Submitted:
14 November 2025
Posted:
14 November 2025
You are already at the latest version
Abstract
Chinese cabbage (Brassica rapa ssp. pekinensis) is a globally important leafy vegetable, but its recalcitrance to Agrobacterium-mediated genetic transformation has severely limited functional genomics research. Here we demonstrate that both Agrobacterium infection and antibiotic selection impose significant inhibition on cotyledonary petiole regeneration, representing one principal bottleneck to high-throughput transformation. Infection with different Agrobacterium strains suppressed regenerated shoot per explant by 30.98-69.16%. Supplying the salicylic-acid-signalling inhibitor tenoxicam in the seed-germination medium raised post-infection regeneration by up to 37.90%. Compared with non-infected controls, the optimal NAA concentration for explant regeneration after infection was higher, and 0.5 mg/L increased post-infection regeneration by 27.66 %. Replacing antibiotic selectable markers with the visual reporters eYGFPuv or RUBY eliminated phytotoxicity, reduced false-positive shoots, and further elevated genetic transformation efficiency to 19.33-20.00% (versus 2.67–6.67% under antibiotic selection). The integrated protocol yielded stable RUBY over-expressing lines whose biomass declined with rising transcript levels. Restricting RUBY expression to the inner head leaves generated a novel germplasm with less yield penalty. This work provides a high-efficiency transformation method that will accelerate gene discovery and genome editing in Chinese cabbage.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Vector Construction and Agrobacterium Strain
2.3. Culture Media
2.4. Cotyledonary Petiole Tissue Culture
2.5. Genetic Transformation
2.6. Assessment of Regeneration and Genetic Transformation Efficiency
2.7. Agronomic Trait Evaluation of Transgenic Lines
2.8. RNA Extraction, RT-PCR, and qRT-PCR Analysis
2.9. Statistical Analysis
3. Result
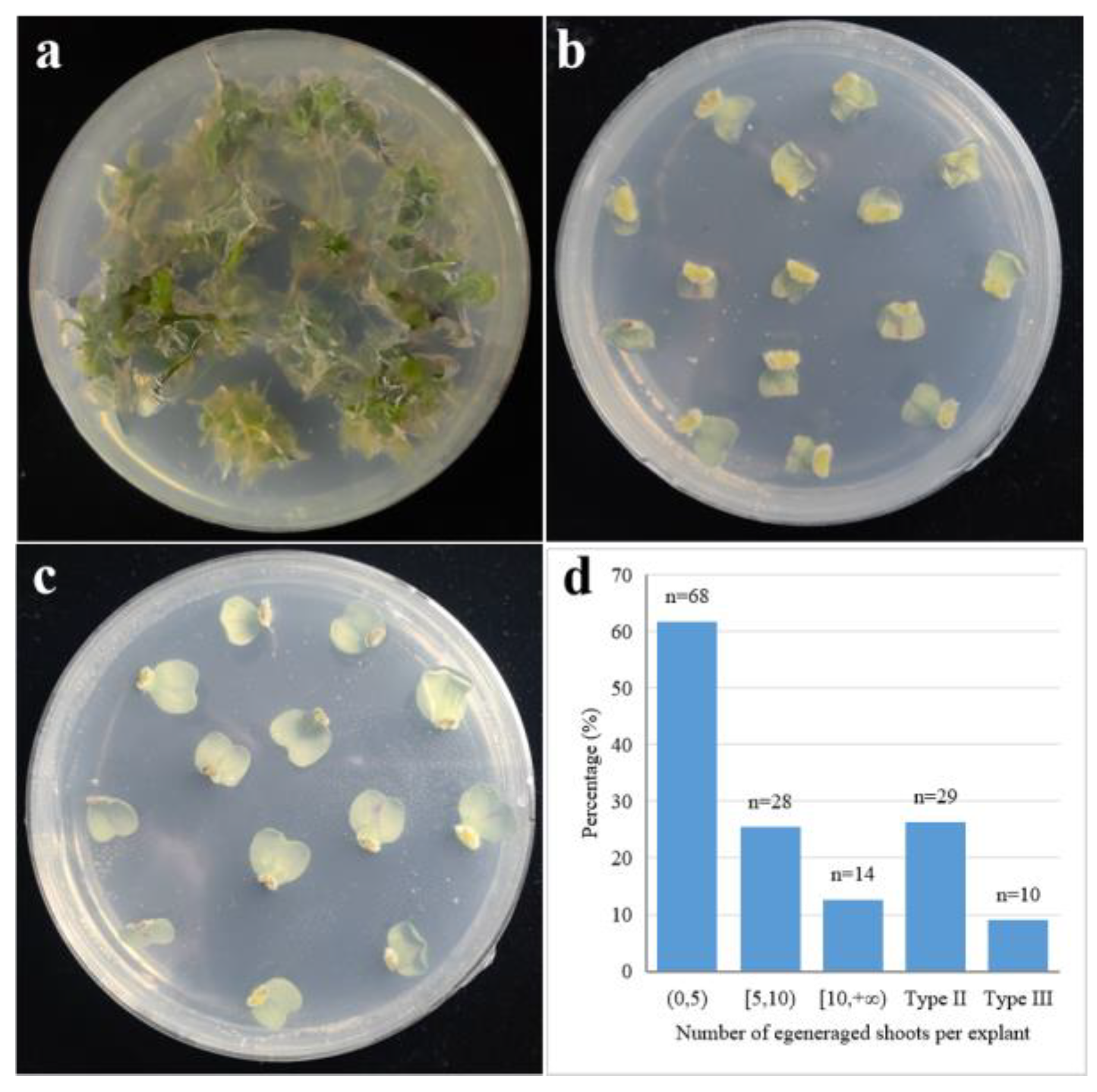
3.1. Variation in Regeneration Efficiency of Cotyledonary Petioles among Different Genotypes
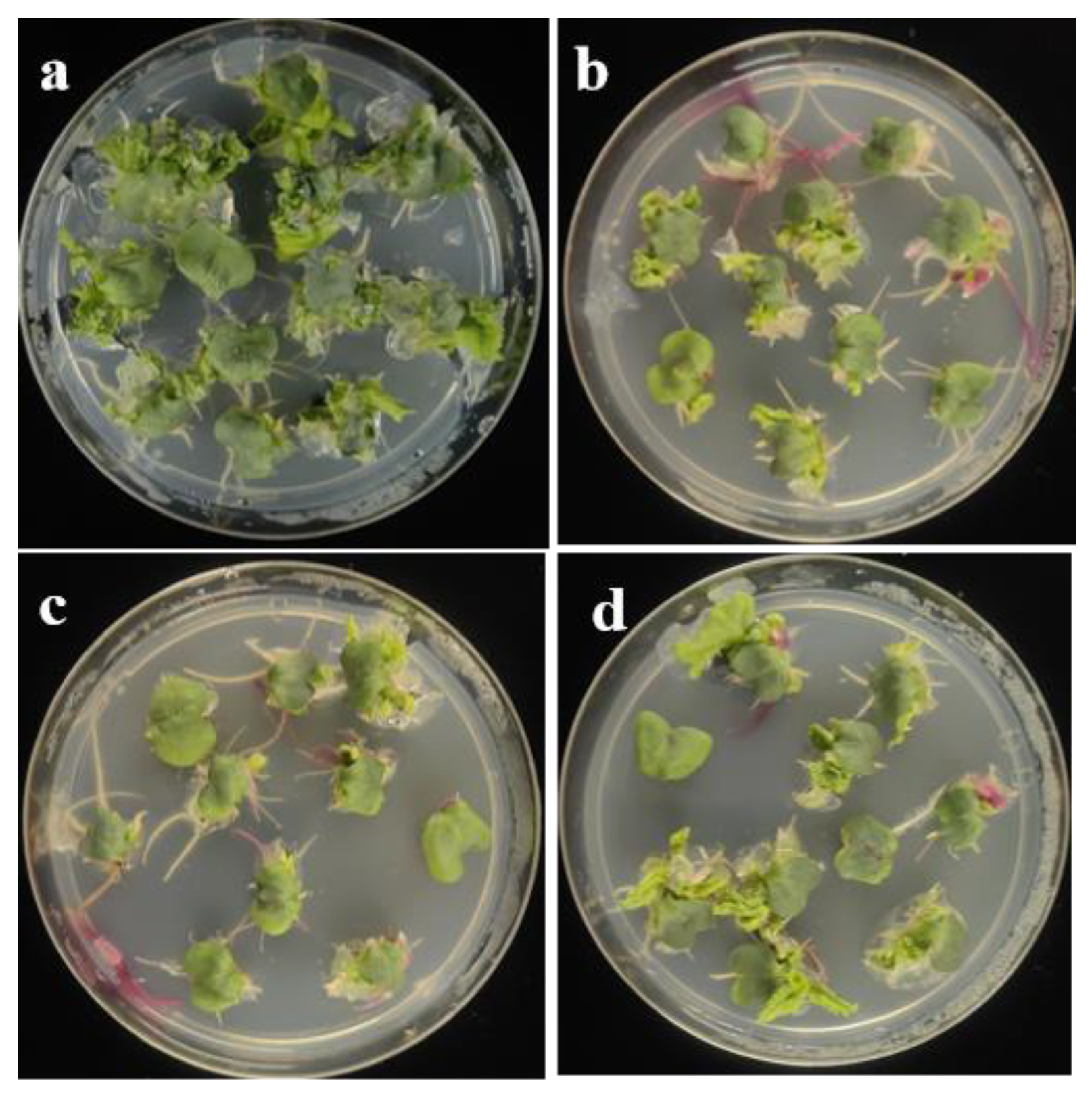
3.2. Impact of Agrobacterium Infection on the Regeneration Capacity of Cotyledonary Petioles
3.3. Screening of Chemical Reagents that Promote Cotyledonary Petiole Regeneration After Agrobacterium Infection
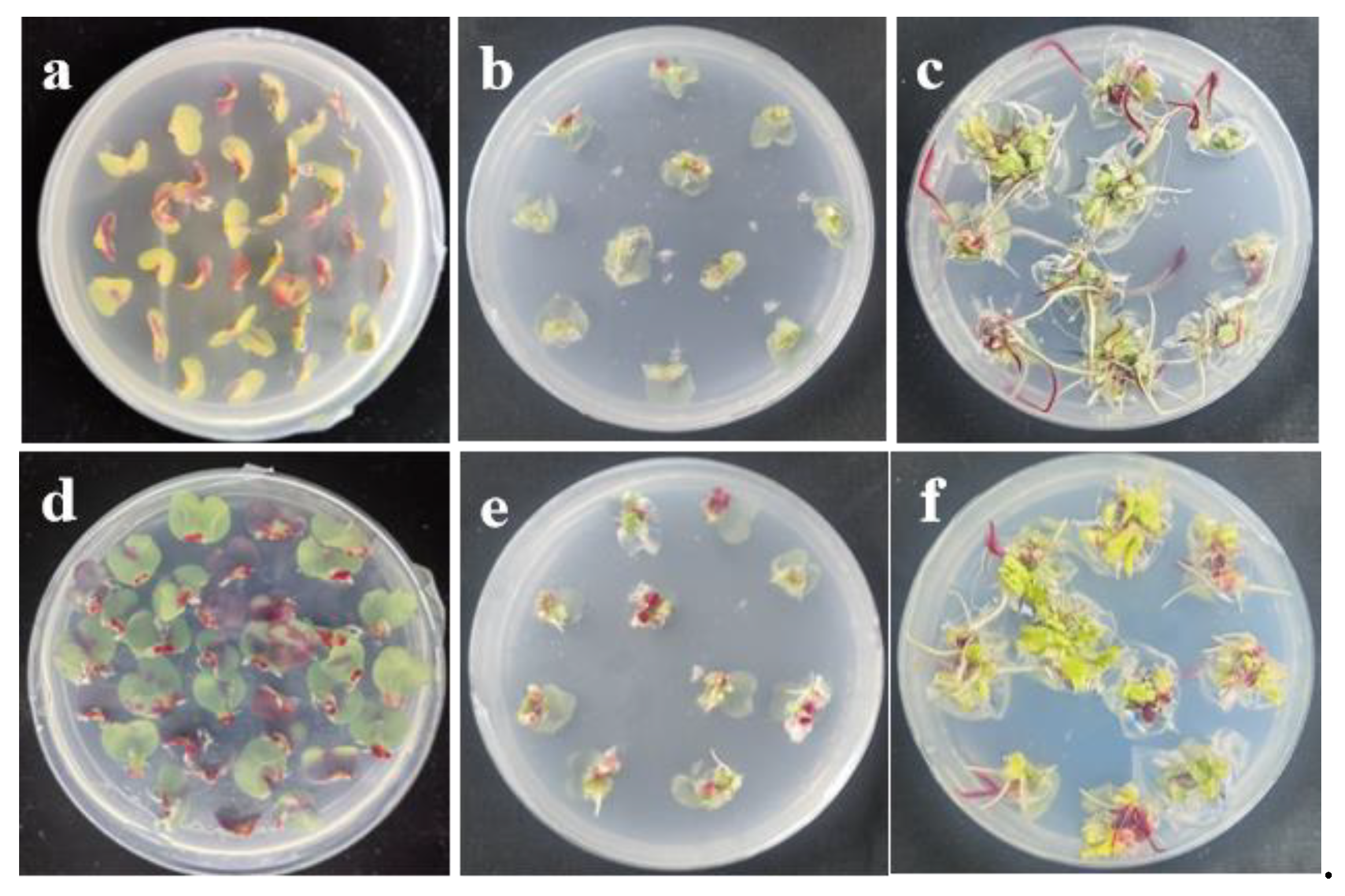
3.4. Effects of 6-BA and NAA Concentrations on the Regeneration Efficiency of Cotyledonary Petioles after Agrobacterium Infection
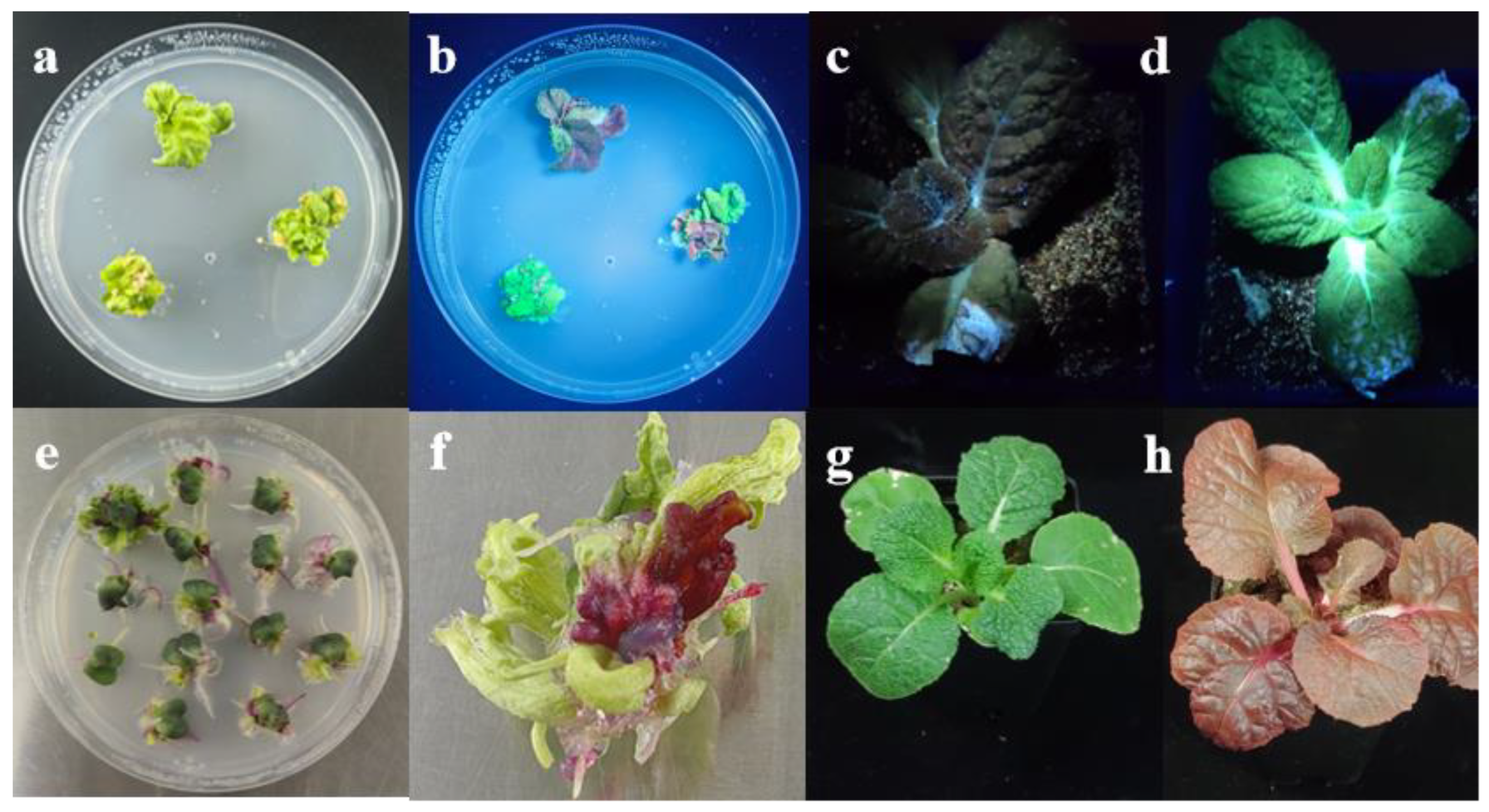
3.5. Comparison of Selectable Markers and Visual Reporters for Genetic Transformation Efficiency
3.6. Genetic Segregation and Phenotypic Characterization of RUBY Overexpression Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Su, T.; Wang, W.; Li, P.; Zhang, B.; Li, P.; Xin, X.; Sun, H.; Yu, Y.; Zhang, D.; Zhao, X.; et al. A Genomic Variation Map Provides Insights into the Genetic Basis of Spring Chinese Cabbage (Brassica rapa ssp. pekinensis) Selection. Mol. Plant. 2018, 11, 1360–1376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Su, T.; Zhang, B.; Yu, S.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, W.; Li, P.; Xin, X. Review and Prospects of Chinese Cabbage Breeding for the Past 70 Years in China. Acta. Hortic. Sin. 2025, 52, 1111–1135. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The Genome of the Mesopolyploid Crop Species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Han, D.; Huang, K.; Du, Y.; Yang, J.; Zhang, J.; Wen, C.; Wu, T. Genome-based Breeding Approaches in Major Vegetable Crops. Theor. Appl. Genet. 2020, 133, 1739–1752. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, P.; Zhao, J.; Hong, Y. Chinese Cabbage: An Emerging Model for Functional Genomics in Leafy Vegetable Crops. Trends Plant Sci. 2023, 28, 515–518. [Google Scholar] [CrossRef]
- Thomson, G.; Dickinson, L.; Jacob, Y. Genomic Consequences Associated with Agrobacterium-Mediated Transformation of Plants. Plant J. 2024, 117, 342–363. [Google Scholar] [CrossRef]
- Cardi, T.; Murovec, J.; Bakhsh, A.; Boniecka, J.; Bruegmann, T.; Bull, S.E.; Eeckhaut, T.; Fladung, M.; Galović, V.; Linkiewicz, A.; et al. CRISPR/Cas-Mediated Plant Genome Editing: Outstanding Challenges a Decade After Implementation. Trends Plant Sci. 2023, 28, 1144–1165. [Google Scholar] [CrossRef]
- Chen, Z.; Debernardi, J.M.; Dubcovsky, J.; Gallavotti, A. Recent Advances in Crop Transformation Technologies. Nat. Plants 2022, 8, 1343–1351. [Google Scholar] [CrossRef]
- Rahman, S.U.; Khan, M.O.; Ullah, R.; Ahmad, F.; Raza, G. Agrobacterium-Mediated Transformation for the Development of Transgenic Crops; Present and Future Prospects. Mol. Biotechnol. 2024, 66, 1836–1852. [Google Scholar] [CrossRef]
- Takasaki, T.; Hatakeyama, K.; Suzuki, G.; Watanabe, M.; Isogai, A.; Hinata, K. The S Receptor Kinase Determines Self-Incompatibility in Brassica Stigma. Nature 2000, 403, 913–916. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Ahn, H.; Ryu, J.; Oh, Y.; Sivanandhan, G.; Won, K.H.; Park, Y.D.; Kim, J.S.; Kim, H.; Lim, Y.P.; et al. Generation of Early-Flowering Chinese Cabbage (Brassica rapa spp. pekinensis) through CRISPR/Cas9-Mediated Genome Editing. Plant Biotechnol. Rep. 2019, 13, 491–499. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, L.; Pei, Z.; Zhang, L.; Liu, Z.; Liu, D.; Hao, X.; Jin, Z.; Pei, Y. Hydrogen Sulfide Promotes Flowering in Heading Chinese Cabbage by S-Sulfhydration of BraFLCs. Hortic. Res. 2021, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ma, W.; Liu, M.; Li, X.; Li, J.; Lu, Y.; Li, G.; Zhang, S.; Feng, D.; Wang, Y.; et al. OCTOPUS Regulates BIN2 to Control Leaf Curvature in Chinese Cabbage. Proc. Natl. Acad. Sci. USA 2022, 119, e2208978119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Su, T.; Xin, X.; Li, P.; Wang, J.; Wang, W.; Yu, Y.; Zhao, X.; Zhang, D.; Li, D.; et al. Wall-Associated Kinase BrWAK1 Confers Resistance to Downy Mildew in Brassica rapa. Plant Biotechnol. J. 2023, 21, 2125–2139. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Siddique, M.I.; Kim, D.S.; Lee, E.S.; Han, K.; Kim, S.G.; Lee, H.E. CRISPR/Cas9-Mediated Gene Editing to Confer Turnip Mosaic Virus (TuMV) Resistance in Chinese Cabbage (Brassica rapa). Hortic. Res. 2023, 10, uhad078. [Google Scholar] [CrossRef]
- Su, T.; Wang, W.; Li, P.; Xin, X.; Yu, Y.; Zhao, X.; Zhang, D.; Yu, S.; Zhang, F. Natural Variations of BrHISN2 Provide a Genetic Basis for Growth-Flavour Trade-Off in Different Brassica rapa Subspecies. New Phytol. 2021, 231, 2186–2199. [Google Scholar] [CrossRef]
- Yuan, J.; Shen, C.; Chen, R.; Qin, Y.; Li, S.; Sun, B.; Feng, C.; Guo, X. BrCNGC12 and BrCNGC16 Mediate Ca2+ Absorption and Transport to Enhance Resistance to Tipburn in Chinese Cabbage. Plant Biotechnol. J. 2025, 23, 2871–2887. [Google Scholar] [CrossRef]
- Shen, C.; Qin, Y.; Li, S.; Chen, R.; Zhang, X.; Zhang, Y.; Feng, C.; Xu, Y.; Yuan, R.; Guo, X.; Yuan, J. Molecular Mechanism by Which the BrFIT2-BrbHLH-BrIRT1 Module Synergistically Regulates Iron Absorption in Brassica rapa ssp. Pekinensis. Plant Biotechnol. J. 2025, in press. [Google Scholar] [CrossRef]
- Sparrow, P.A.C.; Irwin, J.A.; Goldsack, C.M.; Østergaard, L. Brassica Transformation: Commercial Application and Powerful Research Tool. Transgenic Plant J. 2007, 1, 330–339. [Google Scholar]
- Ahmed, N.U.; Park, J.I.; Kim, H.R.; Nou, I.S. Progress in Genetic Manipulation of the Brassicaceae. J. Plant Biotechnol. 2012, 39, 1–12. [Google Scholar] [CrossRef]
- Jun, S.; Kwon, S.Y.; Pack, K.Y.; Paek, K.H. Agrobacterium-Mediated Transformation and Regeneration of Fertile Transgenic Plants of Chinese Cabbage (Brassica campestris ssp. pekinensis cv. ‘Spring Flavor’). Plant Cell Rep. 1995, 14, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, T.; Hatakeyama, K.; Ojima, K.; Watanabe, M.; Toriyama, K.; Hinata, K. Factors Influencing Agrobacterium-Mediated Transformation of Brassica rapa L. Jpn. J. Breed. 1997, 47, 127–134. [Google Scholar] [CrossRef]
- Zhang, F.L.; Takahata, Y.; Watanabe, M.; Xu, J.B. Agrobacterium-Mediated Transformation of Cotyledonary Explants of Chinese Cabbage (Brassica campestris L. ssp. pekinensis). Plant Cell Rep. 2000, 19, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, H.; Zhao, Y.; Zong, P.; Zhan, Z.; Piao, Z. Establishment of a Simple and Efficient Agrobacterium-Mediated Genetic Transformation System to Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Plant J. 2021, 7, 117–128. [Google Scholar] [CrossRef]
- Sivanandhan, G.; Moon, J.; Sung, C.; Bae, S.; Yang, Z.H.; Jeong, S.Y.; Choi, S.R.; Kim, S.G.; Lim, Y.P. L-Cysteine Increases the Transformation Efficiency of Chinese Cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 2021, 12, 767140. [Google Scholar] [CrossRef]
- Maren, N.A.; Duan, H.; Da, K.; Yencho, G.C.; Ranney, T.G.; Liu, W. Genotype-Independent Plant Transformation. Hortic. Res. 2022, 9, uhac047. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, S.; An, X. Functional Mechanisms and the Application of Developmental Regulators for Improving Genetic Transformation in Plants. Plants 2024, 13, 2841. [Google Scholar] [CrossRef]
- Xu, P.; Zhong, Y.; Xu, A.; Liu, B.; Zhang, Y.; Zhao, A.; Yang, X.; Ming, M.; Cao, F.; Fu, F. Application of Developmental Regulators for Enhancing Plant Regeneration and Genetic Transformation. Plants 2024, 13, 1272. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Wang, W.; Wang, Y.; Chen, X.; Wu, H.; Gao, Z.; Xu, H.; Liu, T.; Li, Y.; et al. Efficient Genetic Transformation and Gene Editing of Chinese Cabbage Using Agrobacterium rhizogenes. Plant Physiol. 2025, 197, kiae543. [Google Scholar] [CrossRef]
- Lian, Z.; Nguyen, C.D.; Ruan, Z.; Liu, L.; Wang, G.; Chen, J.; Wang, S.; Yi, G.; Wilson, S.; Ozias-Akins, P.; Gong, H.; Huo, H. Application of Developmental Regulators to Improve in Planta or in Vitro Transformation in Plants. Plant Biotechnol. J. 2022, 20, 1622–1635. [Google Scholar] [CrossRef]
- Pitzschke, A. Agrobacterium Infection and Plant Defense—Transformation Success Hangs by a Thread. Front. Plant Sci. 2013, 4, 519. [Google Scholar] [CrossRef]
- Nonaka, S.; Yuhashi, K.I.; Takada, K.; Sugawara, M.; Minamisawa, K.; Ezura, H. Ethylene Production in Plants During Transformation Suppresses vir Gene Expression in Agrobacterium tumefaciens. New Phytol. 2008, 178, 647–656. [Google Scholar] [CrossRef]
- Dan, Y.; Zhang, S.; Matherly, A. Regulation of Hydrogen Peroxide Accumulation and Death of Agrobacterium-Transformed Cells in Tomato Transformation. Plant Cell Tissue Organ Cult. 2016, 127, 229–236. [Google Scholar] [CrossRef]
- Anand, A.; Uppalapati, S.R.; Ryu, C.M.; Allen, S.N.; Kang, L.; Tang, Y.; Mysore, K.S. Salicylic Acid and Systemic Acquired Resistance Play a Role in Attenuating Crown Gall Disease Caused by Agrobacterium tumefaciens. Plant Physiol. 2008, 146, 703–715. [Google Scholar] [CrossRef]
- Rosas-Díaz, T.; Cana-Quijada, P.; Amorim-Silva, V.; Botella, M.A.; Lozano-Durán, R.; Bejarano, E.R. Arabidopsis NahG Plants as a Suitable and Efficient System for Transient Expression Using Agrobacterium tumefaciens. Mol. Plant 2017, 10, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, S.; Sugawara, M.; Minamisawa, K.; Yuhashi, K.I.; Ezura, H. 1-Aminocyclopropane-1-carboxylate Deaminase Enhances Agrobacterium tumefaciens-Mediated Gene Transfer into Plant Cells. Appl. Environ. Microbiol. 2008, 74, 2526–2528. [Google Scholar] [CrossRef]
- Choi, S.W.; Kumaishi, K.; Motohashi, R.; Enoki, H.; Chacuttayapong, W.; Takamizo, T.; Saika, H.; Endo, M.; Yamada, T.; Hirose, A.; et al. Oxicam-Type Nonsteroidal Anti-Inflammatory Drugs Enhance Agrobacterium-Mediated Transient Transformation in Plants. Plant Biotechnol. 2022, 39, 323–327. [Google Scholar] [CrossRef]
- Liu, S.; Shi, Y.; Liu, F.; Guo, Y.; Lu, M. LaCl3 Treatment Improves Agrobacterium-Mediated Immature Embryo Genetic Transformation Frequency of Maize. Plant Cell Rep. 2022, 41, 1439–1448. [Google Scholar] [CrossRef]
- Sundar, I.K.; Sakthivel, N. Advances in Selectable Marker Genes for Plant Transformation. J. Plant Physiol. 2008, 165, 1698–1716. [Google Scholar] [CrossRef]
- Miki, B.; McHugh, S. Selectable Marker Genes in Transgenic Plants: Applications, Alternatives and Biosafety. J. Biotechnol. 2004, 107, 193–232. [Google Scholar] [CrossRef]
- Penna, S.; Sági, L.; Swennen, R. Positive Selectable Marker Genes for Routine Plant Transformation. In Vitro Cell. Dev. Biol. Plant 2002, 38, 125–128. [Google Scholar] [CrossRef]
- Chiu, W.L.; Niwa, Y.; Zeng, W.; Hirano, T.; Kobayashi, H.; Sheen, J. Engineered GFP as a Vital Reporter in Plants. Curr. Biol. 1996, 6, 325–330. [Google Scholar] [CrossRef]
- Yuan, G.; Lu, H.; Tang, D.; Hassan, M.M.; Li, Y.; Chen, J.G.; Tuskan, G.A.; Yang, X. Expanding the Application of a UV-Visible Reporter for Transient Gene Expression and Stable Transformation in Plants. Hortic. Res. 2021, 8, 234. [Google Scholar] [CrossRef]
- He, Y.; Zhang, T.; Sun, H.; Zhan, H.; Zhao, Y. A Reporter for Noninvasively Monitoring Gene Expression and Plant Transformation. Hortic. Res. 2020, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Sharifova, S.; Prasad, K.V.; Cheema, A.; Reddy, A.S. Genetically Encoded Betalain-Based RUBY Visual Reporters: Noninvasive Monitoring of Biological Processes. Trends Plant Sci. 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Malabadi, R.B.; Teixeira da Silva, J.A.; Nataraja, K. Green Fluorescent Protein in the Genetic Transformation of Plants. Transgenic Plant J. 2008, 2, 86–109. [Google Scholar]
- Saika, H.; Sakamoto, W.; Maekawa, M.; Toki, S. Highly Efficient Visual Selection of Transgenic Rice Plants Using Green Fluorescent Protein or Anthocyanin Synthetic Genes. Plant Biotechnol. 2011, 28, 107–110. [Google Scholar] [CrossRef]
- Komori, T.; Imayama, T.; Kato, N.; Ishida, Y.; Ueki, J.; Komari, T. Current Status of Binary Vectors and Superbinary Vectors. Plant Physiol. 2007, 145, 1155–1160. [Google Scholar] [CrossRef]
- Kay, R.; Chan, A.M.Y.; Daly, M.; McPherson, J. Duplication of CaMV 35S Promoter Sequences Creates a Strong Enhancer for Plant Genes. Science 1987, 236, 1299–1302. [Google Scholar] [CrossRef]
- Yamasaki, S.; Sanada, Y.; Imase, R.; Matsuura, H.; Ueno, D.; Demura, T.; Kato, K. Arabidopsis thaliana Cold-Regulated 47 Gene 5′-Untranslated Region Enables Stable High-Level Expression of Transgenes. J. Biosci. Bioeng. 2018, 125, 124–130. [Google Scholar] [CrossRef]
- Bevan, M.; Barnes, W.M.; Chilton, M.D. Structure and Transcription of the Nopaline Synthase Gene Region of T-DNA. Nucleic Acids Res. 1983, 11, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Hernández-Coronado, M.; Dias Araujo, P.C.; Ip, P.L.; Nunes, C.O.; Rahni, R.; Wudick, M.M.; Lizzio, M.A.; Feijó, J.A.; Birnbaum, K.D. Plant Glutamate Receptors Mediate a Bet-Hedging Strategy between Regeneration and Defense. Dev. Cell 2022, 57, 451–465.e6. [Google Scholar] [CrossRef]
- Liu, C.; Song, G.; Zhao, Y.; Fang, B.; Liu, Z.; Ren, J.; Feng, H. Trichostatin A Induced Microspore Embryogenesis and Promoted Plantlet Regeneration in Ornamental Kale (Brassica oleracea var. acephala). Horticulturae 2022, 8, 790. [Google Scholar] [CrossRef]
- Camacho-Fernández, C.; Corral-Martínez, P.; Calabuig-Serna, A.; Arjona-Mudarra, P.; Sancho-Oviedo, D.; Boutilier, K.; Seguí-Simarro, J.M. The Different Response of Brassica napus Genotypes to Microspore Embryogenesis Induced by Heat Shock and Trichostatin A Is Not Determined by Changes in Cell Wall Structure and Composition but by Different Stress Tolerance. Physiol. Plant. 2024, 176, e14405. [Google Scholar] [CrossRef]
- Tran, S.; Ison, M.; Ferreira Dias, N.C.; Ortega, M.A.; Chen, Y.F.S.; Peper, A.; Hu, L.; Xu, D.; Mozaffari, K.; Severns, P.M.; et al. Endogenous Salicylic Acid Suppresses de novo Root Regeneration from Leaf Explants. PLoS Genet. 2023, 19, e1010636. [Google Scholar] [CrossRef]
- Yi, X.; Wang, C.; Yuan, X.; Zhang, M.; Zhang, C.; Qin, T.; Wang, H.; Xu, L.; Liu, L.; Wang, Y. Exploring an Economic and Highly Efficient Genetic Transformation and Genome-Editing System for Radish Through Developmental Regulators and Visible Reporter. Plant J. 2024, 120, 1682–1692. [Google Scholar] [CrossRef]
- Ge, X.; Wang, P.; Wang, Y.; Wei, X.; Chen, Y.; Li, F. Development of an Eco-Friendly Pink Cotton Germplasm by Engineering Betalain Biosynthesis Pathway. Plant Biotechnol. J. 2022, 21, 674. [Google Scholar] [CrossRef]
- Deng YJ, Duan AQ, Liu H, Wang YH, Zhang RR, Xu ZS, Xiong AS. Generating colorful carrot germplasm through metabolic engineering of betalains pigments. Hortic Res. 2023, 10, uhad024. [CrossRef] [PubMed] [PubMed Central]
- Tan, J.; Wang, Y.; Lin, Z.; Chai, N.; Xue, Y.; Chen, L.; Liu, Y.G.; Zhu, Q. eRUBY Rice: Co-expression of a Feedback-Insensitive TyrA Arogenate Dehydrogenase with RUBY Enhances Endosperm Betalain Levels. Plant Physiol. 2025, 199, kiaf416. [Google Scholar] [CrossRef]
- Zhang, F.L.; Takahata, Y.; Xu, J.B. Medium and Genotype Factors Influencing Shoot Regeneration from Cotyledonary Explants of Chinese Cabbage (Brassica campestris L. ssp. pekinensis). Plant Cell Rep. 1998, 17, 780–786. [Google Scholar] [CrossRef]
- Türkoğlu, A.; Haliloğlu, K.; Demirel, F.; Aydin, M.; Çiçek, S.; Yiğider, E.; Demirel, S.; Piekutowska, M.; Szulc, P.; Niedbała, G. Machine Learning Analysis of the Impact of Silver Nitrate and Silver Nanoparticles on Wheat (Triticum aestivum L.): Callus Induction, Plant Regeneration, and DNA Methylation. Plants 2023, 12, 4151. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, G.L. Evidence of Multiple Complex Patterns of T-DNA Integration into the Rice Genome. Theor. Appl. Genet. 2000, 100, 461–470. [Google Scholar] [CrossRef]
- Veena, V.; Taylor, C.G. Agrobacterium rhizogenes: Recent Developments and Promising Applications. In Vitro Cell. Dev. Biol. Plant 2007, 43, 383–403. [Google Scholar] [CrossRef]

| Accession | Non-infection | GV3101 | LBA4404 | K599 |
|---|---|---|---|---|
| L68 | 12.52 ± 0.39 a | 5.70 ± 0.47 c | 4.28 ± 0.63 d | 6.24 ± 0.68 b |
| L69 | 10.46 ± 0.54 a | 4.52 ± 0.32 c | 5.18 ± 0.58 c | 7.22 ± 0.66 b |
| L131 | 11.69 ± 0.64 a | 4.24 ± 0.64 c | 3.68 ± 0.66 c | 6.54 ± 0.67 b |
| Reagent | Concentration | Treatment Stage | Regenerated Shoots per Explant |
|---|---|---|---|
| CK | / | / | 6.20 ± 0.38 |
| Tenoxicam | 50 μM | Seed germination | 8.55 ± 0.36 * |
| Tenoxicam | 50 μM | Co-cultivation | 6.15 ± 0.39 |
| Tenoxicam | 50 μM | Shoot induction | 5.27 ± 0.45 |
| CNQX | 50 μM | Seed germination | 6.12 ± 0.38 |
| CNQX | 50 μM | Co-cultivation | 6.08 ± 0.68 |
| CNQX | 50 μM | Shoot induction | 6.05 ± 0.30 |
| LaCl3 | 10 mM | Seed germination | 6.14 ± 0.43 |
| LaCl3 | 10 mM | Co-cultivation | 6.06 ± 0.33 |
| LaCl3 | 10 mM | Shoot induction | 4.77 ± 0.30 * |
| Trichostatin A | 1 μM | Seed germination | 6.10 ± 0.63 |
| Trichostatin A | 1 μM | Co-cultivation | 5.98 ± 0.33 |
| Trichostatin A | 1 μM | Shoot induction | 5.95 ± 0.45 |
| 6-BA (mg/L) | NAA (mg/L) | Non-infection | Infection |
|---|---|---|---|
| 4 | 0.0 | 0.22 ± 0.24 f | 0.14 ± 0.21 e |
| 4 | 0.2 | 12.05 ± 0.73 a | 5.82 ± 0.64 c |
| 4 | 0.4 | 9.12 ± 0.64 b | 6.63 ± 0.55 b |
| 4 | 0.5 | 6.45 ± 0.49 c | 7.43 ± 0.49 a |
| 4 | 0.6 | 3.15 ± 0.41 e | 5.88 ± 0.38 c |
| 4 | 0.2 | 4.86 ± 0.48 d | 4.86 ± 0.48 d |
| 2 | 0.2 | 8.76 ± 0.58 b | 4.20 ± 0.52 d |
| 6 | 0.2 | 9.15 ± 0.57 b | 4.55 ± 0.55 d |
| Selection Method | Vector | Regenerated Shoots per Explant | False Positive Rate (%) |
Genetic Transformation Efficiency (%) |
|---|---|---|---|---|
| CK (no infection, no selection) | NA | 12.03 ± 0.62 a | NA | NA |
| Kanamycin | pCAMBIA2300-eYGFPuv | 0.45 ± 0.14 c | 82.81 ± 12.31 b | 6.67 ± 3.06 b |
| Hygromycin | pCMABIA1300-eYGFPuv | 0.19 ± 0.03 c | 84.72 ± 16.67 b | 2.67 ± 3.06 b |
| eYGFPuv | pCMABIA2300-eYGFPuv | 8.55 ± 0.72 b | 97.72 ± 0.56 a | 19.33 ± 4.16 a |
| RUBY | pCMABIA2300-RUBY | 9.77 ± 0.44 b | 97.95 ± 0.46 a | 20.00 ± 4.62 a |
| Line | Growth Period (days) |
Gross Weight (kg) |
Net Weight (kg) |
Head Height (cm) |
Head Width (cm) |
Number of Head Leaves |
|---|---|---|---|---|---|---|
| L68 | 61.40 ± 6.22 a | 1.98 ± 0.13 a | 1.49 ± 0.08 a | 25.51 ± 1.66 a | 13.43 ± 0.83 a | 27.33 ± 1.08 a |
| RB16 | 53.87 ± 3.15 b | 1.32 ± 0.11 c | 0.92 ± 0.11 c | 16.60 ± 0.90 c | 8.91 ± 0.39 c | 24.73 ± 0.60 a |
| RB25 | 54.53 ± 2.01 b | 1.72 ± 0.11 b | 1.32 ± 0.13 b | 22.79 ± 1.04 b | 12.45 ± 0.18 b | 25.00 ± 0.90 a |
| RB52 | 47.93 ± 1.12 c | 0.91 ± 0.11 d | 0.72 ± 0.12 c | 11.87 ± 0.51 d | 6.65 ± 0.19 d | 26.80 ± 0.69 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




