-step closure of the Miocene Indian Ocean Gateway to the Mediterranean. Scientific Reports 9: 8842.
- Breusing, C., M. Genetti, S. L. Russell, R. B. Corbett-Detig, and R. A. Beinart. 2022. Horizontal transmission enables flexible associations with locally adapted symbiont strains in deep-sea hydrothermal vent symbioses. Proceedings of the National Academy of Sciences 119: e2115608119.
- Breusing, C., S. B. Johnson, S. Mitarai, R. A. Beinart, and V. Tunnicliffe. 2023. Differential patterns of connectivity in Western Pacific hydrothermal vent metapopulations: A comparison of biophysical and genetic models. Evolutionary Applications 16: 22–35.
- Campbell, K. A. 2006. Hydrocarbon seep and hydrothermal vent paleoenvironments and paleontology: Past developments and future research directions. Palaeogeography, Palaeoclimatology, Palaeoecology 232: 362–407.
- Cavanaugh, C. M., Z. P. McKiness, I. L. G. Newton, and F. J. Stewart. 2006. Marine Chemosynthetic Symbioses. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt [eds.], The Prokaryotes, 475–507. Springer New York, New York, NY.
- Chagas, A. T. D. A., S. Ludwig, J. D. S. M. Pimentel, N. L. De Abreu, D. L. Nunez-Rodriguez, H. G. Leal, and E. Kalapothakis. 2020. Use of complete mitochondrial genome sequences to identify barcoding markers for groups with low genetic distance. Mitochondrial DNA Part A 31: 139–146.
- Colgan, D. J., A. McLauchlan, G. D. F. Wilson, S. P. Livingston, G. D. Edgecombe, J. Macaranas, G. Cassis, and M. R. Gray. 1998. Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Australian Journal of Zoology 46: 419–437.
- Connelly, D. P., J. T. Copley, B. J. Murton, K. Stansfield, P. A. Tyler, C. R. German, C. L. Van Dover, et al. 2012. Hydrothermal vent fields and chemosynthetic biota on the world’s deepest seafloor spreading centre. Nature Communications 3: 620.
- Dai, Q., T. Xu, Y. Li, Y. Sun, Y. Lin, T. Yahagi, M. Perez, et al. 2025. Comparative Population Genetics of Two Alvinocaridid Shrimp Species in Chemosynthetic Ecosystems of the Western Pacific. Integrative Zoology n/a.
- Dierckxsens, N., P. Mardulyn, and G. Smits. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research 45: e18.
- Dubilier, N., C. Bergin, and C. Lott. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nature Reviews Microbiology 6: 725–740.
- Excoffier, L., and H. E. L. Lischer. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567.
- Fisher, C., K. Takai, and N. Le Bris. 2007. Hydrothermal Vent Ecosystems. Oceanography 20: 14–23.
- Folmer, O., M. Black, W. Hoeh, R. Lutz, and R. Vrijenhoek. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
- Fuchs, S., M. D. Hannington, and S. Petersen. 2019. Divining gold in seafloor polymetallic massive sulfide systems. Mineralium Deposita 54: 789–820.
- German, C. R., S. Petersen, and M. D. Hannington. 2016. Hydrothermal exploration of mid-ocean ridges: Where might the largest sulfide deposits be forming? Chemical Geology 420: 114–126.
- Hebert, P. D. N., A. Cywinska, S. L. Ball, and J. R. deWaard. 2003a. Biological identifications through DNA barcodes. Proceedings. Biological Sciences 270: 313–321.
- Hebert, P. D. N., S. Ratnasingham, and J. R. deWaard. 2003b. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings. Biological Sciences 270 Suppl 1: S96-99.
- Hinsinger, D., R. Debruyne, M. Thomas, G. Denys, M. I. Mennesson, J. Utage, and A. Dettaï. 2015. Fishing for barcodes in the Torrent: from COI to complete mitogenomes on NGS platforms. DNA Barcodes 3: 170.
- Hou, Z., and S. Li. 2018. Tethyan changes shaped aquatic diversification. Biological Reviews 93: 874–896.
- Hrbek, T., and A. Meyer. 2003. Closing of the Tethys Sea and the phylogeny of Eurasian killifishes (Cyprinodontiformes: Cyprinodontidae). Journal of Evolutionary Biology 16: 17–36.
- Irwin, D. E. 2000. SONG VARIATION IN AN AVIAN RING SPECIES. Evolution 54: 998–1010.
- Irwin, D. E., S. Bensch, J. H. Irwin, and T. D. Price. 2005. Speciation by Distance in a Ring Species. Science 307: 414–416.
- Jan, C., J. M. Petersen, J. Werner, H. Teeling, S. Huang, F. O. Glöckner, O. V. Golyshina, et al. 2014. The gill chamber epibiosis of deep-sea shrimp Rimicaris exoculata: an in-depth metagenomic investigation and discovery of Zetaproteobacteria. Environmental Microbiology 16: 2723–2738.
- Kiel, S. 2010. The Fossil Record of Vent and Seep Mollusks. In S. Kiel [ed.], The Vent and Seep Biota, Topics in Geobiology, 255–277. Springer Netherlands, Dordrecht.
- Komai, T., and T. Giguère. 2019. A new species of alvinocaridid shrimp Rimicaris Williams & Rona, 1986 (Decapoda: Caridea) from hydrothermal vents on the Mariana Back Arc Spreading Center, northwestern Pacific. Journal of Crustacean Biology 39: 640–650.
- Komai, T., L. Menot, and M. Segonzac. 2016. New records of caridean shrimp (Crustacea: Decapoda) from hydrothermally influenced fields off Futuna Island, Southwest Pacific, with description of a new species assigned to the genus Alvinocaridinides Komai & Chan, 2010 (Alvinocarididae). Zootaxa 4098: 298–310.
- Komai, T., and M. Segonzac. 2005. A revision of the genus Alvinocaris Williams and Chace (Crustacea: Decapoda: Caridea: Alvinocarididae), with descriptions of a new genus and a new species of Alvinocaris. Journal of Natural History 39: 1111–1175.
- Komai, T., and S. Tsuchida. 2015. New records of Alvinocarididae (Crustacea: Decapoda: Caridea) from the southwestern Pacific hydrothermal vents, with descriptions of one new genus and three new species. Journal of Natural History 49: 1789–1824.
- Lee, W.-K., S. K. Juniper, M. Perez, S.-J. Ju, and S.-J. Kim. 2021. Diversity and characterization of bacterial communities of five co-occurring species at a hydrothermal vent on the Tonga Arc. Ecology and Evolution 11: 4481–4493.
- Lee, W.-K., S.-J. Kim, B. K. Hou, C. L. Van Dover, and S.-J. Ju. 2019. Population genetic differentiation of the hydrothermal vent crab Austinograea alayseae (Crustacea: Bythograeidae) in the Southwest Pacific Ocean J. A. Eble [ed.],. PLOS ONE 14: e0215829.
- Librado, P., and J. Rozas. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
- Lins, L. S. F., S. Y. W. Ho, G. D. F. Wilson, and N. Lo. 2012. Evidence for Permo-Triassic colonization of the deep sea by isopods. Biology Letters 8: 979–982.
- Little, C. T. S., R. J. Herrington, V. V. Maslennikov, and V. V. Zaykov. 1998. The fossil record of hydrothermal vent communities. Geological Society, London, Special Publications 148: 259–270.
- Liu, H., S. Li, A. Ugolini, F. Momtazi, and Z. Hou. 2018. Tethyan closure drove tropical marine biodiversity: Vicariant diversification of intertidal crustaceans. Journal of Biogeography 45: 941–951.
- Mao, Q., C. Chen, J. Sun, J. Liang, Y. Sun, Y. Wang, C. Zeng, et al. 2025. Genetic Structure in a Trans-Oceanic Hot Vent Mussel Reveals Four Metapopulations With Implications for Conservation. Journal of Biogeography n/a: e70017.
- Martin, J. W., and T. A. Haney. 2005. Decapod crustaceans from hydrothermal vents and cold seeps: a review through 2005. Zoological Journal of the Linnean Society 145: 445–522.
- Matzen da Silva, J., S. Creer, A. dos Santos, A. C. Costa, M. R. Cunha, F. O. Costa, and G. R. Carvalho. 2011. Systematic and Evolutionary Insights Derived from mtDNA COI Barcode Diversity in the Decapoda (Crustacea: Malacostraca). PLOS ONE 6: e19449.
- McArthur, A. G., and V. Tunnicliffe. 1998. Relics and antiquity revisited in the modern vent fauna. Geological Society, London, Special Publications 148: 271–291.
- McClain, C. R., and S. M. Hardy. 2010. The dynamics of biogeographic ranges in the deep sea. Proceedings of the Royal Society B: Biological Sciences 277: 3533–3546.
- Methou, P., C. Chen, and T. Komai. 2024a. Revision of the alvinocaridid shrimp genus Rimicaris Williams & Rona, 1986 (Decapoda: Caridea) with description of a new species from the Mariana Arc hydrothermal vents. Zootaxa 5406: 501–518.
- Methou, P., V. Cueff-Gauchard, L. N. Michel, N. Gayet, F. Pradillon, and M. Cambon-Bonavita. 2023. Symbioses of alvinocaridid shrimps from the South West Pacific: No chemosymbiotic diets but conserved gut microbiomes. Environmental Microbiology Reports 15: 614–630.
- Methou, P., M. Guéganton, J. T. Copley, H. Kayama Watanabe, F. Pradillon, M.-A. Cambon-Bonavita, and C. Chen. 2024b. Distinct development trajectories and symbiosis modes in vent shrimps M. Kronforst, and M. Zelditch [eds.],. Evolution 78: 413–422.
- Methou, P., I. Hernández-Ávila, C. Cathalot, M. Cambon-Bonavita, and F. Pradillon. 2022a. Population structure and environmental niches of Rimicaris shrimps from the Mid-Atlantic Ridge. Marine Ecology Progress Series 684: 1–20.
- Methou, P., M. Hikosaka, C. Chen, H. K. Watanabe, N. Miyamoto, H. Makita, Y. Takahashi, and R. G. Jenkins. 2022b. Symbiont Community Composition in Rimicaris kairei Shrimps from Indian Ocean Vents with Notes on Mineralogy K. N. Johnson [ed.],. Applied and Environmental Microbiology 88: e00185-22.
- Mitarai, S., H. Watanabe, Y. Nakajima, A. F. Shchepetkin, and J. C. McWilliams. 2016. Quantifying dispersal from hydrothermal vent fields in the western Pacific Ocean. Proceedings of the National Academy of Sciences 113: 2976–2981.
- Moalic, Y., D. Desbruyères, C. M. Duarte, A. F. Rozenfeld, C. Bachraty, and S. Arnaud-Haond. 2012. Biogeography Revisited with Network Theory: Retracing the History of Hydrothermal Vent Communities. Systematic Biology 61: 127.
- van der Most, N., P.-Y. Qian, Y. Gao, and S. Gollner. 2023. Active hydrothermal vent ecosystems in the Indian Ocean are in need of protection. Frontiers in Marine Science 9.
- Mullineaux, L. S., A. Metaxas, S. E. Beaulieu, M. Bright, S. Gollner, B. M. Grupe, S. Herrera, et al. 2018. Exploring the Ecology of Deep-Sea Hydrothermal Vents in a Metacommunity Framework. Frontiers in Marine Science 5: 49.
- Mullineaux, L. S., S. W. Mills, N. Le Bris, S. E. Beaulieu, S. M. Sievert, and L. N. Dykman. 2020. Prolonged recovery time after eruptive disturbance of a deep-sea hydrothermal vent community. Proceedings of the Royal Society B: Biological Sciences 287: 20202070.
- Nye, V., J. Copley, and S. Plouviez. 2012. A new species of Rimicaris (Crustacea: Decapoda: Caridea: Alvinocarididae) from hydrothermal vent fields on the Mid-Cayman Spreading Centre, Caribbean. Journal of the Marine Biological Association of the United Kingdom 92: 1057–1072.
- Nymoen, A. R., J. A. Kongsrud, E. Willassen, and T. Bakken. 2024. When standard DNA barcodes do not work for species identification: intermixed mitochondrial haplotypes in the Jaera albifrons complex (Crustacea: Isopoda). Marine Biodiversity 54: 43.
- Palumbi, S. R. 1991. The simple fool’s guide to PCR. Version 2.0. Dept. of Zoology and Kewalo Marine Laboratory, University of Hawaii, Honolulu, HI.
- Peakall, R., and P. E. Smouse. 2006. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295.
- Perez, M., J. Sun, Q. Xu, and P.-Y. Qian. 2021. Structure and Connectivity of Hydrothermal Vent Communities Along the Mid-Ocean Ridges in the West Indian Ocean: A Review. Frontiers in Marine Science 8: 744874.
- Plouviez, S., A. L. LaBella, D. W. Weisrock, F. A. B. Von Meijenfeldt, B. Ball, J. E. Neigel, and C. L. Van Dover. 2019. Amplicon sequencing of 42 nuclear loci supports directional gene flow between South Pacific populations of a hydrothermal vent limpet. Ecology and Evolution 9: 6568–6580.
- Ramirez-Llodra, E., T. M. Shank, and C. R. German. 2007. Biodiversity and Biogeography of Hydrothermal Vent Species: Thirty Years of Discovery and Investigations. Oceanography 20: 30–41.
- Ranasinghe, U. G. S. L., J. Eberle, J. Thormann, C. Bohacz, S. P. Benjamin, and D. Ahrens. 2022. Multiple species delimitation approaches with COI barcodes poorly fit each other and morphospecies – An integrative taxonomy case of Sri Lankan Sericini chafers (Coleoptera: Scarabaeidae). Ecology and Evolution 12: e8942.
- Shearer, T. L., and M. A. Coffroth. 2008. DNA BARCODING: Barcoding corals: limited by interspecific divergence, not intraspecific variation. Molecular Ecology Resources 8: 247–255.
- Sogin, E. M., M. Kleiner, C. Borowski, H. R. Gruber-Vodicka, and N. Dubilier. 2021. Life in the Dark: Phylogenetic and Physiological Diversity of Chemosynthetic Symbioses. Annual Review of Microbiology 75: 695–718.
- Suh, Y. J., M.-S. Kim, S.-J. Kim, D. Kim, and S.-J. Ju. 2022a. Carbon sources and trophic interactions of vent fauna in the Onnuri Vent Field, Indian Ocean, inferred from stable isotopes. Deep Sea Research Part I: Oceanographic Research Papers 182: 103683.
- Suh, Y. J., M.-S. Kim, W.-K. Lee, H. Yoon, I. Moon, J. Jung, and S.-J. Ju. 2022b. Niche partitioning of hydrothermal vent fauna in the North Fiji Basin, Southwest Pacific inferred from stable isotopes. Marine Biology 169: 149.
- Sun, S., Z. Sha, and Y. Wang. 2018. Phylogenetic position of Alvinocarididae (Crustacea: Decapoda: Caridea): New insights into the origin and evolutionary history of the hydrothermal vent alvinocarid shrimps. Deep Sea Research Part I: Oceanographic Research Papers 141: 93–105.
- Sun, Y., W. Liu, J. Chen, J. Li, Y. Ye, and K. Xu. 2024. Sequence comparison of the mitochondrial genomes of five caridean shrimps of the infraorder Caridea: phylogenetic implications and divergence time estimation. BMC Genomics 25: 968.
- Tamura, K., G. Stecher, and S. Kumar. 2021. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38: 3022–3027.
- Teixeira, S., K. Olu, C. Decker, R. L. Cunha, S. Fuchs, S. Hourdez, E. A. Serrão, and S. Arnaud-Haond. 2013. High connectivity across the fragmented chemosynthetic ecosystems of the deep Atlantic Equatorial Belt: efficient dispersal mechanisms or questionable endemism? Molecular Ecology 22: 4663–4680.
- Templeton, A. R., K. A. Crandall, and C. F. Sing. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633.
- Thaler, A. D., S. Plouviez, W. Saleu, F. Alei, A. Jacobson, E. A. Boyle, T. F. Schultz, et al. 2014. Comparative Population Structure of Two Deep-Sea Hydrothermal-Vent-Associated Decapods (Chorocaris sp. 2 and Munidopsis lauensis) from Southwestern Pacific Back-Arc Basins E. Sotka [ed.],. PLoS ONE 9: e101345.
- Thaler, A. D., K. Zelnio, W. Saleu, T. F. Schultz, J. Carlsson, C. Cunningham, R. C. Vrijenhoek, and C. L. Van Dover. 2011. The spatial scale of genetic subdivision in populations of Ifremeria nautilei, a hydrothermal-vent gastropod from the southwest Pacific. BMC Evolutionary Biology 11: 372.
- Thomas, E. A., A. Molloy, N. B. Hanson, M. Böhm, M. Seddon, and J. D. Sigwart. 2021. A Global Red List for Hydrothermal Vent Molluscs. Frontiers in Marine Science 8.
- Tunnicliffe, V., C. Chen, T. Giguère, A. A. Rowden, H. K. Watanabe, and O. Brunner. 2024. Hydrothermal vent fauna of the western Pacific Ocean: Distribution patterns and biogeographic networks. Diversity and Distributions 30: e13794.
- Van Dover, C. L. 2012. Hydrothermal Vent Ecosystems and Conservation. Oceanography 25: 313–316.
- Van Dover, C. L. 2014. Impacts of anthropogenic disturbances at deep-sea hydrothermal vent ecosystems: A review. Marine Environmental Research 102: 59–72.
- Van Dover, C. L. 2002. Trophic relationships among invertebrates at the Kairei hydrothermal vent field (Central Indian Ridge). Marine Biology 141: 761–772.
- Van Dover, C. L., S. Arnaud-Haond, M. Gianni, S. Helmreich, J. A. Huber, A. L. Jaeckel, A. Metaxas, et al. 2018. Scientific rationale and international obligations for protection of active hydrothermal vent ecosystems from deep-sea mining. Marine Policy 90: 20–28.
- Van Dover, C. L., C. R. German, K. G. Speer, L. M. Parson, and R. C. Vrijenhoek. 2002. Evolution and Biogeography of Deep-Sea Vent and Seep Invertebrates. Science 295: 1253–1257.
- Vences, M., S. Patmanidis, J.-C. Schmidt, M. Matschiner, A. Miralles, and S. S. Renner. 2024. Hapsolutely: a user-friendly tool integrating haplotype phasing, network construction, and haploweb calculation. Bioinformatics Advances 4: vbae083.
- Vereshchaka, A. L., D. N. Kulagin, and A. A. Lunina. 2015. Phylogeny and New Classification of Hydrothermal Vent and Seep Shrimps of the Family Alvinocarididae (Decapoda) A. Hejnol [ed.],. PLOS ONE 10: e0129975.
- Versteegh, E. A. A., C. L. Van Dover, L. Van Audenhaege, and M. Coleman. 2023. Multiple nutritional strategies of hydrothermal vent shrimp (Rimicaris hybisae) assemblages at the Mid-Cayman Rise. Deep Sea Research Part I: Oceanographic Research Papers 192: 103915.
- Vrijenhoek, R. C. 2010. Genetic diversity and connectivity of deep-sea hydrothermal vent metapopulations: HYDROTHERMAL VENT METAPOPULATIONS. Molecular Ecology 19: 4391–4411.
- Watabe, H., and J. Hashimoto. 2002. A New Species of the Genus Rimicaris (Alvinocarididae: Caridea: Decapoda) from the Active Hydrothermal Vent Field, “Kairei Field,” on the Central Indian Ridge, the Indian Ocean. Zoological Science 19: 1167–1174.
- Watanabe, H. K., C. Chen, D. P. Marie, K. Takai, K. Fujikura, and B. K. K. Chan. 2018. Phylogeography of hydrothermal vent stalked barnacles: a new species fills a gap in the Indian Ocean ‘dispersal corridor’ hypothesis. Royal Society Open Science 5: 172408.
- Williams, A. B., and P. A. Rona. 1986. Two New Caridean Shrimps (Bresiliidae) From a Hydrothermal Field on the Mid-atlantic Ridge. Journal of Crustacean Biology 6: 446–462.
- Wolff, T. 2005. Composition and endemism of the deep-sea hydrothermal vent fauna. CBM-Cahiers de Biologie Marine. 46: 97–104.
- Won, Y., S. J. Hallam, G. D. O’Mullan, and R. C. Vrijenhoek. 2003. Cytonuclear disequilibrium in a hybrid zone involving deep-sea hydrothermal vent mussels of the genus Bathymodiolus. Molecular Ecology 12: 3185–3190.
- WoRMS Editorial Board. 2025. World Register of Marine Species. Available from https://www.marinespecies.org at VLIZ. Accessed 2025-09-09.
- Yearsley, J. M., D. M. Salmanidou, J. Carlsson, D. Burns, and C. L. Van Dover. 2020. Biophysical models of persistent connectivity and barriers on the northern Mid-Atlantic Ridge. Deep Sea Research Part II: Topical Studies in Oceanography 180: 104819.
- Zhang, Z. 2022. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genomics, Proteomics & Bioinformatics 20: 536–540.
- Zhou, Y., C. Chen, D. Zhang, Y. Wang, H. K. Watanabe, J. Sun, D. Bissessur, et al. 2022. Delineating biogeographic regions in Indian Ocean deep-sea vents and implications for conservation. Diversity and Distributions 28: 2858–2870.
|
Species (no., %)† |
R. chacei (5, 0.00) |
R. hybisae (6, 0.08) |
R. exoculata (10, 0.09) |
R. kairei (1, –) |
R. variabilis (90, 0.13) |
R. cf. variabilis (9, 0.25) |
|
| Clade I |
R. chacei (167, 0.19) |
0.04 | 0.30 | 0.50 | 0.50 | 0.61 | |
|
R. hybisae (197, 0.19) |
0.35 | 0.34 | 0.54 | 0.55 | 0.65 | ||
| Clade II |
R. exoculata (246, 0.35) |
7.47 | 7.70 | 0.30 | 0.79 | 0.90 | |
|
R. kairei (112, 0.33) |
7.09 | 6.97 | 1.90 | 0.99 | 1.10 | ||
| Clade III |
R. variabilis (196, 1.47) |
8.60 | 8.74 | 7.47 | 8.05 | 0.18 | |
|
R. cf. variabilis (9, 0.98) |
8.59 | 8.73 | 6.95 | 7.75 | 1.34 | ||
| Gene | Species | N | S | H | Hd | Nd (%) | D | FS | Pairwise FST† | |
| COI | Clade Ⅰ | R. chacei | 167 | 18 | 17 | 0.56 | 0.19 | –2.02* | –14.99* | – |
| R. hybisae | 197 | 23 | 24 | 0.61 | 0.19 | –2.18* | –28.13* | – | ||
| Overall | 364 | 35 | 37 | 0.75 | 0.27 | –2.16* | –27.75* | 0.472* | ||
| Clade II | R. exoculata | 246 | 36 | 38 | 0.83 | 0.35 | –2.13* | –27.10* | – | |
| R. kairei | 112 | 36 | 37 | 0.79 | 0.33 | –2.43* | –28.56* | – | ||
| Overall | 358 | 56 | 75 | 0.90 | 1.02 | –1.45* | –25.15* | 0.819* | ||
| Clade III | R. variabilis | 196 | 93 | 128 | 0.96 | 1.47 | –1.89* | –24.86* | – | |
| R. cf. variabilis | 9 | 15 | 9 | 1.00 | 0.98 | –1.21 | –5.58* | – | ||
| Overall | 205 | 95 | 136 | 0.96 | 1.46 | –1.91* | –24.82* | 0.100* | ||
| 16S | Clade III | R. variabilis | 90 | 12 | 13 | 0.36 | 0.13 | –2.07* | –13.35* | – |
| R. cf. variabilis | 9 | 4 | 5 | 0.81 | 0.25 | –1.15 | –2.36* | – | ||
| Overall | 99 | 16 | 17 | 0.41 | 0.14 | –2.25* | –20.51* | 0.089* | ||
| Gene | Clade II(no. of mitogenomes) | Clade III (no. of mitogenomes) | ||||||||
| R. exoculata(1) vs. R. kairei (1) | R. variabilis(4) vs. R. cf. variabilis (1) | |||||||||
| Nucleotide | Amino acid |
Substitution ratio (Ka/Ks) |
Nucleotide | Amino acid | Substitution ratio (Ka/Ks)* |
|||||
|
Length (bp)† |
Similarity (%) |
Length (no.) |
Similarity (%) |
Length (bp)†, * |
Similarity (%)* |
Length (no.)* |
Similarity (%)* | |||
| ATP6 | 672/672 | 97.62 | 224/224 | 99.55 | 0.06 | 672/672 | 99.00 | 224/224 | 100.00 | 0.00 |
| ATP8 | 156/156 | 98.72 | 52/52 | 100.00 | 0.00 | 156/156 | 100.00 | 52/52 | 100.00 | 0.00 |
| COXI | 1536/1536 | 98.24 | 512/512 | 100.00 | 0.00 | 1536/1536 | 98.23 | 512/512 | 100.00 | 0.00 |
| COXII | 690/690 | 98.99 | 230/230 | 100.00 | 0.00 | 690/690 | 99.35 | 230/230 | 100.00 | 0.00 |
| COXIII | 786/786 | 99.11 | 262/262 | 100.00 | 0.00 | 786/786 | 99.75 | 262/262 | 100.00 | 0.00 |
| CYTB | 1134/1134 | 98.59 | 378/378 | 99.47 | 0.06 | 1134/1134 | 98.48 | 378/378 | 99.60 | 0.00 |
| ND1 | 939/939 | 97.76 | 313/313 | 100.00 | 0.00 | 939/939 | 98.90 | 313/313 | 99.60 | 0.03 |
| ND2 | 993/993 | 97.89 | 331/331 | 99.40 | 0.05 | 993/993 | 98.36 | 331/331 | 99.62 | 0.03 |
| ND3 | 351/351 | 99.15 | 117/117 | 99.15 | 0.16 | 351/351 | 99.86 | 117/117 | 100.00 | 0.00 |
| ND4 | 1338/1338 | 97.82 | 446/446 | 99.55 | 0.02 | 1338/1338 | 98.41 | 446/446 | 99.78 | 0.02 |
| ND4L | 297/297 | 99.00 | 99/99 | 100.00 | 0.00 | 297/297 | 99.50 | 99/99 | 100.00 | 0.00 |
| ND5 | 1728/1728 | 97.14 | 576/576 | 99.31 | 0.02 | 1728/1728 | 98.24 | 576/576 | 99.44 | 0.04 |
| ND6 | 513/513 | 96.78 | 171/171 | 97.69 | 0.11 | 513/513 | 98.78 | 171/171 | 99.71 | 0.04 |
| 13 PCGs | 11133/11133 | 98.04 | 3711/3711 | 99.57 | 0.03 | 11133/11133 | 98.70 | 3711/3711 | 99.76 | 0.03 |
| 12S rRNA | 865/865 | 99.42 | – | – | – | 866/866 | 99.25 | – | – | – |
| 16S rRNA | 1310/1310 | 99.47 | – | – | – | 1310/1309 | 99.62 | – | – | – |
| Control Region | 1005/1004 | 93.84 | – | – | – | 1008/1008 | 97.07 | – | – | – |
| Species | Distribution | Density* | Cephalothorax | Reference | |||
| Volume | Symbiotic diet | Symbiont† | |||||
| Clade I | R. chacei | MAR | Low | Non-enlarged | Partially dependent | C > G | Apremont et al. (2018) Methou et al. (2024b) |
| R. hybisae | MCSC | High or low | Enlarged | Dependent | C | Assié (2016) Versteegh et al. (2023) Methou et al. (2024b) |
|
| Clade II | R. exoculata | MAR | High | Enlarged | Dependent | C > G | Williams and Rona (1986) Jan et al. (2014) Methou et al. (2022a) Methou et al. (2024b) |
| R. kairei | CR-CIR | High | Enlarged | Dependent | C > D > B | Watabe and Hashimoto (2002) Van Dover (2002) Methou et al. (2022b) |
|
| Clade III | R. variabilis | SWP | High or low | Non-enlarged | Partially dependent | G > C | Komai and Tsuchida (2015) Lee et al. (2021) Suh et al. (2022b) Methou et al. (2023) |
| R. cf. variabilis | CIR | Low | Non-enlarged | Dependent | N/A | Suh et al. (2022a) This study |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





