I. Introduction
Multicellular organisms use a variety of signals to orchestrate the processes that keep them alive. Plants coordinate their development, reproduction and environmental response through phytohormones. These organic molecules are perceived by proteins to initiate signaling cascades that result in cellular and systemic outcomes. During the 20th century, the studied phytohormones were molecules with simple structures (1). It wasn’t until the 90s that the first plant peptide hormone, named Systemin, was discovered in tomato plants (2). From there on, knowledge on these peptides only expanded, and their essential roles in plants became increasingly prominent (3).
The RAPID ALKALINIZATION FACTORS (RALFs) form a family of peptide hormones present throughout land plants. The number of RALFs varies among different plant species. In eudicots and monocots, the average number of RALF genes per species is approximately 20 and 14, respectively (4). Arabidopsis thaliana possesses a markedly high number of RALF peptides, totaling 37. Evidence suggests that this elevated quantity is the result of tandem gene duplications followed by neofunctionalization (5–7). Curiously, RALF-like genes are also found in phytopathogenic fungi, nematodes and bacteria (4,8,9).
RALFs are involved in plant development, fertilization and response to biotic and abiotic stresses (6). Since their discovery in 2001, the molecular mechanisms behind RALF-induced responses are being unraveled (6,10). The diversity of proteins involved in RALFs’ signaling pathways demonstrates an intricate regulation occurring at multiple levels, including protein interaction and phosphorylation, transcription, transcript processing, translation and post-translational modifications (11–13). These peptides are secreted to the apoplast, where they bind to protein receptors and pectin, inducing intra- and extracellular responses (10).
Topics II to X of this review focus on Arabidopsis thaliana, while Topic XI describes RALF roles in other plant species.
II. Structure
The
RALF genes are translated into precursor peptides (proRALFs or preproRALFs) with domains that dictate their processing and secretion. These newly synthesized RALFs have a length that varies between 80 and 120 amino acids (5). The precursors of all
Arabidopsis mature RALFs have a signal peptide in their N-terminal that directs them to the secretory pathway (5,10). PreproRALFs, however, have an additional domain to be processed, known as the pro-domain. At the C-terminal boundary of this domain is the RRXL motif, which is recognized and cleaved by subtilases (5,14). For example, preproAtRALF22 and preproAtRALF23 are processed by the subtilase SITE-1 PROTEASE (S1P), and this step is necessary for their secretion (15–17). Notably, the most studied RALFs from
A. thaliana, namely RALF1,4,19,22,23,34, possess a pro-domain (5). After processing, mature RALFs have a length of around 50 amino acids (5), and at least mature RALF23 is endogenously present at the 10s-100s of nanomolar range (13).
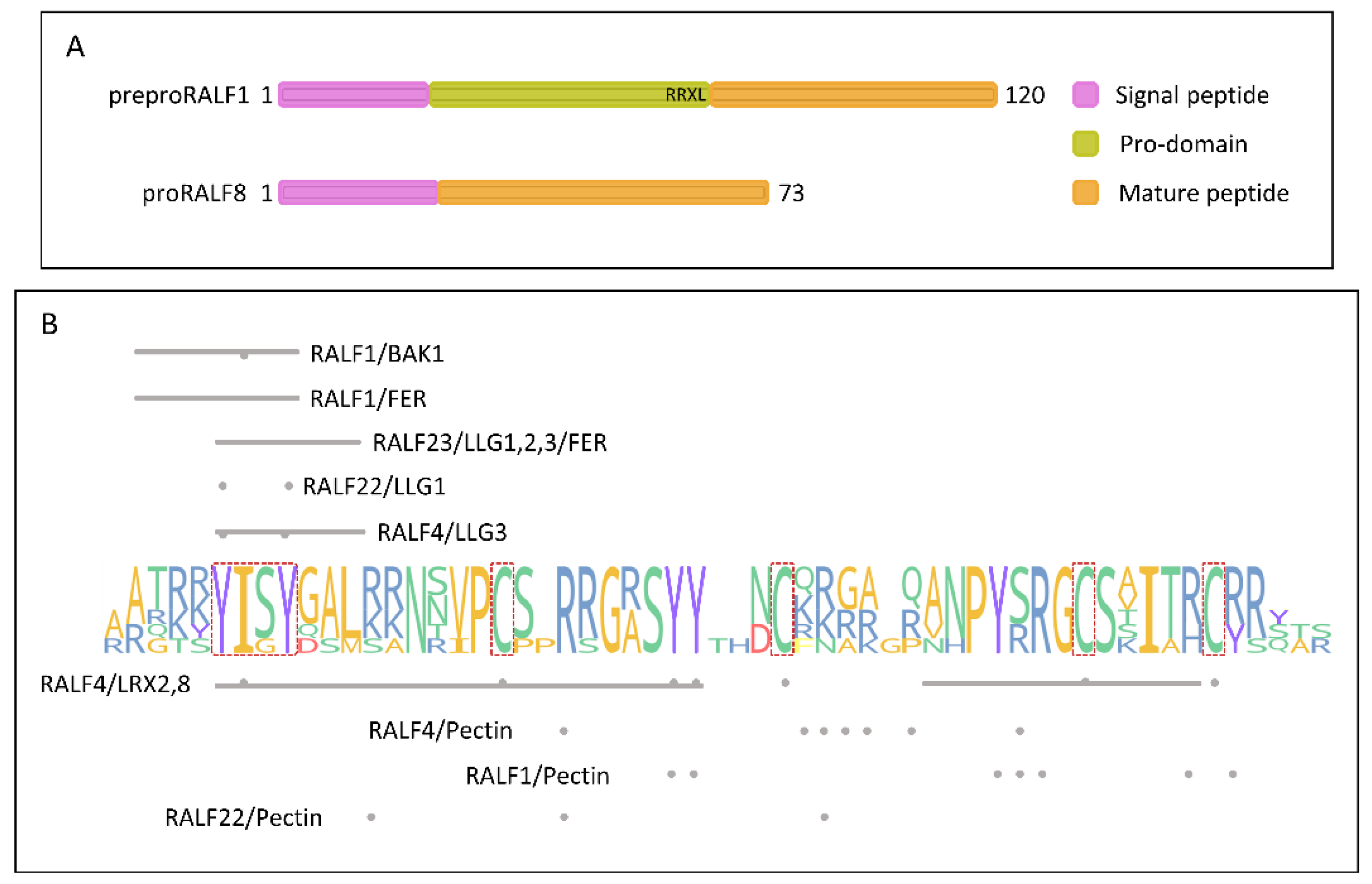
Figure 1A exemplifies the primary structures of a preproRALF and a proRALF.
These peptides can be divided into four clades. The RALFs whose precursor possesses the pro-domain are members of clades I, II, and III. Peptides from these three clades share the YISY motif and four cysteines residues (4). The YISY motif is responsible for RALFs binding to their receptors, while the cysteine pairs form disulfide bridges that structure the peptides and strengthen these interactions (18–22). On the other hand, RALFs whose precursors lack the pro-domain belong to clade IV, do not possess the YISY motif, and often miss one of the four cysteines (4). Despite their structural differences, RALFs from the four clades can share receptors, as evidenced by the fact that, in the reproductive context, at least one member of each clade functionally binds to FERONIA (FER) (23–25). Curiously, in silico studies suggest different binding conformations of various RALFs to FER, even among closely related peptides (26). In addition to their protein receptors, some RALF peptides are capable of binding to pectin (27–29).
Figure 1B illustrates regions and specific residues experimentally shown to be important for RALFs’ interactions with their binding partners.
Figure 1.
Primary structure of RALF peptides.
Figure 1.
Primary structure of RALF peptides.
(A) Examples of RALF precursors from clade I (preproRALF1) and clade IV (proRALF8), highlighting their domains.
(B) Alignment logo of the six most studied RALFs (RALF1, RALF4, RALF19, RALF22, RALF23, and RALF34), showing regions or residues important for peptide binding to their receptors or to pectin.
Red outline: YISY motif and cysteines; Gray bars: important regions for binding; Gray points: important residues for binding.
III. Receptors
The RALF peptides family has an intimate connection with three receptor families:
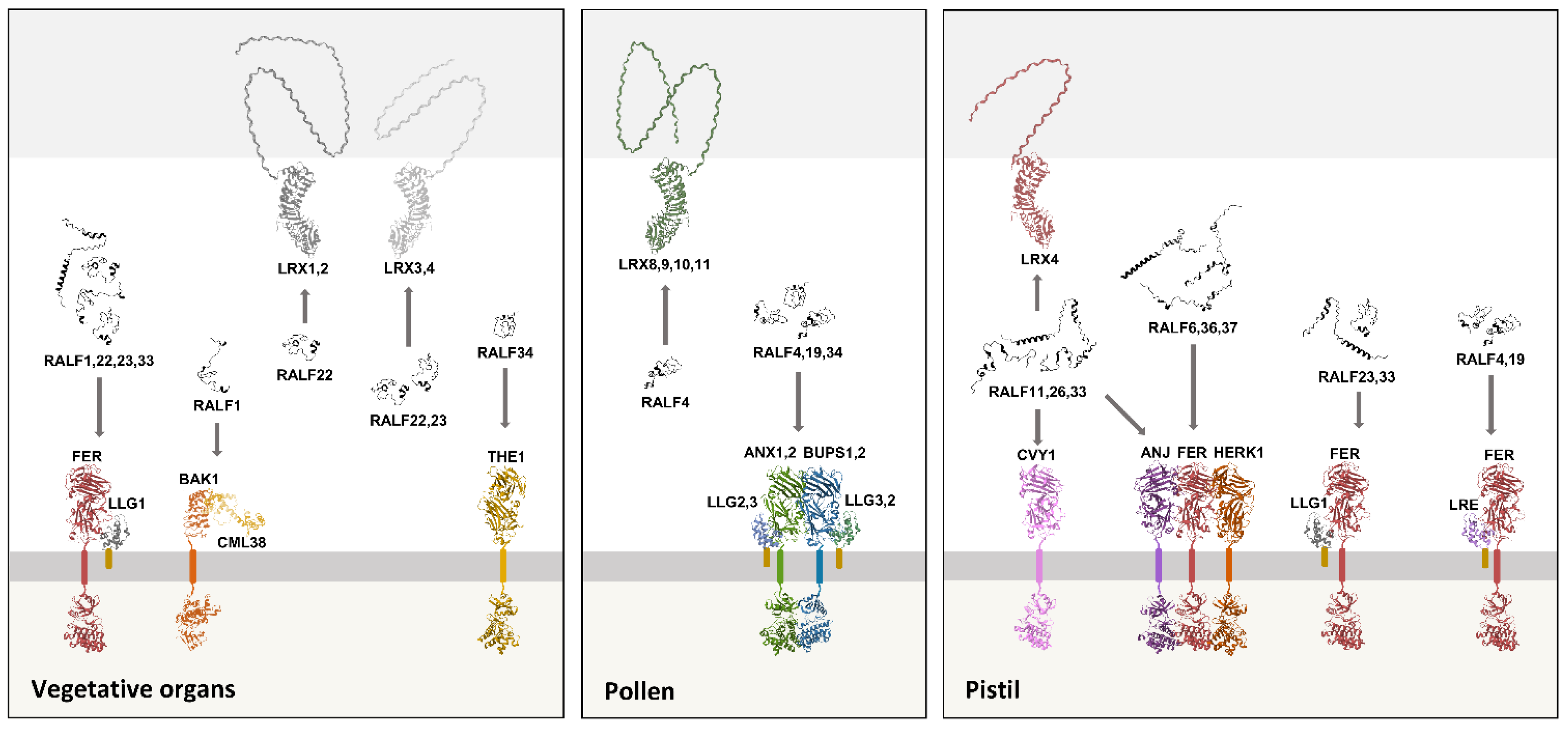
Catharanthus roseus RECEPTOR-LIKE KINASE 1-LIKE (CrRLK1Ls), LORELEI-LIKE-GPI-ANCHORED PROTEIN (LLGs) and LEUCINE-RICH REPEAT EXTENSIN (LRXs). The CrRLK1Ls family comprises 17 members in
A. thaliana (30). Among these, FERONIA, THESEUS1 (THE1), ANJEA (ANJ1,2), HERCULES1 (HERK1), BUDDHA'S PAPER SEAL (BUPS1,2), ANXUR (ANX1,2), and CURVY1 (CVY1) stand out for being RALF receptors (20,21,24,25,31,32). The extracellular portion of CrRLK1Ls consists of two malectin-like domains, responsible for binding to RALFs and pectin (21,33). The intracellular portion harbors a kinase domain, and upon RALF perception, a series of auto- and transphosphorylations activate the receptor (34). Acting as CrRLK1Ls’ chaperones and coreceptors, LORELEI (LRE) and LLG1,2,3 form a family of proteins anchored to the plasma membrane by a glycosylphosphatidylinositol (GPI), a lipid attached to LLGs posttranslationally (35–37). Members of the third receptor family, the LRXs, are characterized by an extensin domain, key to cell wall association, and a LEUCINE-RICH REPEAT (LRR) domain, responsible for their binding to RALF peptides (22,28,29,38–40). The LRX family comprises 11 paralogues in
A. thaliana, which are expressed in tissue-specific groups with functionally redundant members (40). Intriguingly, the dissociation constants (Kd) for the RALF/CrRLK1L and RALF/LLG interactions are around 0.2–5 µM, while the Kd for RALF/LRX interactions is around 3–5 nM, indicating a remarkably stronger association in the latter (16,18,22,40,41). In addition to these three receptor families, a protein complex formed by BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) and CALMODULIN-LIKE 38 (CML38) also perceives RALF1 in roots, with a Kd of ~4.5 µM (19,42).
Figure 2 summarizes the receptors whose RALFs binding was experimentally shown to have biological function.
IV. Rapid Cellular Responses
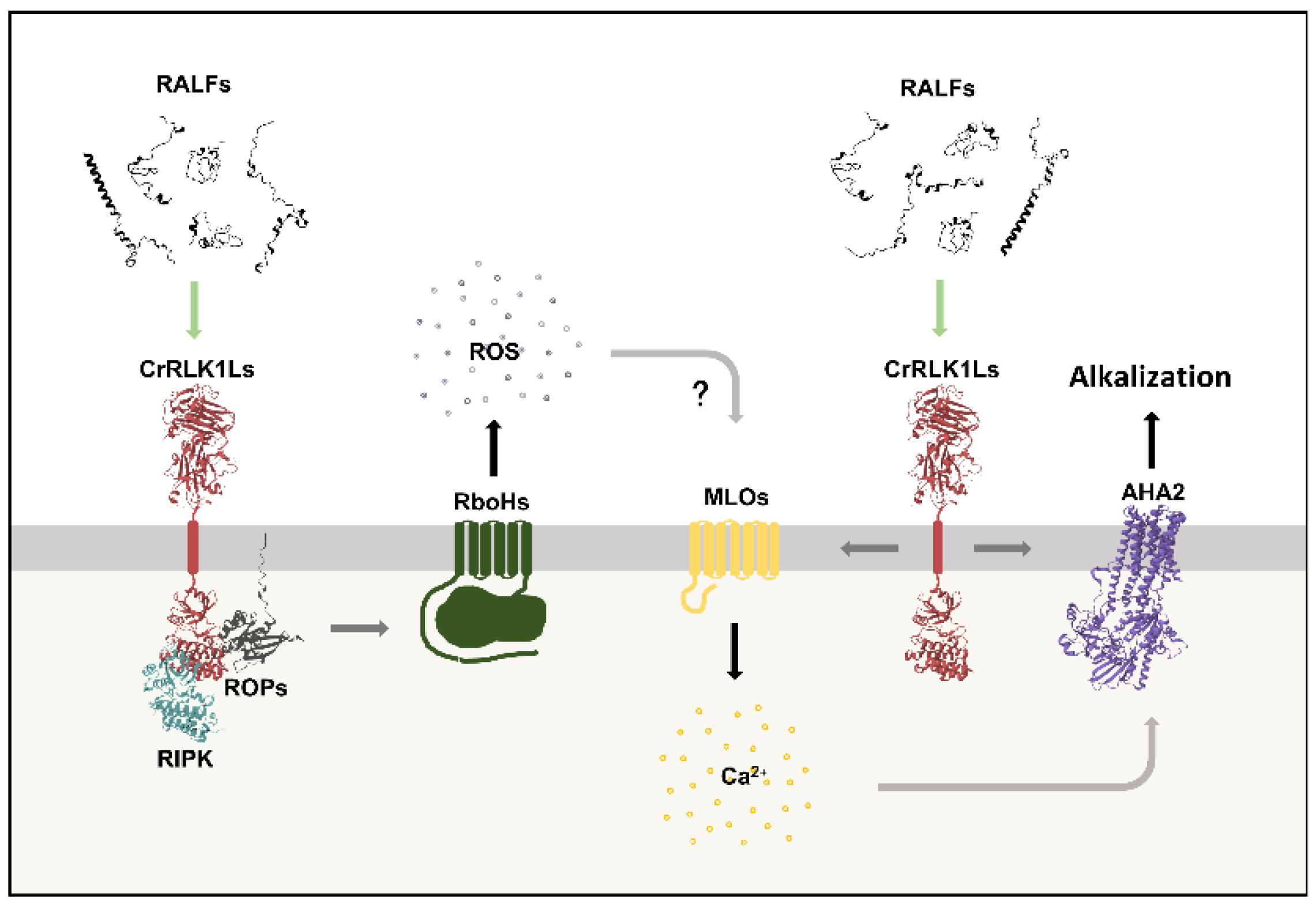
RALF peptides can elicit three rapid changes in the cell’s signaling status: a rise in apoplastic pH, the production of reactive oxygen species (ROS), and an influx of calcium. These three responses are mediated by the peptides’ canonical CrRLK1L receptors.
For apoplastic alkalinization, RALF1 binds FER to induce phosphorylation and inactivation of the proton pump H(+)-ATPASE 2 (AHA2) (21). The involvement of AHAs in this response was recently questioned (43). However, at the stigma, RALF23 and RALF33 binding to FER and ANJ leads to the protonation of AHA2 at its inactivation residue S899 (44), as seen for RALF1/FER in vegetative tissues (21). RALF1 treatment also induces AHA2 internalization, providing additional evidence for the peptide’s influence on the pumps’ activity (27). In addition to RALF1, treatment with most RALFs increases the apoplastic pH, and receptors other than FER may be involved (5). For example, THE1 is RALF34’s canonical receptor, responsible for transducing the peptide-triggered alkalinization (31). Furthermore, mutants lacking FER show extracellular alkalinization when treated with RALF36, although reduced, suggesting a second receptor (45).
The RALF-induced alkalinization contributes to root growth inhibition (43). The Acid Growth Theory postulates that apoplastic acidification triggered by the phytohormone auxin induces cell wall flexibility, leading to cellular expansion (46). Auxin activates proton pumps, which lower the apoplast pH and alter the activity of cell wall remodeling proteins. The proton gradient also propels water and solute intake, raising turgor pressure, and therefore aiding in cell expansion (47). On the other hand, apoplastic alkalinization by RALF1 is likely responsible for repressing cell expansion, as it is temporally correlated with root growth arrest (43). Moreover, the growth promoting PLANT PEPTIDE CONTAINING SULPHATED TYROSINE 1 (PSY1) binding to PSY1-RECEPTOR (PSY1R) activates AHA1 and AHA2, thereby lowering the apoplastic pH (48). PSY1/PSY1R positively regulate the transcription of RALF22,33,36, whose protein products arrest root growth (45). This feedback loop between peptides with opposing effects on root acidification and growth reinforces the Acid Growth Theory in this context.
RALF peptides induce a rise in cytosolic calcium concentrations ([Ca2+]cyt). This ion acts as a secondary messenger, and specific changes in [Ca2+]cyt are perceived by calcium-binding proteins that transduce the signal (49,50). Calcium signaling also occurs in waves that travel through plant tissues (51). RALF1 treatment raises [Ca2+]cyt in root cells through both cellular intake and mobilization from intracellular reserves (52). The RALF1-induced Ca2+ burst is mainly dependent on FER, but also on THE1 (21,27,31). In turn, THE1 is the main responsible for the RALF34-triggered calcium burst (31). Moreover, roots treated with RALF33 or RALF36 show specific changes in cellular and systemic [Ca2+]cyt profiles in a FER-dependent or -independent manner, respectively (45). RALF8 treatment also induces a rise in [Ca2+]cyt (53). In pollen tubes, members of the MILDEW RESISTANCE LOCUS O (MLO) channel family are responsible for RALF4- and RALF19-induced calcium intake following their binding to ANXs and BUPSs (54). In the synergids, RALF4,19 bind to the FER/LRE complex and activate the calcium channel NORTIA (NTA), another MLO (23). Collectively, these results suggest that CrRLK1L receptors and MLO channels modulate specific calcium responses for each RALF or group of RALFs, in different tissues.
In the presence of RALFs, calcium intake precedes apoplast alkalinization. The Ca2+ spike occurs in less than 60 seconds after RALF treatment, while the pH reaches its maximum after a few minutes (21,31,45,55). Plants pre-treated with La³⁺, a calcium channel blocker, show no alkalinization following RALF treatment, suggesting that the increase in apoplastic pH depends on the secondary messenger (45).
Another important change in plant cell’s signaling status is the ROS production by the NADPH oxidases RESPIRATORY BURST OXIDASE HOMOLOG (RboHs). In roots, RALF1/FER signaling is part of the basal ROS production, and exogenous application of the peptide enhances this response (27,56). Similarly, treating leaf discs with different RALFs raises ROS levels in a FER-dependent manner (5). In pollen tubes, RALF4,19 perception by the ANX/BUPS/LLG complexes regulates ROS oscillation (35). ROS are also central to the immune response, where treatments with RALF23 or RALF33 inhibit ROS production, while treatment with RALF17 or RALF22 stimulates it (16,57,58). The cytoplasmic kinase RPM1-INDUCED PROTEIN KINASE (RIPK), a well-described mediator of the immune ROS burst (59), is important for the RALF22-induced response (57). Members of the RHO-RELATED PROTEIN FROM PLANTS (ROP) family are GDP/GTP ligands activated by GUANINE NUCLEOTIDE EXCHANGE FACTOR (GEFs) (60). During pollen/stigma interactions, control of root microbiota and osmotic stress response, RALF/CrRLK1L signaling modulates ROS levels through ROPs (32,61,62).
In conclusion, the interaction of RALF peptides with CrRLK1L receptors modulates ROS, calcium, and pH levels (
Figure 3). In some contexts, an increase in apoplastic ROS induces calcium influx, and in systemic Ca
2+ and ROS waves, these responses stimulate each other (58). Therefore, the ROS production triggered by RALFs may contribute to Ca
2+ influx, which in turn is necessary for apoplastic alkalinization.
The RALF/CrRLK1L binding modulates: ROS production by RboHs through ROPs and/or RIPK activation; calcium influx through MLO channels; and apoplast alkalinization through phosphorylation and inactivation of AHA2 and/or another proton pump. RALF-triggered ROS production potentially contributes to calcium influx, which in turn precedes the peptide-induced apoplastic alkalinization.
For all figures in this review, green arrows indicate the initial stimulus; gray arrows, steps of the pathway; yellow arrows, phosphorylation; brown arrows, dephosphorylation; and black arrows indicate the ultimate effect of the pathway. Question marks (?) indicate potential steps yet to be proven experimentally.
V. Plant Development
RALFs, auxin and brassinosteroids crosstalk
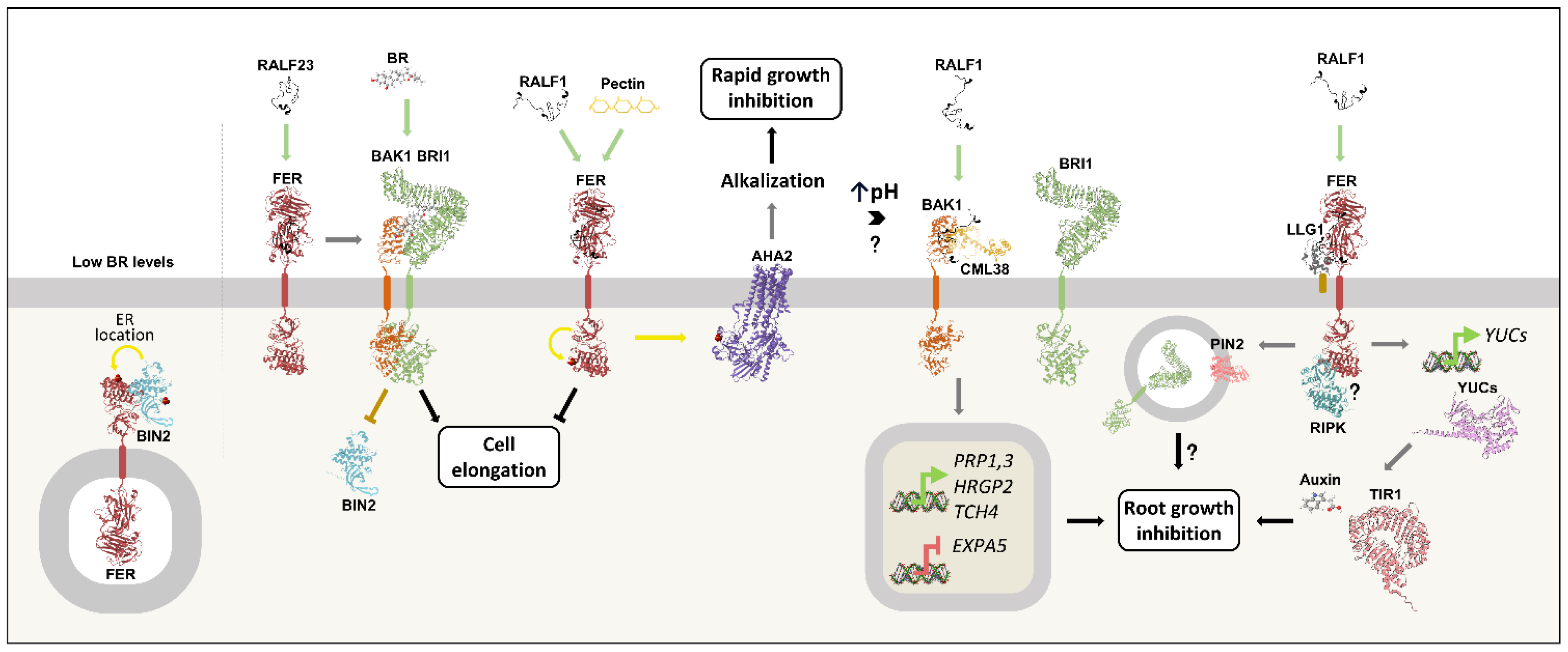
RALF peptides inhibit root and hypocotyl growth by limiting cell expansion (21,63). RALF1-induced growth inhibition involves crosstalk with the phytohormones brassinosteroids (BRs) and auxin (19,43,63). Auxin is a central regulator of plant development and modulates cell expansion, leading to differential growth in roots and hypocotyls (64–66). The auxin indole-3-acetic acid (IAA) is distributed through plant tissues in gradients that are precisely controlled by polar transport and local biosynthesis. Among the enzymes responsible for IAA biosynthesis, the YUCCA (YUCs) stand out for their role in the final reaction that produces the active hormone (67). Meanwhile, auxin polar flux is mediated by the membrane-bound PINs and AUXIN RESISTANT (AUXs) transporters (64). Ultimately, auxin responses rely on changes in gene expression triggered by the degradation of transcriptional repressors, a process where TRANSPORT INHIBITOR RESPONSE 1 (TIR1) is pivotal (47).
The root growth inhibition induced by RALF1 can be divided into rapid and slow phases. The rapid inhibition is reversible and occurs within 1 minute to 1 hour. It is caused by FER-mediated apoplastic alkalinization due to AHA2 phosphorylation and inactivation (21). Alternatively, FER might modulate the activity of a yet undiscovered proton transporter (43). In turn, the RALF-induced slow inhibition phase relies on transcriptional changes. It starts after one hour of RALF1 or RALF22 treatment and is maintained by a FER-mediated induction of the YUC genes, which increases endogenous auxin levels and activates TIR1. The resulting transcriptional changes are responsible for the sustained apoplastic alkalinization and root growth inhibition observed in the presence of RALFs (43). Remarkably, root growth inhibition induced by auxin or RALF1 is dependent on FER-mediated activation of RIPK, making it a potential player in the crosstalk between these hormones (68). Furthermore, plant mutants lacking LLG1 are insensitive to RALF1-induced root growth arrest and to auxin-triggered ROS production (36). Given the FER/LLG1 complex formation (18,27,29,36), it is possible that the coreceptor also contributes to the interplay between these hormones.
FERONIA's involvement in auxin-mediated growth regulation extends beyond the above-mentioned. IAA-triggered apoplast alkalinization is FER-dependent (69). FER also modulates PIN2 localization, and the resulting auxin distribution is necessary for root gravitropism (70). Similarly, root nutation relies on auxin gradients created by PIN2 and AUX1, as well as on calcium intake, both disrupted in the loss-of-function mutant fer-4 (71). Moreover, FER is an important player in the blue light-induced hypocotyl phototropic growth. In this response, FER maintains a high pH on the illuminated side and induces PIN3 accumulation and a high auxin concentration on the shaded side, resulting in differential growth towards light (72). The involvement of RALFs in these mechanisms has not yet been investigated. However, it is worth noting that RALF1 treatment induces PIN2 internalization in a FER-dependent manner (73), which aligns with a possible role for these peptides in auxin-triggered, FER-mediated tropism.
Brassinosteroids are central players in plant growth regulation. These steroid hormones are perceived by the receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) and its coreceptor BAK1, starting a signaling cascade that culminates in the activation of two transcription factors, BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS SUPPRESSOR 1 (BES1). The repressor kinase BR INSENSITIVE 2 (BIN2) is a key component in this signaling, being dephosphorylated and degraded upon BR binding to BRI1/BAK1, consequently releasing BZR1 and BES1 from repression. The resulting change in thousands of genes' expression underlies the brassinosteroid responses (74,75).
RALF1 opposes BR-induced root and hypocotyl growth, as attested by the diminished sensitivity to brassinolide (BL) in RALF1 overexpression lines (63,76). The RALF1-triggered root growth inhibition depends on BAK1 and CML38, to which the peptide binds directly (19,42). In turn, the BR/BRI1/BAK1 complex formation is strictly dependent on acidic pH (77). Thus, a potential mechanism may take place where RALF1/FER-induced apoplast alkalinization favors BAK1 recruitment to the RALF1/BAK1/CML38 complex to the detriment of BR/BRI1/BAK1 complex formation, thereby reducing BR signaling (19,42,77). RALF1 also induces BRI1 internalization in a FER-dependent manner, potentially adding to the peptide antagonism (27,73). Remarkably, the BAK1/CML38 complex is not involved in RALF1-induced alkalinization or in root inhibition by RALF23 or RALF34 (19,42). Through BAK1 and CML38, RALF1 also alters gene expression. RALF1 treatment represses EXPA5, which plays a positive role in cell expansion and is upregulated by BR (19,42,63,78). The peptide also upregulates TCH4, HRGP2, and PRP1,3, which are involved in cell wall rearrangement (19,42,79).
FER is central to BR-induced cell elongation in roots and hypocotyls, a process that requires weakening of cell wall matrix connections and synthesis of new wall components, and thus needs to be tightly coordinated to maintain cell wall integrity (CWI) (80). Under low BR levels, the BIN2 kinase is active and phosphorylates FER, leading to its retention in the endoplasmic reticulum (ER). In the presence of BR, BIN2 is inactivated, enabling greater FER localization at the plasma membrane (76), where it senses CWI (33,81). The BR-induced cell expansion generates pectin fragments that act as FER activators and, importantly, RALF1 and pectin exert additive effects on the receptor's signaling. As a result, the activated FER represses the BR stimulus, fine-tuning cellular elongation to prevent CW damage and rupture (76).
Other RALF peptides are also involved in FER-BR crosstalk. The FER/LLG1 complex regulates anisotropic growth of hypocotyl epidermal cells, controlling cell wall pectin content and modifications through PME activity. The
RALF1,22,23,33 genes are expressed in hypocotyls, and the
ralf1/22/23/33 quadruple mutant phenocopies the abnormal growth and CW content phenotype observed in FER and LLG1 mutants. Importantly, hypocotyls of the quadruple
RALF mutant and of plants lacking FER or LLG1 are hypersensitive to BL-induced growth and have diminished levels of the BR-repressed
CPD and
DWF4 transcripts. Further investigation focusing on RALF23 revealed that the peptide promotes BRI1/BAK1 complex formation and BR signaling (82). Therefore, distinct RALFs differentially influence BR signaling, as demonstrated by the opposing effects of RALF1 and RALF23 (76,82). In synthesis, RALF peptides are essential for hypocotyl growth by remodeling the cell wall and mediating FER regulation of the brassinosteroid pathway.
Figure 4 summarizes the crosstalk between RALFs, auxin and brassinosteroids.
RALF1-induced root growth inhibition involves crosstalk with the brassinosteroid (BR) and auxin signaling pathways. Under low BR levels, BIN2 is active and phosphorylates FER, which is retained in the endoplasmic reticulum (ER). In the presence of BR, BIN2 is inactivated by dephosphorylation, resulting in greater FER localization at the plasma membrane. There, FER senses CWI to counterbalance BR-induced cell expansion, thus preventing cell wall damage and rupture. The receptor achieves this through apoplast alkalinization, which is stimulated by RALF1 and pectin. RALF1-induced alkalinization leads to rapid root growth arrest and relies on AHA2 phosphorylation and inactivation in a FER-dependent manner. Apoplast alkalinization also leads to BR/BRI1/BAK1 complex dissociation, potentially contributing to the RALF1/BAK1/CML38 complex formation. Through this complex, RALF1 triggers transcriptional changes that lead to the inhibition of cell expansion. In contrast, RALF23/FER binding promotes BRI1/BAK1 complex formation in the hypocotyl, thus contributing to BR signaling. Furthermore, the RALF1-induced inhibition of root growth involves the transcription of YUCCA genes in a FER-dependent manner, leading to increased auxin production and activation of the hormone’s signaling pathway, which includes TIR1. In addition, RALF1/FER induces BRI1 and PIN2 endocytosis, potentially contributing to growth arrest. Finally, RIPK and LLG1 are essential for both RALF1- and auxin-induced responses and may act in the crosstalk between these hormones.
Root cellular signaling
The signaling pathways triggered by RALFs to regulate root development are well characterized. They involve protein interactions and phosphorylation, as well as transcriptional, post-transcriptional, and protein synthesis regulation. A first example is the phosphorylation cascades of MITOGEN-ACTIVATED PROTEIN KINASE (MAPKs), a common molecular mechanism to amplify intracellular signals (83). RALF1 treatment induces the phosphorylation of MAPK3,4,6, leading to a yet unknown outcome (84). RALF1 also triggers RIPK phosphorylation. In this response, RALF1 binding to FER induces FER/RIPK interaction and transphosphorylation, thus transducing the peptide’s signal to inhibit root growth (68).
The EUKARYOTIC TRANSLATION INITIATION FACTOR 4 (eIF4) complex is responsible for promoting protein synthesis (85). Upon RALF1 binding, FER undergoes autophosphorylation and interacts with eIF4E1, phosphorylating and activating it. EIF4E1 then binds to eIF4G and to the 5’ cap of diverse mRNAs, such as ERBB-3 BINDING PROTEIN 1 (EBP1), inducing their translation. Plant mutants with non-phosphorylatable eIF4E1 are insensitive to RALF1-triggered growth arrest, attesting the importance of eIF4E1 activation for the peptide response (12).
RALF1 also regulates protein synthesis through the tRNA-binding proteins YUELAO (YL1,2,3). The RALF1/FER complex triggers direct phosphorylation of YL1,2,3, leading to tRNA methylation, which reduces the production of specific tRNA fragments (tRFs) (86). Some tRFs are global translation regulators (87), and the RALF1/FER/YL-triggered tRF modifications promote protein synthesis. Remarkably, YL mutants are less sensitive to RALF1-induced root growth inhibition, further supporting the importance of translational control for the peptide response (86).
EBP1 is a DNA- and RNA-binding protein capable of regulating both transcription and translation (88). RALF1 stimulates EBP1 expression and protein synthesis (12,84), and the RALF1/FER ligation also induces the receptor's interaction with EBP1 and phosphorylation of both proteins. Once phosphorylated, EBP1 is directed to the nucleus, where it triggers transcriptional changes, such as the direct repression of CML38 (84). Notably, EBP1 negatively regulates RALF1-triggered root growth inhibition, apoplast alkalinization, and MAPK phosphorylation (12,84). Hence, RALF1 and EBP1 are part of a negative feedback loop.
RALF1 promotes alternative splicing through GLYCINE-RICH RNA-BINDING PROTEIN 7 (GRP7), an RNA-binding protein that regulates the transcription and splicing of other genes (89,90). The RALF1/FER ligation induces GRP7 phosphorylation and directs the protein to the nucleus, where it interacts with the spliceosome component U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA (U170K). Accordingly, the RALF1/FER/GRP7 pathway elicits changes in the cell’s splicing profile, contributing to the peptide's root growth inhibition response. Plant mutants lacking SM-LIKE 8 (LSM8) or ARGININE/SERINE-RICH 45 (SR45), two spliceosome components, exhibit reduced sensitivity to RALF1 and to RALF23 treatment (11). Thus, alternative splicing has an essential role in RALF-triggered root growth inhibition.
The Golgi-located ADENYLPYROPHOSPHATASE (APYs) form a family of membrane-bound proteins that can hydrolyze nucleotides (91). APY7 is expressed in roots, and the loss-of-function apy7 mutant exhibits longer roots due to elongated cells. The mutant also has an altered cell wall polysaccharide composition and a higher apoplastic pH. Remarkably, apy7 plants are insensitive to RALF1-induced root growth inhibition (92). How this endomembrane protein affects the peptide signaling, however, remains to be determined.
The transduction of RALF1 signal relies on the peptide’s extracellular dynamics. An important factor is RALF1’s interaction with cell wall pectin in its negatively charged, de-esterified state, a product of PECTIN METHYLESTERASE (PMEs) catalysis (27,93,94). The RALF1-triggered root growth inhibition, apoplast alkalinization and FER internalization responses are absent when PME activity is abolished, either pharmacologically or genetically. In addition, two newly described RALF1-induced phenotypes of cell wall swelling and plasma membrane invagination are reversed in the absence of PME activity, when an excess of de-esterified pectin fragments out-titrates RALF1, and in fer-4 mutants. Therefore, pectin is a central part of the
RALF1/FER-triggered responses, and may act as a signaling scaffold for the peptide (93). Notably, the RALF/LRX complex binds to pectin in root hairs and pollen tubes (28,29). However, quintuple mutants lacking LRX1-5 resemble wild-type plants under RALF1 treatment, indicating that LRXs are not part of the aforementioned responses (93).
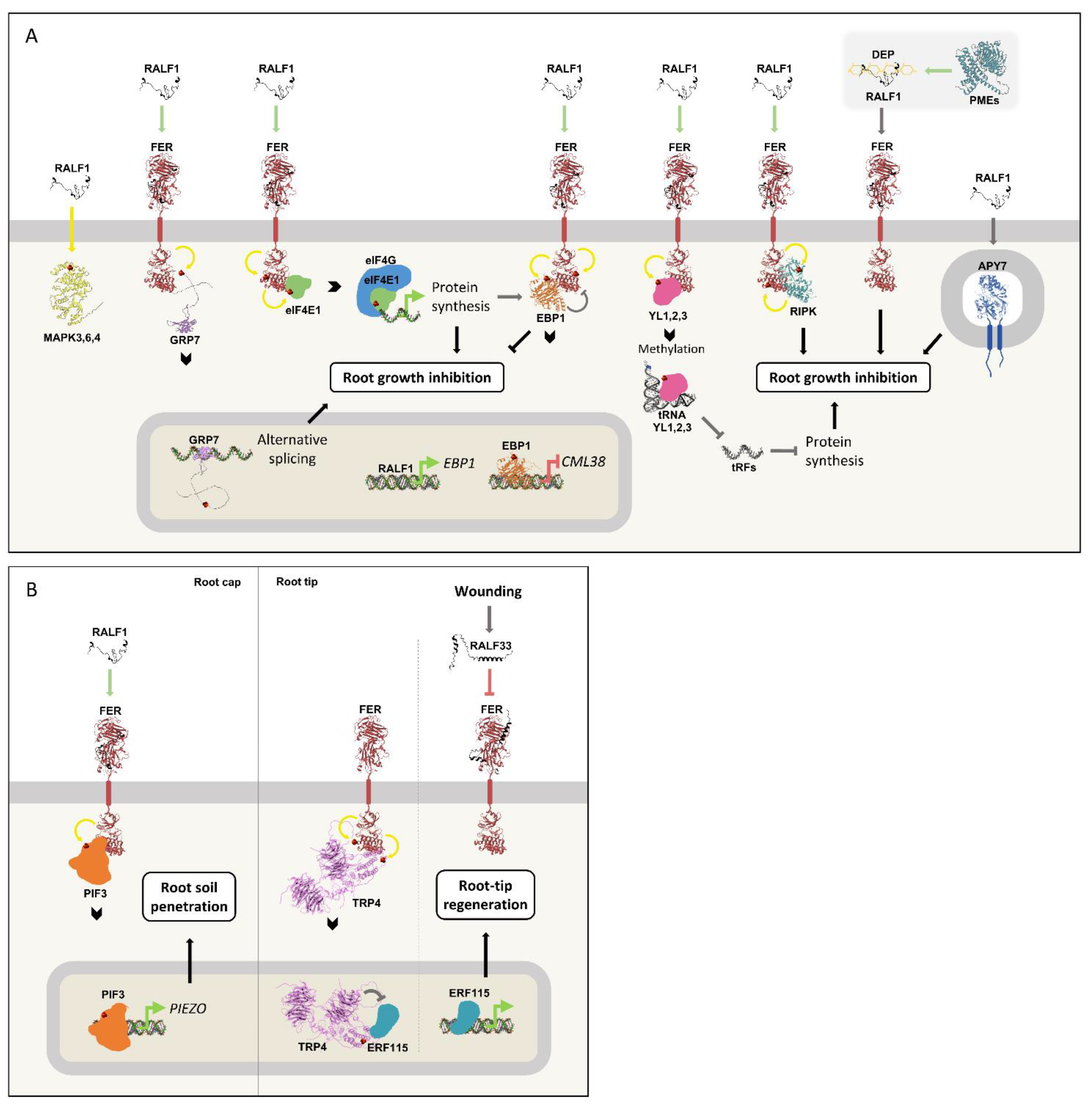
Figure 5A summarizes the complex intracellular signaling elicited by RALF1.
FER signaling is essential for root soil penetration, and involves the transcription factor (TF) PHYTOCHROME-INTERACTING FACTOR 3 (PIF3), better known for its role in etiolated cotyledons' soil emergence (95,96). In root cap cells, FER phosphorylates PIF3, contributing to its stabilization and abundance, which leads to the TF’s direct induction of PIEZO expression, a mechanosensitive ion channel that modulates calcium flux (95,97). The FER-PIF3-PIEZO pathway leads to proper soil penetration and is induced by RALF1, but not by RALF23. Remarkably, RALF1 treatment also triggers the expression of dozens of genes in columella root cap cells in a FER-dependent manner (95).
Under normal root conditions, FER phosphorylates the transcriptional corepressor TOPLESS-RELATED 4 (TPR4), which translocates to the nucleus, where it directly represses ETHYLENE RESPONSE FACTOR115 (ERF115) (98,99), a TF involved in root tip regeneration (100). After root wounding, RALF33 levels rise to 200 nM and bind to FER, reducing the receptor’s autophosphorylation at its activation residue Y648 (34,98). Accordingly, the RALF33/FER interaction lowers TPR4 phosphorylation and nuclear localization. Thus, RALF33 inhibits FER repression of ERF115 through TPR4, allowing the TF to modulate gene expression leading to root tip regeneration (98).
Figure 5B illustrates RALF-regulated responses promoting proper root development and integrity.
Figure 5.
RALF peptides elicit complex signaling in roots.
Figure 5.
RALF peptides elicit complex signaling in roots.
(A) RALF1 induces the phosphorylation of MAPK3,4,6. RALF1 also binds to FER, and the receptor's kinase domain phosphorylates GRP7, eIF4E1, EBP1, YL1,2,3 and RIPK, leading to root growth inhibition. Phosphorylated GRP7 accumulates in the nucleus, where it regulates alternative splicing. The phosphorylation of eIF4E1 increases its affinity for eIF4G and mRNAs, inducing protein synthesis. One of the translated proteins is EBP1, whose phosphorylation and transcription are also induced by RALF1. After its phosphorylation by RALF1/FER, EBP1 accumulates in the nucleus, where it regulates gene transcription, notably inhibiting CML38 expression. In summary, RALF1 induces EBP1 production and activation, which in turn represses RALF1 signaling via a negative feedback loop. Moreover, RALF1 induces the phosphorylation of YL1,2,3 through FER, leading to tRNA methylation and a consequent decrease in tRF production, which reduces protein synthesis and inhibits root growth. The PME enzymatic activity generates de-esterified pectin (DEP), and RALF1 interaction with the negatively charged polysaccharide is essential to root growth inhibition. The Golgi-located APY7 is also necessary for RALF1-induced root growth inhibition.
(B) At the root cap, FER directly phosphorylates PIF3, stabilizing it and increasing its abundance in the nucleus, leading to the upregulation of PIEZO. This signaling pathway is essential for root soil penetration and is induced by RALF1 treatment. At the root tip, FER autophosphorylates and phosphorylates TPR4, which translocates to the nucleus where it represses ERF115. After root wounding, RALF33 levels rise, and its binding to FER represses TPR4 phosphorylation, allowing ERF115 to initiate a transcriptional response that culminates in root tip regeneration.
Lateral roots
RALF peptides are involved in lateral root (LR) formation. RALF34 is expressed in LR primordia, and its transcription is regulated by auxin and the transcription factors ETHYLENE RESPONSE FACTOR (ERF4,9) (101). RALF34 binds the receptor kinase THE1 to regulate LR emergence. Curiously, mutants lacking RALF34 or THE1, as well as plants overexpressing either gene, exhibit a similar phenotype of increased LR primordia and incorrect primordia patterning, pointing to a finely tuned regulation (31). Furthermore, plants treated with RALF33 or RALF36 have higher LR density despite having smaller primary roots (45). In contrast, mutants with reduced RALF1 expression exhibit higher LR density and longer primary roots than wild-type plants (63). Together, these results suggest that RALF peptides regulate lateral root emergence and development through yet underexplored signaling pathways.
Root hairs
Root hairs (RH) are extremely elongated extensions of epidermal cells responsible for water and nutrient acquisition, anchoring, and interaction with microorganisms. RH's rapid expansion is sustained by the exocytosis of plasma membrane and cell wall components at the hair’s apex (102). RALF peptides are involved in the emergence and development of these structures. Among the family members, RALF22 is the most expressed in RH, where it binds to the receptor complex FER/LLG1 and to LRX1,2, promoting cell wall integrity (CWI) and growth (29). Furthermore, lines overexpressing RALF8 or plants treated with RALF33 or RALF36 exhibit longer RH with increased density (45,103). Similarly, RALF1 treatment induces longer RH in a FER-dependent manner, while RALF1 knockout mutants show the opposite phenotype (12,68).
During root hair expansion, RALF22 functions both as a signaling molecule and as a cell wall component. The peptide binds to the FER/LLG1 complex, promoting calcium influx, apoplast alkalinization, and cell wall esterified pectin (EP) accumulation. RALF22 also binds to LRX1 and LRX2, forming heterotetrameric complexes composed of two RALFs and two LRXs (29). In pollen tubes, RALF4/LRX8 ligation exposes the peptide's basic residues, which increases its affinity to negatively charged de-esterified pectin (DEP) (22,28). The RALF22/LRX1,2 complexes formed in RHs also interacts with DEP, potentially through the same configuration. This RALF22/LRX1,2/DEP assembly expels water molecules from the pectin layer of RH’s cell wall, thereby stiffening it and aiding in CWI during tip growth. Remarkably, root hair apical expansion occurs in an oscillatory manner, where growth arrests are synchronized with the cyclical incorporation of RALF22 into the cell wall. This process results in the formation of RALF22/LRX1,2/DEP rings around the root hair flanks (29). Based on this evidence and the observation that PMEs' enzymatic activity peaks at an alkaline pH (104), Schoenaers and collaborators proposed the following mechanism for RH growth: 1) RALF22 and LRX1,2, along with esterified pectin and PMEs, are secreted at the root hair’s apex; 2) RALF22 binds to FER/LLG1 and induces calcium influx, cell wall’s EP accumulation, and apoplast alkalinization, activating PMEs and thereby increasing the levels of DEP; 3) RALF22/LRX1,2 complexes then bind to the newly synthesized DEP, forming a ring that stiffens the cell wall, marking the end of a growth cycle; 4) A new round of secretion takes place and free RALF22 binds to FER/LLG1, initiating a new cycle (29).
It is important to mention that sucrose induces RALF22 expression and that, in media not supplemented with this sugar, the ralf22 mutant exhibits hair growth comparable to the wild-type plants (105). Thus, RALF22-mediated RH growth appears to be dependent on sucrose, opening the possibility of other RALFs acting under normal conditions.
RALF1 changes the root hair’s protein composition through eIF4E1, contributing to its growth. RALF1 binding to FER induces the receptor autophosphorylation and transphosphorylation of eIF4E. Phosphorylated eIF4E1 exhibits increased affinity for eIF4EG and the 5’ cap of various transcripts, notably
EBP1,
ROP2 and
ROOT HAIR DEFECTIVE 6-LIKE 4 (
RSL4), thereby increasing their protein levels (12). EBP1 is a negative regulator of RALF1-triggered root growth inhibition (84). ROPs’ regulation of ROS production is essential for RH growth, and FER induces this response in a ROP2-dependent manner (12,106–108). RSL4 is a transcription factor whose protein levels during RH expansion directly influence the structure's final size (109,110). Moreover, RSL4 binds to
RALF1 promoter and represses its expression, thus forming a negative feedback loop (12). Single-cell RNA-seq of roots treated with RALF1 further demonstrates its impact on RH transcription in a FER-dependent manner (95). In summary, RALF1/FER regulates the RH translation through eIF4E1 and its transcription through RSL4. Remarkably, treating seedlings with RALF22 results in RH growth inhibition, in contrast to the growth induction caused by RALF1, RALF33, or RALF36 treatments (12,29,45). These opposing effects confirm the existence of at least two distinct RALF-regulated mechanisms governing root hair cellular expansion.
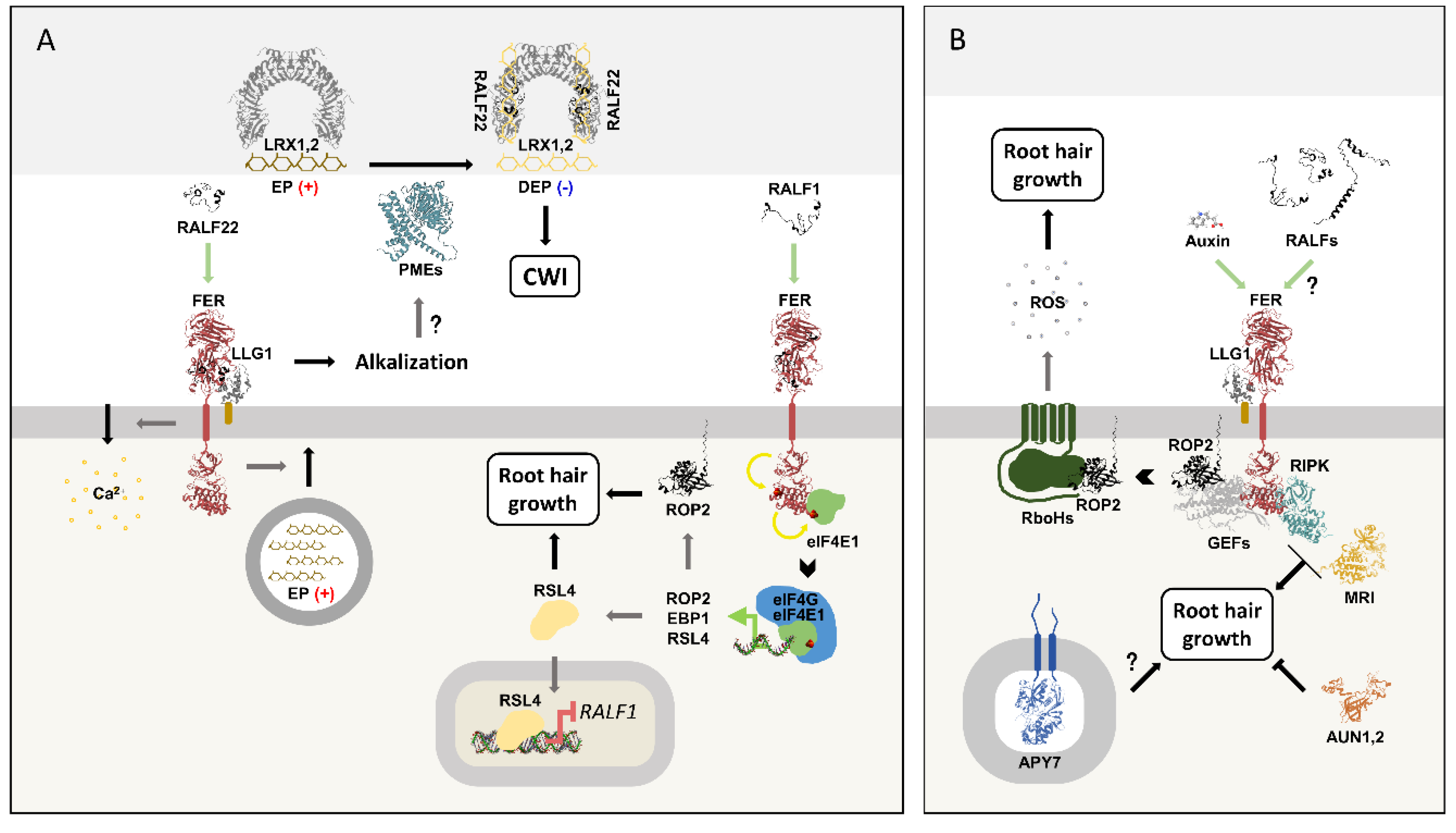
Figure 6A summarizes the roles of RALF1 and RALF22 in RH growth.
Root hair development is promoted by auxin (111), and the FER/LLG1 complex is indispensable for auxin-triggered RH initiation and elongation (36,108). In this context, FER induces ROS production through ROP2 and GEF1,4,10 (108,112). In addition, the fer-4 mutant is deficient in RH formation, while overexpression of RIPK in the fer-4 background reverses this phenotype, pointing to a positive role for RIPK downstream of FER (68). Remarkably, IAA-induced RH growth is absent in the ralf22 knockout mutant, indicating that RALF22 is also involved in auxin signaling (105). Together, this information suggests a potential crosstalk between RALFs and auxin in RHs, as seen in primary roots (43).
APY7 is bound to the Golgi membrane and is required for RALF1-induced inhibition of primary root growth. The apy7 mutant was found to reverse the lack of root hairs in the lrx1/2 double mutant and partially reverse this phenotype in the fer-4 background (92). These results raise the possibility that APY7 acts downstream of RALFs, LRXs, and/or FER in root hair development, which requires further investigation.
Similar to root hairs, pollen tubes (PTs) exhibit rapid apical growth. In both structures, different RALFs, CrRLK1Ls, and LRXs members are involved in cellular expansion (28,29,41), pointing to a conserved molecular mechanism driving the coevolution of these protein families. A key intracellular aspect of this mechanism investigated in PTs has not yet been fully explored in RHs. In PTs, the cytoplasmic kinase MARIS (MRI) acts downstream of the RALF4,19/ANXs/BUPSs complexes to maintain CWI (38,54). In contrast, the cytoplasmic phosphatases ATUNIS (AUN1,2) oppose RALF4,19’s and LRX8-11’s positive regulation of CWI maintenance (113). MRI and AUN1,2 roles in pollen tubes extend to root hairs, where they act downstream of FER and LRX1,2. Accordingly, MRI positively regulates, while AUN1,2 negatively regulate the CWI of RHs (40,114), and the involvement of RALFs in this pathway has not been investigated. This and other potential components of the RALF-induced root hair expansion pathway are summarized in
Figure 6B.
Figure 6.
RALF1 and RALF22 regulate root hair growth.
Figure 6.
RALF1 and RALF22 regulate root hair growth.
(A) RALF22 binds to FER/LLG1, inducing apoplast alkalinization, calcium influx, and the accumulation of esterified pectin (EP) in the cell wall. RALF22 also forms heterotetrameric complexes with LRX1,2, which bind to de-esterified pectin (DEP) and maintain cell wall integrity (CWI). The alkalinization induced by RALF22/FER/LLG1 potentially activates PMEs and contributes to the RALF22/LRX1,2/DEP assembly. RALF1 binds to FER, inducing eIF4E1 phosphorylation, thereby increasing the synthesis of ROP2, EBP1, and RSL4. ROPs are important players in RH growth. RSL4 regulates RH transcription, inducing its growth. RSL4 also represses RALF1 expression, forming a negative feedback loop.
(B) Steps that may be part of the RALF-mediated RH growth pathway. The FER/LLG1 complex is required for auxin-induced ROS production through ROPs/GEFs, which is essential to RH growth. RIPK, MRI, and AUN1,2 modulate RH growth downstream of FER. APY7 is necessary for RALF1-induced primary root growth inhibition and is involved in the FER and LRX root hair development pathways.
VI. Membrane Dynamics
Membrane components are in constant cycling, and endocytosis can be mediated by clathrin (CME) or not (CMI) (115). FER undergoes constitutive endocytosis through both CME and CMI pathways; however, in the presence of RALF1, FER is internalized mainly through CME. Consistently, CME-defective double mutants lacking CLATHRIN LIGHT CHAIN (CLC1,3) are less sensitive to RALF1-induced root growth inhibition, although they remain sensitive to the RALF1-induced transcription changes and MAPK activation. Moreover, RALF1 treatment decreases FER diffusion and increases its enrichment in microdomains (73).
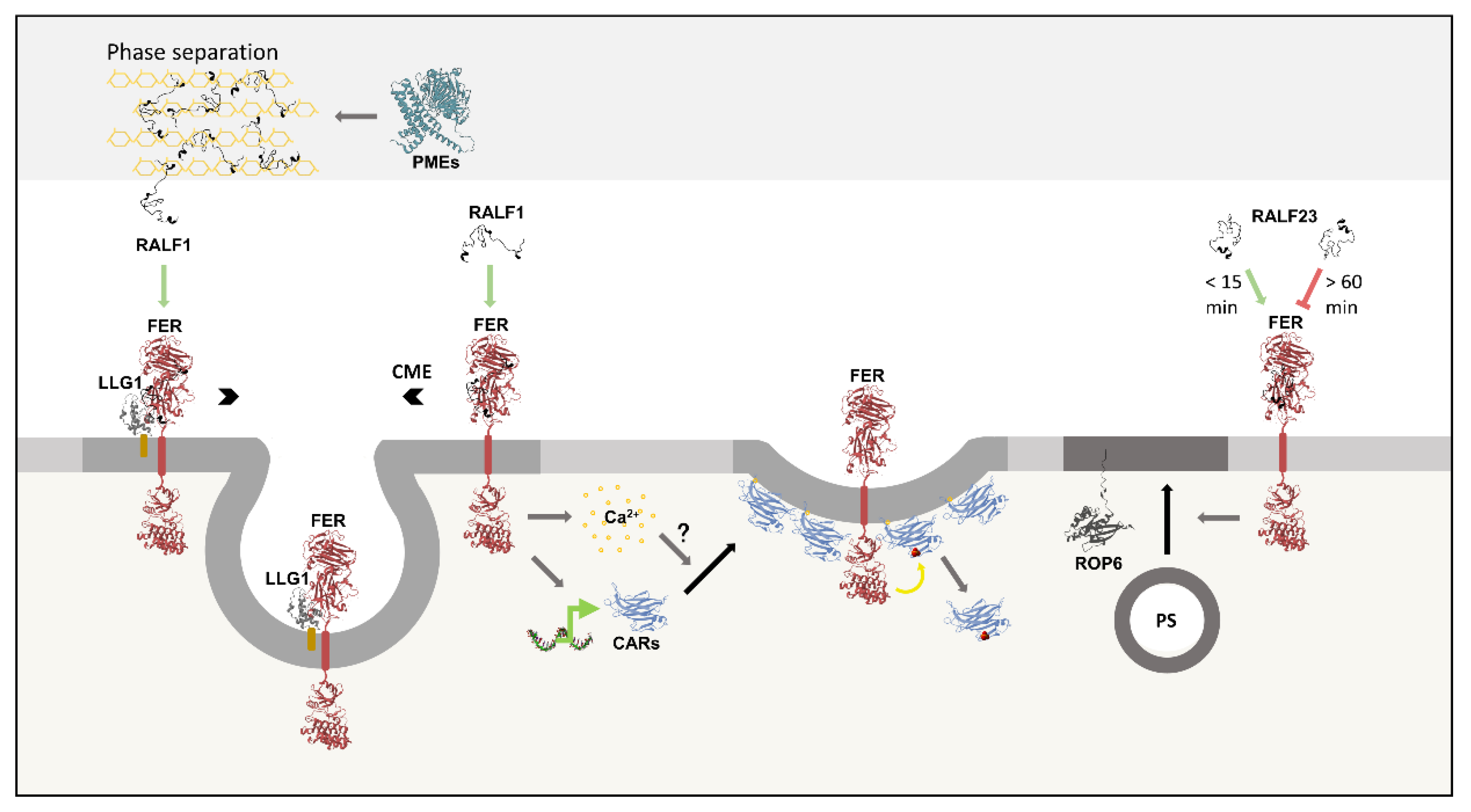
The plasma membrane is a heterogeneous structure with micro and nanodomains that exhibit diverse lipid and protein compositions, as well as mobility characteristics (116). RALFs peptides and FER can induce the formation of membrane domains through at least three different mechanisms: RALF1 and pectin phase separation (27); synthesis and recruitment of C2-DOMAIN ABA-RELATED (CAR) proteins (117); and accumulation of the phospholipid phosphatidylserine (PS) in the membrane (61).
Phase separation is the aggregation of macromolecules into two solutions with distinct and stable compositions (118). In the apoplast, RALF1 interacts with de-esterified pectin, leading to phase separation. At the plasma membrane in contact with this aggregate, FER and LLG1 are enriched due to their interaction with RALF1. Other receptors, such as FLS2 and BRI1, are also recruited by unknown means. This molecular organization creates micro and nanodomains with a high density of membrane-bound proteins, where endocytosis is intense. Curiously, RALF1 is not internalized with its receptors. RALF1/DEP phase separation is necessary for the maintenance of root’s basal ROS levels in a FER/LLG1-dependent manner, as well as for the rise in ROS production and Ca2+ influx triggered by an increase in RALF1 concentrations. Furthermore, pharmacological or genetic inhibition of PME activity suppresses RALF1-induced phase separation and endocytosis, highlighting the importance of these enzymes in the peptide’s responses. Notably, RALF1/DEP/FER/LLG1 aggregates are present during regular plant development, serving as a constitutive mechanism. However, salt and heat stresses significantly increase the extent of this arrangement (27).
In addition to the changes in membrane organization driven from the apoplast, RALF1 signaling also induces domain formation from the cell’s interior. CAR proteins bind to Ca2+ through their C2 domain and interact with negatively charged phospholipids, creating a membrane curvature (119). RALF1 binding to FER induces the translation of CAR1,4,5,6,9,10, along with rapid formation of CAR nanodomains (117). The calcium influx elicited by RALF1/FER is potentially part of this response (21,117). Once in the nanodomains, FER phosphorylates the CARs’ C2 domain, reducing the protein's affinity for phospholipids, likely due to charge repulsion. Thus, the nanodomain formation by RALF1/FER/CARs is an autoregulated process (117).
FER controls the accumulation of phosphatidylserine in the plasma membrane. PS accounts for 10-15% of the total plasma membrane lipids in eukaryotic cells and is kept in the cytoplasmic-facing leaflet (120). In plants, PS nanodomains stabilizes ROP6 membrane dynamics, which is essential for ROS production under osmotic stress (61,121,122). Short-term (<15 min) RALF23 treatment positively regulates PS nanodomain formation and osmotic stress response through FER. In contrast, long-term (>60 min) RALF23 treatment decreases PS membrane abundance and nanodomain formation, reducing ROP6 signaling and the consequent ROS production under osmotic stress (61). Together, these three RALF/FER-induced mechanisms modulating membrane dynamics add an important layer to our understanding of RALF peptide signaling (
Figure 7).
RALF1 interacts with de-esterified pectin (depicted in yellow) in the cell wall, inducing phase separation. At the adjacent plasma membrane, RALF1 binds to FER/LLG1, forming micro- and nanodomains where endocytosis takes place. Notably, the PMEs activity is essential to this response. RALF1 binding to FER also decreases the receptor diffusion and increases its enrichment in microdomains, where it undergoes clathrin-mediated endocytosis (CME). Furthermore, RALF1/FER induces the synthesis of CAR proteins, which bind to membrane phospholipids, forming curved nanodomains. The RALF1/FER-induced calcium influx potentially contributes to this response. Once in these nanodomains, FER phosphorylates CARs, decreasing their membrane affinity. FER is also involved in the incorporation of phosphatidylserine (PS) into the membrane and its organization in micro- and nanodomains, which stabilize ROP6 mobility. Short-term (<15 min) RALF23 treatment increases this response, while long-term (>60 min) treatment suppresses it.
VII. RALF-ABA Crosstalk
During their life cycle, plants receive diverse environmental stimuli, which are integrated into responses that favor development or defense. The phytohormone abscisic acid (ABA) plays a central role in this process, leading to outputs that include the control of growth and stomatal movement (123). In a default state, ABA signaling components remain suppressed due to the action of the type 2C protein phosphatases ABA INSENSITIVE (ABI1,2). The perception of ABA by its receptors, including PYRABACTIN RESISTANCE 1-LIKE (PYLs), inhibits these phosphatases to activate the hormone’s signaling pathway, which includes transcription factors such as ABI5 and ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTOR (ABFs) (124).
RALF1 and ABA signaling pathways regulate each other through distinct mechanisms. RALF1 binding to FER induces the receptor phosphorylation, while ABI2 dephosphorylates FER, decreasing the RALF1-triggered root growth inhibition response. (125). Evidence points to a FER-stimulated activation of ABI2, which may serve as a self-regulating mechanism (126). The presence of ABA favors FER phosphorylation by inactivating ABI2 in a PYL-dependent manner. Accordingly, plant mutants lacking PYL receptors exhibit reduced sensitivity to RALF1-triggered root growth inhibition, indicating an agonistic role of ABA in the peptide’s signaling. In contrast, RALF1 decreases ABA-induced root growth inhibition, supporting an antagonistic role for the peptide in the ABA signaling pathway (125). This negative effect is further supported by the observation that plant mutants lacking FER, LLG1, or RIPK, key proteins for RALF1 responses, are hypersensitive to ABA-triggered root growth inhibition (36,68,126). In addition, mutants lacking EBP1, a negative regulator of RALF1’s pathway, are less sensitive to the ABA-induced response (84). Collectively, these findings point to a negative feedback loop where ABA presence favors RALF1 signaling, which in turn negatively regulates the phytohormone responses.
A recently discovered mechanism of RALF1-mediated repression of ABA signaling involves the protein FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1) (127), which was initially described for its role in vesicle sorting (128). FREE1 was later characterized as a negative regulator of ABA signaling by interacting with ABI5 and ABF4 in the nucleus, reducing their transcriptional activity (129). RALF1 binding to FER induces the receptor’s direct phosphorylation of FREE1, which then translocates to the nucleus. There, phosphorylated FREE1 negatively regulates the expression of ABA-responsive genes induced by ABI5 or ABF4, reinforcing a RALF1/FER role as antagonists of ABA signaling (127).
Another mechanism of reciprocal modulation between these hormones involves the RNA-binding protein GRP7. RALF1/FER ligation induces direct GRP7 phosphorylation, which subsequently binds to the
ABF1 mRNA (11). ABF1 is a transcription factor in the ABA pathway, and its transcript exists in two splicing variants,
ABF1.1 and
ABF1.2 (11,124). GRP7 activation by RALF1/FER favors the
ABF1.2 splicing variant, which dampens ABA’s responses. In turn, ABA induces
GRP7 alternative splicing, generating a premature stop codon and therefore decreasing its levels (11). This elegant mechanism illustrates how the crosstalk between RALF1 and ABA extends to post-transcriptional regulation.
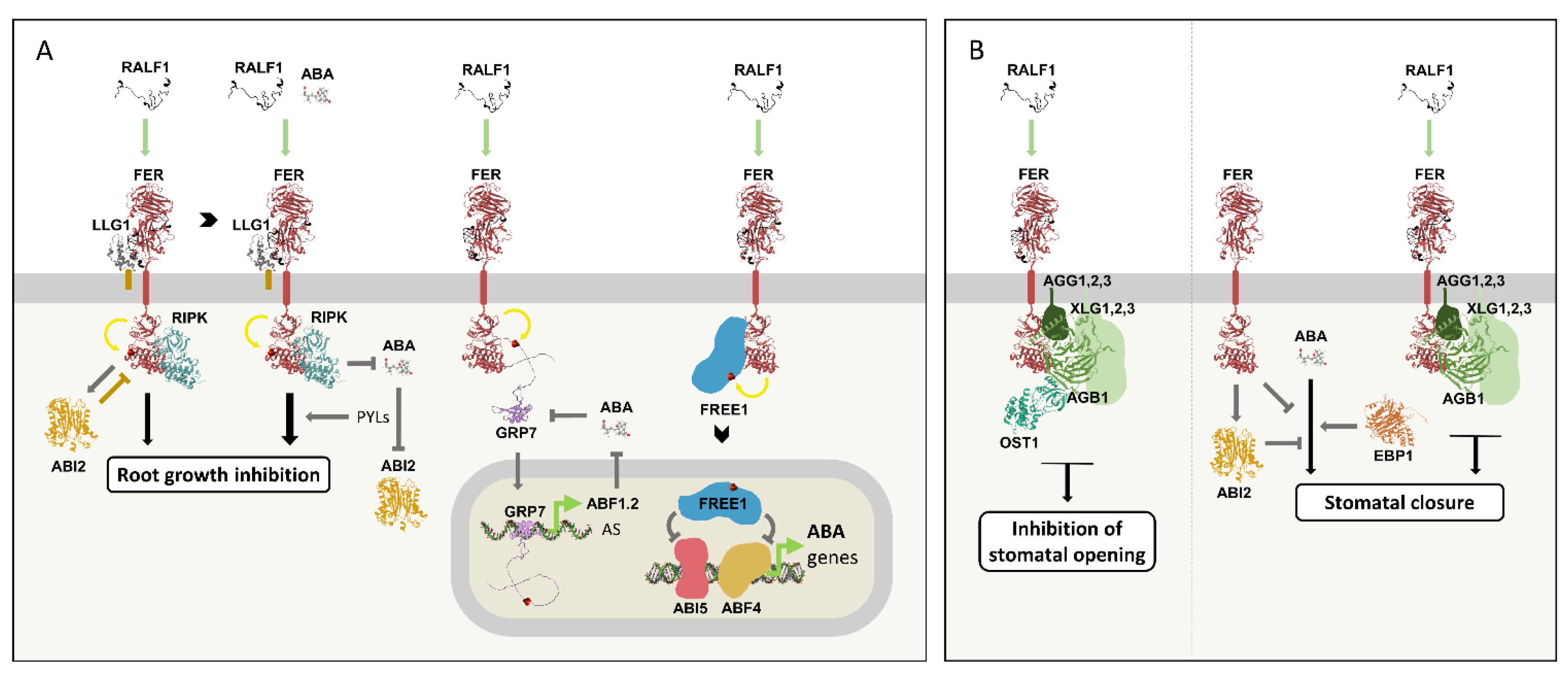
Figure 8A summarizes the known interactions between the RALF1 and ABA signaling pathways during root growth regulation.
Stomatal closure plays a fundamental role in regulating gas exchange and preventing water loss through the leaves (123). G proteins act as heterotrimers composed of the subunits Gα, Gβ, and Gγ, and are well-known players in the transduction of multiple signals in eukaryotes. Plants possess an additional type of Gα subunit, the EXTRA-LARGE G PROTEINs (XLGs) (130). FER binds to G PROTEIN SUBUNIT BETA 1 (AGB1) and this interaction depends on G PROTEIN SUBUNIT GAMMA (AGG1,2,3). RALF1 treatment inhibits stomatal aperture and induces its closure, and these responses are absent in mutants lacking FER, AGB1, AGG1,2,3, or XLG1,2,3 (131). Therefore, RALF1’s control of stomatal movement is mediated by FER and the G proteins. Moreover, overexpression of the kinase-dead FERK565R in a fer-4 background only partially restores the RALF1-induced stomatal regulation absent in fer-4, suggesting a secondary role for the receptor's phosphorylation activity in this response (132).
The RALF1- and ABA-mediated pathways controlling stomatal movement share proteins. Plants lacking OPEN STOMATA 1 (OST1), an ABA signaling kinase that interacts with AGB1, are insensitive to RALF1’s inhibition of stomatal opening (131). In contrast, the
fer-4 mutant is hypersensitive to ABA's inhibition of stomatal opening (125). This hypersensitivity is opposed by an ABI2 gain-of-function mutation, in which the phosphatase activity is resistant to ABA inhibition, suggesting that FER’s negative role in ABA signaling may result from the activation of ABI2 (125,126). Lastly,
EBP1 knockout mutants are less sensitive to ABA-induced stomatal closure, suggesting a positive role in ABA response for this negative regulator of RALF1 signaling (84). Collectively, these results point to an integration of RALF1’s and ABA’s pathways that extends to the regulation of stomatal movement (
Figure 8B).
Lastly, the loss-of-function mutants fer-4 and lrx345, as well as lines overexpressing RALF22 or RALF23, have increased ABA levels compared to wild-type plants. These results suggest that some RALFs and their canonical receptors also regulate ABA homeostasis (133), which further supports a close relation between ABA and this peptide hormone family.
Figure 8.
Crosstalk between the RALF1 and ABA signaling pathways.
Figure 8.
Crosstalk between the RALF1 and ABA signaling pathways.
A) RALF1 binding to FER/LLG1 induces the receptor phosphorylation, leading to root growth inhibition in a RIPK-dependent manner. This response is counteracted by ABI2 dephosphorylating FER, which, in turn, activates ABI2. The presence of ABA increases FER phosphorylation by suppressing ABI2 activity in a PYL-dependent manner, thereby favoring RALF1 signaling. In contrast, the RALF1/FER/LLG1/RIPK complex represses the ABA pathway. RALF1/FER phosphorylates GRP7, which binds to ABF1 mRNA, favoring the splicing variant ABF1.2 and consequently dampening ABA signaling. In turn, ABA induces GRP7 alternative splicing (AS), resulting in a premature stop codon variant and therefore reducing GRP7 protein levels. RALF1/FER also phosphorylates FREE1, which translocates to the nucleus and represses ABI5- and ABF4-induced transcriptional responses, thereby negatively regulating ABA signaling.
B) RALF1/FER inhibits stomatal opening through the G proteins (AGG1,2,3, AGB1, and XLG1,2,3), and OST1. RALF1/FER also induces stomatal closure through the G proteins. Furthermore, FER negatively regulates ABA-induced stomatal closure, potentially by activating ABI2. In contrast, EBP1 positively regulates ABA induced stomatal closure.
VIII. Abiotic Stresses
Salinity stress
RALF peptides and their receptors are involved in the response to high salinity. The excess of sodium chloride (NaCl) is particularly harmful to plants, as it leads to both ionic and osmotic stresses (134). On top of that, ROS production is an essential part of salt tolerance and needs to be precisely regulated to avoid an additional burden, the oxidative stress (135).
FER plays a central role in regulating salt stress response, as attested by the salt-hypersensitive phenotype of the fer-4 mutant (33,56). Under NaCl treatment, FER and AGB1 positively regulate short-term (minutes) and negatively regulate long-term (hours) ROS production (56). Additionally, FER directly regulates the dynamic phosphorylation of COMPANION OF CELLULOSE SYNTHASE 1 (CC1) under salt stress, potentially influencing microtubule organization (136). Salt treatment also inhibits FER’s autophosphorylation and its direct phosphorylation of phytochrome B (phyB). The latter increases phyB abundance in the nucleus, which in turn promotes PIF5 degradation, an important step in a controlled salt-stress response (137).
Salt treatment enhances PME activity, which is required for the salt-induced transcriptional response and phosphorylation of MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6). These characteristic responses are increased in
fer-4, and chemical inhibition of PME activity alleviates the mutant hypersensitivity, suggesting that FER may sense salt-induced cell wall modifications to regulate subsequent intracellular signaling (138). FER’s role as a CW sensor in BR-induced growth, and its regulation by RALFs, further support this hypothesis (76). Moreover, during the recovery phase from salinity stress, a rise in [Ca
2+]
cyt mediated by FER is necessary for maintaining CWI (33). FER-regulated salt-stress responses with unconfirmed RALF involvement are summarized in
Figure 9A.
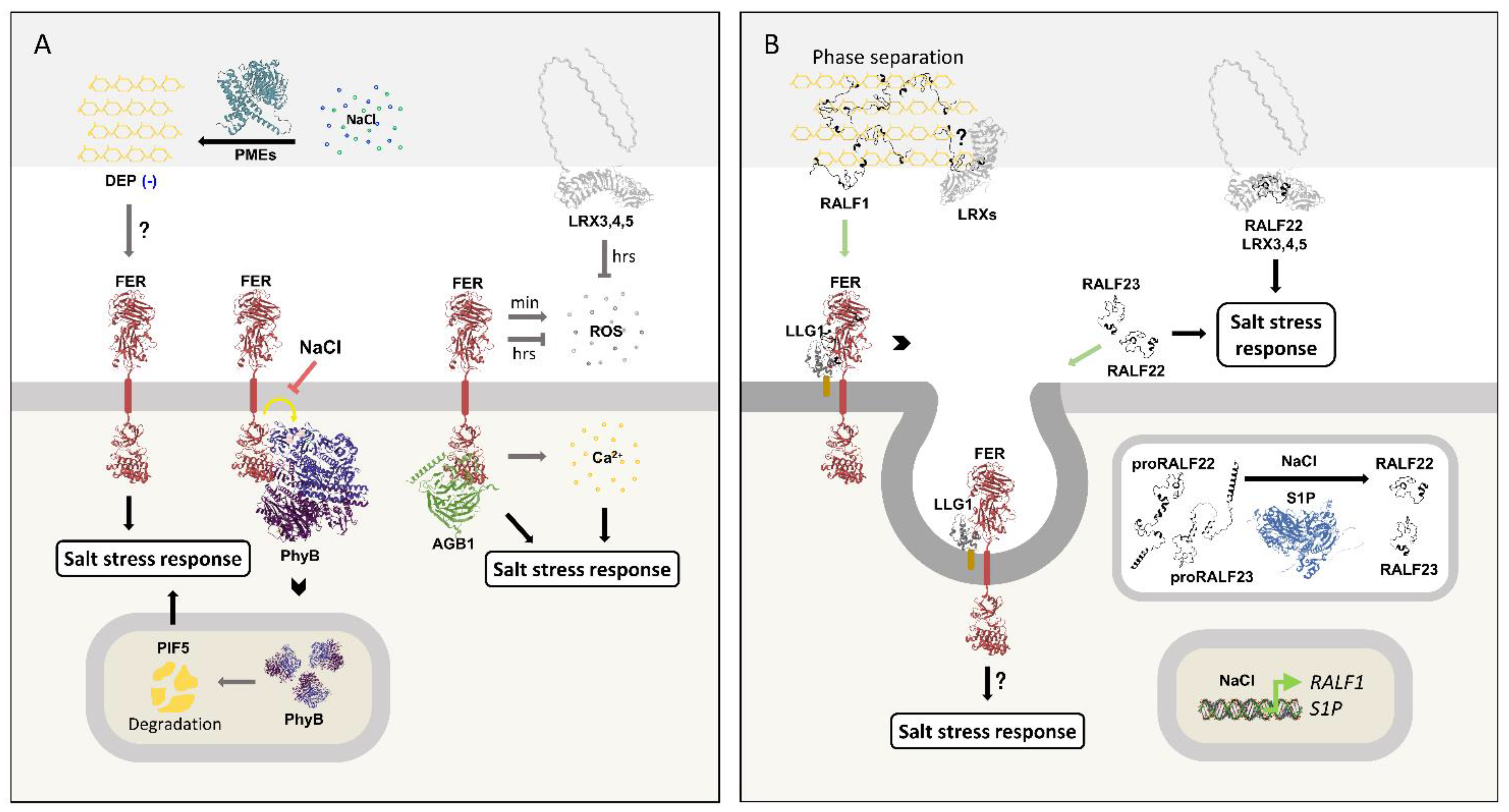
Salt induces RALF1 transcription and raises de-esterified pectin levels in the cell wall. These changes increase RALF1 and DEP phase separation, leading to the formation of membrane micro- and nanodomains enriched with FER, LLG1, FLS2 and BRI1 that undergo intense endocytosis. This mechanism may act as a general regulator that integrates and coordinates the biotic and abiotic stresses responses. This assembly also regulates FER/LLG1-induced ROS production under normal conditions and potentially during salt stress (27). Notably, the combined treatment of NaCl and RALF22 or RALF23 also promotes FER internalization (17).
The cell wall-associated proteins LRX3,4,5 are expressed in vegetative tissues and are essential to salt stress tolerance. Part of their role arises from the negative regulation of long-term (hours) ROS production during salinity response, as seen with FER (133). Another part involves their binding to RALF22 and RALF23 in a still poorly understood dynamic (17). NaCl induces S1P subtilase transcription and processing of proRALF22 and proRALF23 (17,27). In turn, plants overexpressing RALF22 or RALF23 and the lrx345 triple mutant are hypersensitive to NaCl treatment. Strikingly, this phenotype is partially reversed in the quadruple mutants s1p/lrx345 and ralf22/lrx345 (17), suggesting a complex mechanism, discussed below. The salt-hypersensitivity of both lrx345 and fer-4 is also reversed by a phyB loss-of-function mutant and a PIF5 overexpressor line, further supporting the joint action of these RALF receptors (137). RALF involvement in the phyB/PIF-mediated response, however, remains to be investigated.
The findings mentioned above should be carefully analyzed. As described, RALF1/DEP phase separation may play a positive role in salinity stress tolerance (27). Nonetheless, RALF1 and NaCl combined treatments disrupt the plant's Na
+/K
+ ionic homeostasis in a FER-dependent manner, leading to salt hypersensitivity (56). Thus, the negative effects of RALF1 treatment and
RALF22 or
RALF23 overexpression on salinity tolerance might stem from an imbalanced response, that otherwise has a positive effect when the peptides are present at endogenous levels. The salt-induced transcription of
RALF1 and processing of proRALF22 and proRALF23 contribute to this assumption (17,27). Alternatively, RALF peptides might be part of a negative feedback loop that finely regulates the stress response. In line with both possibilities, the partial reversal of
lrx345 mutant salt hypersensitivity phenotype by diminishing RALF’s presence (as seen in the
s1p/lrx345 and
ralf22/lrx345 mutants) points to a response that relies on an equilibrium involving RALFs and LRXs. A single split luciferase complementation assay showed that the RALF22/LRX3 interaction is disrupted by NaCl (17). This binding dynamic might be central to an apoplast sensing mechanism, although it must be confirmed by other approaches. In root hairs, RALF22 interacts with LRXs and DEP (29). Given the RALF1/DEP phase separation induction by NaCl, and the importance of LRXs for salt stress tolerance, there is a potential role for the RALF/LRX/DEP assembly in this context. In summary, the involvement of RALF peptides in the salinity stress response still holds many questions (
Figure 9B).
Figure 9.
RALF peptides and their receptors are involved in the salinity stress response.
Figure 9.
RALF peptides and their receptors are involved in the salinity stress response.
A) FER plays a central role in regulating the salt stress response. Salt treatment enhances PME activity, which triggers intracellular signaling. FER regulates this response, potentially by sensing changes in cell wall composition, particularly the PME-generated de-esterified pectin (DEP; depicted in yellow). Salt also inhibits FER-mediated phosphorylation of phyB, thereby increasing phyB accumulation in the nucleus. This promotes PIF5 degradation and contributes to tolerance. Furthermore, in the presence of NaCl, FER positively regulates short-term (minutes) ROS production, while FER and LRX3,4,5 negatively regulate long-term (hours) ROS production. FER is also essential for cell recovery after salt stress by modulating calcium influx. Finally, FER and AGB1 have a synergistic effect on NaCl tolerance, but, as with the above-mentioned responses, RALF involvement remains unexplored.
B) Salinity stress induces phase separation of RALF1 and de-esterified pectin, creating membrane micro- and nanodomains enriched in FER/LLG1, where endocytosis is intense. This response may be part of FER-mediated ROS regulation and potentially serves as a mechanism to regulate the salt stress response. RALF22 and RALF23 also induce FER internalization in the presence of NaCl. RALF22 binds to LRX3,4,5, and this interaction regulates salinity stress tolerance by a yet poorly understood mechanism. One possibility is that LRXs are part of the RALF/DEP phase separation. Additionally, the presence of NaCl induces RALF1 and S1P transcription, as well as proRALF22 and RALF23 processing by the subtilase.
Nitrogen deficiency
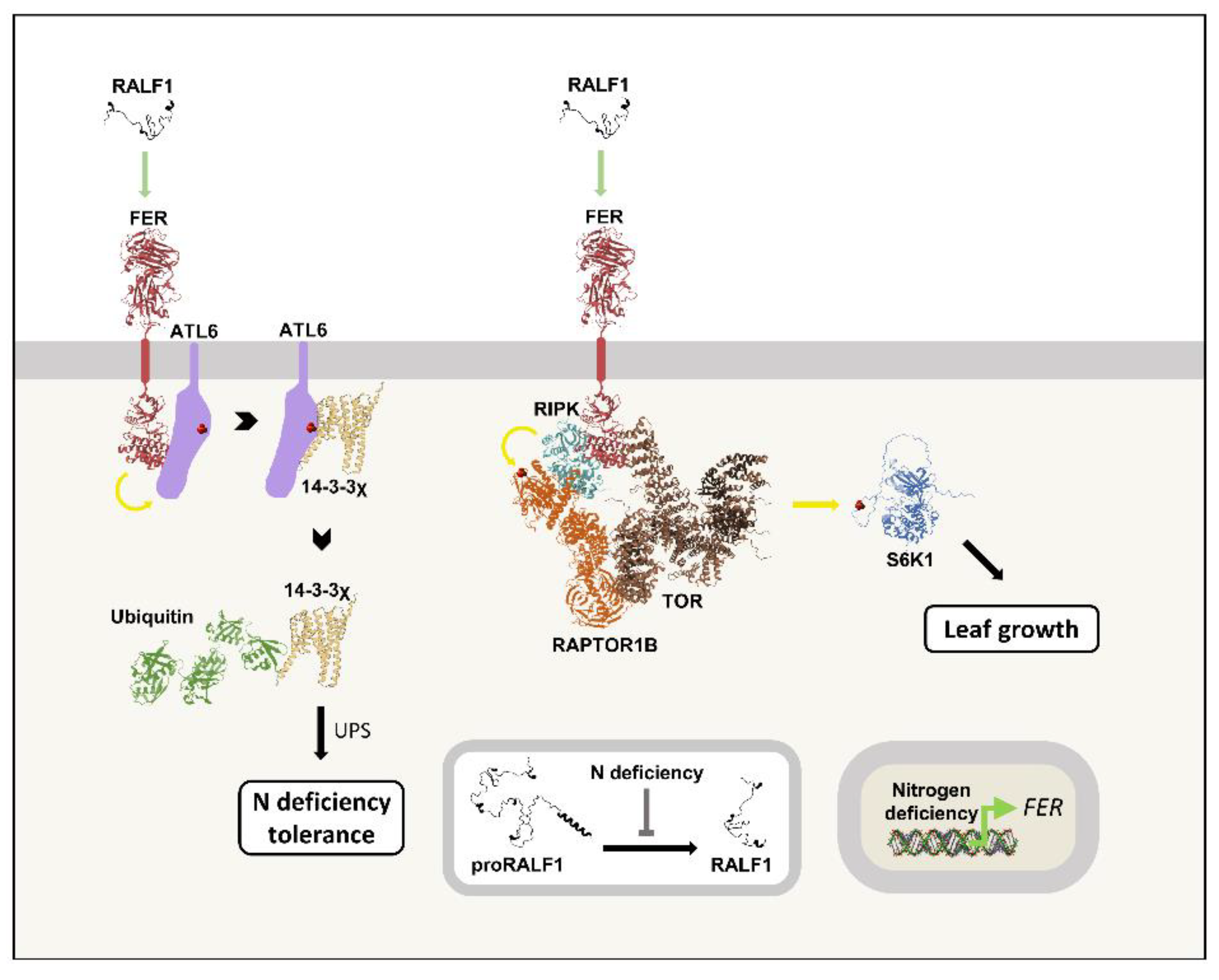
A fine balance between carbon and nitrogen levels (C/N) is of great importance for plant metabolism. The ubiquitin-proteasome system (UPS) contributes to this balance through the degradation of 14-3-3 proteins, which are targeted to the system by the ubiquitin ligases ARABIDOPSIS TOXICOSIS IN YEAST (ATLs) (139). FER is part of this pathway, and its expression is induced in high C/N environments, hereafter also referred to as nitrogen deficiency. FER phosphorylates ATL6, which promotes ATL6/14-3-3χ interaction, leading to the ubiquitination of the latter and its subsequent degradation by the UPS. RALF1 stimulates 14-3-3χ degradation in a FER-dependent manner, and plants overexpressing RALF1 are more resistant to nitrogen deficiency than wild-type plants (140). In summary, RALF1 and FER are important for the adaptation to nitrogen-depleted media by modulating 14-3-3χ levels through ATL6.
The kinase TARGET OF RAPAMYCIN (TOR) is a master regulator of cellular nutrient status. TOR’s control of plant growth involves its canonical partner REGULATORY-ASSOCIATED PROTEIN OF TOR 1 (RAPTOR1) and the kinase PROTEIN-SERINE KINASE 6 (S6K1) (141). Under nitrogen deficiency, RALF1 treatment promotes RAPTOR1 phosphorylation and its interaction with TOR in a FER-dependent manner, potentially through RIPK (68,142). The TOR/RAPTOR interaction induces S6K1 phosphorylation, leading to an increase in shoot biomass (142). Therefore, in this pathway, RALF1 promotes growth, opposing its canonical role as a growth inhibitor (10,142). Curiously, nitrogen-depleted media negatively regulate both proRALF1 processing and FER phosphorylation (142). This signal to decrease mature RALF1 presence, along with its positive role in nitrogen deficiency, indicates a finely regulated response. Notably, under normal conditions, FER activates TOR to induce growth and repress autophagy in roots (143), but RALF1 involvement was not investigated.
Figure 10 summarizes RALF1's role in the nitrogen deficiency response.
IX. Biotic Stresses
Crosstalk with biotic stress phytohormones
The phytohormones jasmonic acid (JA) and salicylic acid (SA) play crucial roles in the defense against pathogens (144). Plant lines that overexpress RALF22 or RALF23, and the loss-of-function mutants fer-4 and lrx345, have considerably higher levels of SA and, especially, JA. Accordingly, these mutants display an increased expression of the JA- and SA-responsive genes PDF1.2 and PDF1.3, as well as PR1 and PR5. The JASMONATE-ZIM-DOMAIN (JAZ) proteins are repressors of the JA signaling pathway that get degraded in the presence of the hormone (144). RALF22 treatment induces the degradation of JAZ1 and JAZ9 in a FER- and LRX3,4,5-dependent manner (133).
The transcription factor MYC2 plays a pivotal role in JA signaling (144). FER phosphorylates MYC2, leading to a decrease in its protein levels. The consequent reduction of JA signaling confers resistance to the bacterium
Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). The binding of RALF23 to FER inhibits MYC2 phosphorylation, increasing the TF's stability and its mobilization to the nucleus, where MYC2 activates JA signaling. Consequently, plants treated with RALF23 become susceptible to Pst DC3000 (145). Collectively, these results point to a crosstalk between RALFs, jasmonic acid, and salicylic acid, which relies on FER and LRX3,4,5 (
Figure 11B).
Pattern-triggered immunity
In order to deal with the threats posed by numerous pathogens, plants first need to recognize them. To achieve this, the membrane-associated Pattern Recognition Receptors (PRRs) perceive molecules derived from infectious organisms, known as Pathogen-Associated Molecular Patterns (PAMPs) (146). Two of the most studied PRRs are FLAGELLIN-SENSITIVE 2 (FLS2) and ELONGATION FACTOR TU RECEPTOR (EFR), the receptors of bacteria-derived flagellin (flg22) and translation factor EF-Tu (elf18), respectively. Both FLS2 and EFR function alongside the coreceptor BAK1 (147). PAMP perception by PRRs elicits the Pattern-Triggered Immunity (PTI), and a key initial step of this immune response is the controlled production of ROS by NADPH oxidases (148).
RALF peptides have opposite effects on immunity depending on their concentrations. At high levels (µM range), these peptides negatively regulate PTI by altering the PRRs’ membrane dynamics. Flg22 and elf18 treatment induces the formation of FLS2/BAK1 and EFR/BAK1 complexes, respectively. FER's extracellular domain stabilizes these complexes, leading to ROS production and resistance to Pst DC3000 (16,149). Upon 1 µM treatment, RALF23 inhibits the PTI response by binding to FER and compromising its role in stabilizing the PRR complexes, which then dissociate (16). Also through FER, high concentrations of RALF23 promote opposite membrane mobility behaviors of FLS2 and BAK1, which may either result from or contribute to the peptide-induced FLS2/BAK1 dissociation. Notably, RALF23's negative control of PTI also depends on FER’s kinase activity, and is not restricted to the receptor’s extracellular interactions with PRRs (149).
The FER homolog ANX1, mainly known for its RALF-regulated role in reproduction, is also expressed in vegetative tissues (41,150,151). Contrary to FER, ANX1 binds to BAK1 and reduces the flg22/FLS2/BAK1 complex formation, negatively regulating PTI. ANX1 also suppress the Effector-Triggered Immunity (ETI) (151). Whether RALFs participate in these ANX1-modulated responses remains unknown.
The coreceptor LLG1 is essential for the RALF23/FER inhibition of elf18-induced ROS production (18). LLG1 also interacts with EFR and FLS2, participating in the PAMP-induced, BAK1-mediated phosphorylation of the cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1), which triggers ROS burst (152). Since LLG1 is involved in both PTI induction by PAMPs and its repression by RALF23, it may function as a molecular switch, determining the signaling output. Alternatively, LLG1 may function solely as a coreceptor in both contexts.
LRX3,4,5 are also involved in PTI regulation. Like FER, these cell wall-associated proteins are important for stabilizing the flg22/FLS2/BAK1 complex, leading to ROS production. RALF23's antagonism on this response also depends on LRX3,4,5, although the mechanism is still obscure (149). The similar roles of FER and LRXs in PTI become even more intriguing given their ability to bind each other (40,149). It is worth noting that RALF23 does not modulate this interaction (149), raising questions about its biological significance.
RALF1 also negatively regulates the PTI response when administered at 1 µM. RALF1/FER binding induces CARs' nanodomain formation, which contributes to the FER-mediated stabilization of the flg22/FLS2/BAK1 complex. Like RALF23, RALF1 promotes the dissociation of this PTI complex, a process further facilitated by CARs (117). Remarkably, the RALF1, RALF23 and de-esterified pectin phase separation induces the formation of membrane domains enriched in FER, LLG1, and FLS2, where endocytosis is intense (27). Considering the players involved, RALF-triggered endocytosis may contribute to PTI modulation.
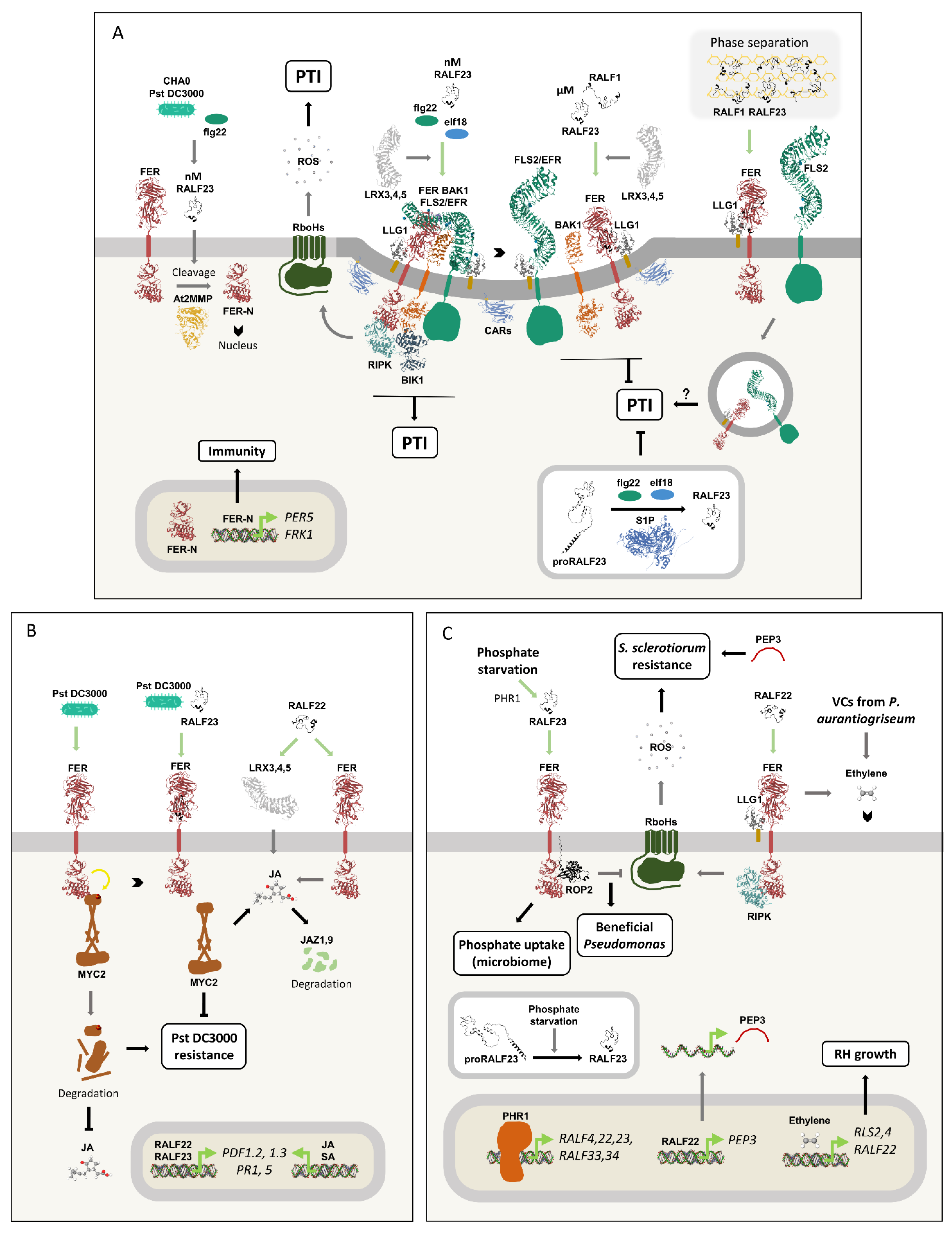
It wasn’t until recently that Feng Yu’s lab showed that RALF23’s effect on immunity is concentration-dependent. Treatment with flg22 and infection with Pst DC3000 or the commensal rhizobacterium P. protegens strain CHA0 induce an increase in mature RALF23, reaching endogenous concentrations of 100–200 nM. At these levels, RALF23 promote cleavage of FER's intracellular domain by MATRIX METALLOPROTEINASE 2 (At2MMP), releasing the N-terminal portion of FER (FER-N) from the plasma membrane. FER-N then translocates to the nucleus, inducing the expression of the immunity genes PER5 and FRK1, enhancing flg22-induced FLS2/BAK1 complex formation and increasing ROS production (13). On the other hand, treatment with 1 µM RALF23 resulted in the opposite responses (13,16,149).
The processing of proRALFs is part of PTI regulation. Both elf18 treatment and Pst DC3000 infection induce proRALF23 processing by the subtilase S1P, and this step is essential for the peptide-triggered PTI inhibition. RALF33 and RALF34 also possess a putative S1P cleavage site and inhibit elf18-induced ROS production. In contrast, RALF17, a peptide without the site, induces PTI (16). Accordingly, a family-wide study of RALFs from A. thaliana pointed to a strong tendency for peptides with the predicted S1P cleavage site to inhibit elf18-induced ROS production and peptides without the site to increase it (5). Nevertheless, exceptions like RALF22 exist, which possess the cleavage site and induce ROS (57).
Other proteins act downstream of RALFs in the PTI modulation. RALF1’s negative effect on flg22-induced ROS production is compromised in a
GRP7 interference mutant (11). In turn, mutants lacking EBP1, a negative regulator of RALF1 signaling, exhibit an even greater reduction in ROS levels under simultaneous treatment with flg22 and RALF1 (84). Furthermore, ROS burst triggered by RALF22 is reduced in
RBOHD,
LLG1,
BIK1 or
RIPK knockout mutants (57). Collectively, these results indicate that some proteins involved in the RALF-induced growth inhibition signaling are also part of the peptides' immunity modulation.
Figure 11A depicts the different aspects of RALFs’ regulation of PTI.
Plant-microorganism interactions
The biological and evolutionary basis of RALFs’ role in immunity modulation is now being unraveled. Under normal growth conditions, RALF23 is expressed in the root stele. In the presence of the pathogenic Pst DC3000 or the commensal Pseudomonas CHA0, RALF23 expression and propeptide processing extend to the outer cell layers of the transition and elongation zones. The mature peptide induces the previously cited cleavage of FER’s intracellular domain and its translocation to the nucleus, where it upregulates immunity genes, resulting in a localized immune response in the root (13). More broadly, FER generates basal root ROS levels through ROP2 and RboHD, which are detrimental to beneficial bacteria of the genus Pseudomonas. This response is opposed by RALF23 at high concentrations, leading to an enrichment of these pseudomonads in the root microbiome (62).
RALF23 also modulates the microbiome composition under phosphate (Pi) starvation through the transcription factor PHOSPHATE STARVATION RESPONSE 1 (PHR1). In response to Pi deprivation, PHR1 binds to the promoters of RALF4,22,23,33,34, upregulating their expression. In addition, phosphate scarcity induces proRALF23 processing, and the increased levels of mature RALF23 reduce the immune response and alters the microbiota composition by favoring the enrichment of bacteria beneficial for Pi uptake (153). In this response, RALF23 acts in a FER-dependent manner through the PRR complex disassembly mechanism described earlier (16,153). The effects of RALF23 under different microbial interactions provide examples of how modulation of peptide concentration, and not just its presence, achieves context-specific immunity.
RALF22 positively regulates the immune defense against the necrotrophic fungi Sclerotinia sclerotiorum. RALF22 treatment triggers ROS production and reduces fungal colonization in a FER/LLG1-dependent manner. RALF22 also promotes the transcription and translation of PEP3, a peptide that amplifies the immune response. Furthermore, RALF22 treatment increases resistance to S. sclerotiorum in different species of the Brassica genus, indicating a conserved response (57).
Plant-interacting microorganisms emit small volatile compounds (VCs, <300 Da) that trigger plant reactions. VCs produced by the pathogenic fungus
Penicillium aurantiogriseum induce changes in Arabidopsis root architecture, such as the hyper-elongation of RHs (154). Notably, these compounds upregulate
RALF22 transcription and both
ralf22 and
fer-4 mutants are less sensitive to VC-triggered RH elongation. Ethylene is the primary VC candidate to promote these root changes, as its precursor ACC induces RH growth in wild-type plants but not in
ralf22 or
fer-4. Consistently, VCs upregulate the ethylene-inducible, RH growth-promoting genes
RSL2 and RLS4 in a RALF22/FER-dependent manner. Taken together, these results suggest that, upon
P. aurantiogriseum infection, the VC-induced RH growth is mediated by ethylene in a signaling pathway that depends on RALF22/FER and RSL2,4 (105).
Figure 11C summarizes the first examples of how plants use RALFs to mediate complex interactions with both beneficial and pathogenic microorganisms.
Figure 11.
RALF peptides modulate immune responses.
Figure 11.
RALF peptides modulate immune responses.
A) The presence of Pseudomonas DC3000 and CHA0, as well as flg22, raises mature RALF23 concentrations to 100–200 nM, which in turn triggers FER’s N-terminal cleavage by AtMMP. FER-N translocates to the nucleus, where it upregulates the immune-related genes PER5 and FRK1, thus promoting immunity. At the membrane, FER stabilizes the PRR complexes in micro- and nanodomains assembled by CAR proteins. Active PRR complexes trigger ROS production by RboHs in a RIPK- and BIK1-dependent manner. At high concentrations (µM), the binding of RALF1 or RALF23 to FER induces the dissociation of these complexes, thereby inhibiting the PTI response. LRX3,4,5 are also involved in this mechanism. In addition, RALF1 and RALF23 bind to de-esterified pectin (depicted in yellow) and induce phase separation, forming membrane microdomains where FER, LLG1, and FLS2 endocytosis is intense, potentially contributing to the modulation of PTI. Notably, flg22 or elf18 treatment induce proRALF23 processing by S1P.
B) FER negatively regulates the jasmonic acid (JA) signaling pathway by phosphorylating MYC2, leading to its degradation. This increases resistance against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). RALF23 binds to FER and inhibits MYC2 phosphorylation, thereby preserving its integrity, favoring the JA pathway, and consequently leading to plant susceptibility to Pst DC3000. RALF22 increases JA levels and induces JAZ1 and JAZ9 degradation in a FER- and LRX3,4,5-dependent manner. Moreover, RALF22, RALF23, JA, and salicylic acid (SA) induce the transcription of PDFs and PRs.
C) Phosphate starvation induces the expression of RALF genes through the transcription factor PHR1, as well as the processing of proRALF23. Mature RALF23 presence decreases the PTI response, enriching the root microbiome with microorganisms that promote phosphate uptake. FER triggers basal ROS production by RboHs through ROP2. This response is repressed by RALF23 ligation, enriching the microbiome with beneficial Pseudomonas. Moreover, RALF22 induces PTI response and the defense peptide PEP3 transcription and translation, leading to resistance against the fungus Sclerotinia sclerotiorum. Ethylene is among the volatile compounds (VCs) secreted by the pathogenic fungus Penicillium aurantiogriseum, and this phytohormone upregulates the expression of RSL2, RSL4, and RALF22, leading to RH growth. RALF22 and FER are essential positive regulators of this response.
Certain fungal species possess genes homologous to plant RALFs, and the polyphyletic origin of these RALF-like genes suggests an acquisition through lateral transfer (8). At least some of them are functional, as attested by FgRALF from Fusarium graminearum, expressed during wheat (Triticum aestivum) infection. The peptide FgRALF is capable of binding to Arabidopsis' FER and its wheat homologue TaFER, suppressing the PTI response in both species (155). Likewise, FoRALF, a peptide from the related species Fusarium oxysporum, negatively regulates immunity in tomato plants (Solanum lycopersicum) and is less effective at infecting the Arabidopsis fer-4 mutant than the wild-type (156). Strikingly, the root-knot nematode (Meloidogyne incognita) also possesses RALF-like genes (MiRALF), four in total. Treatment of Arabidopsis with MiRALF1 or MiRALF3 induces MYC2 degradation and suppresses flg22-induced ROS production in a FER-dependent manner. As expected, these two peptides are secreted during M. incognita infection, bind to FER and enhance the nematode pathogenicity (9).
The results described above suggest a similar mechanism of action between RALF23 and the RALF-like peptides, which dampen the host defense to function as effectors of pathogenicity. Remarkably, the endophytic fungus Colletotrichum tofieldiae also produces a RALF-like peptide that represses Arabidopsis’ immune response in a FER-dependent manner, facilitating its root colonization. However, this interaction benefits the plant by contributing to root growth under Pi starvation (157), illustrating the versatility with which different microorganisms employ RALF-like peptides.
X. Reproduction
Regulation of flowering time
RALF peptides are intimately involved in Arabidopsis reproduction. Among the events regulated by these peptides is flowering time. This complex process involves the convergence of diverse signals in the production of florigens, such as the protein FLOWERING LOCUS T (158). The transcription factor FLOWERING LOCUS C (FLC) and its homologues, MADS AFFECTING FLOWERING (MAFs), suppress flowering by repressing the transcription of FLOWERING LOCUS T (159,160). RALF1 negatively regulates flowering in a FER-dependent manner by inducing the expression and alternative splicing (AS) of FLC, as well as the AS of MAF1, MAF2, and MAF3. FER also contributes to floral transition by positively regulating the transcription of circadian clock genes (161).
Pollen germination and penetration in the stigma
RALF peptides play crucial roles in various steps of the pollen path leading to fertilization. Pollen grains (PG) adhere to the stigma, where they germinate and initiate pollen tube (PT) growth. The PG's hydration is essential for its germination and is mediated by the exchange of signals with the female tissue (162). RALF23 and RALF33 are secreted by the stigma, where they induce ROS production, which represses PG hydration. To this end, these RALFs binds to the FER/ANJ/LLG1 complex, activating the NADPH oxidase RboHD through ROP2 (32). The RALF23,33/FER/ANJ complex also induces the phosphorylation of the proton pump AHA2 at its inhibitory residue, thus retaining H⁺ at the cytosol (44). Aquaporin gating is regulated by protonation (163), and the low intracellular pH maintained by RALF23,33 signaling results in the protonation of PLASMA MEMBRANE INTRINSIC PROTEINs (PIP1;1, 2;2, 2;5) and their subsequent closure (44). Upon pollen grain arrival, the pollen-produced peptide POLLEN COAT PROTEIN-Bγ (PCP-Bγ) outcompetes RALF23,33 for binding to the FER/ANJ/LLG1 complex (32,44). The newly formed PCP/FER/ANJ complex triggers phosphorylation at AHA2 activation sites, which pumps H+ outside the cell, releasing PIPs protonation and opening the water channel (44). The dislodging of RALF peptides by PCP-Bγ also leads to a decrease in ROS production by stigmatic cells, which, together with water efflux, results in PG hydration (32,44).
Plants have strategies to avoid interspecific pollination. The stigma produces RALF1,22,23,33 (sRALFs), which act as autocrine signals perceived by CrRLK1L complexes formed by FER, CVY1, ANJ, and/or HERK1. Meanwhile, the pollen tubes produce RALF10,11,12,13,25,26,30 (pRALFs), which function in a partially redundant manner. The binding of sRALFs to CrRLK1L complexes creates a blockage of PT penetration in the stigma through an unknown mechanism that does not involve ROS. The stigmatic proteins LRX3,4,5 bind to sRALFs and are also necessary for this blockage (24). Once the pollen arrives, pRALFs outcompete and dislodge sRALFs from the CrRLK1L complexes due to a higher binding affinity, 'unblocking' the PT penetration (24,26). This mechanism is responsible for an intergeneric hybridization barrier that prevents the penetration of PTs from distantly related
Brassicaceae species into the stigma of
A. thaliana. This barrier arises from the inability of
Brassicaceae pRALFs to dislodge Arabidopsis sRALFs from their receptors (24). The role of RALF peptides in pollen germination and penetration in the stigma is summarized in
Figure 12A.
Pollen tube growth
Pollen tubes are cells that undergo rapid, unidirectional growth, and are responsible for delivering male gametes to the ovule. At the PT’s tip, exocytosis of plasma membrane and cell wall material sustains rapid cellular expansion (162). Among the secreted cell wall components, pectins are of special importance due to their initial esterified state, which provides malleability to the growing apex, and their subsequent de-esterification, which rigidifies the PT flanks (164). At the PT’s apical region, ROPs/GEFs regulate RboHs, creating apoplastic ROS oscillations that modulate Ca²⁺ influx, and vice versa, a crucial dynamic for PT expansion (162,165).
RALF4 and RALF19 act together with their receptors CrRLK1Ls, LLGs, and LRXs to maintain cell wall integrity (CWI) and regulate PT growth. These proteins act throughout the entire PT, but their signaling activity occurs primarily at the tip, where a complex protein spatial organization and vesicle trafficking take place (20,28,41,166). BUPS1,2, and ANX1,2 interact among themselves and with LLG2,3, forming receptor complexes that transduce the RALF4,19 signal (20,35,41). This pathway regulates the composition of pectin and callose in the CW, as well as the Ca²⁺ influx (35,54). The RALF/BUPS/ANX/LLG complexes also control ROS oscillation, and at least ANXs do so through RboHH,J (35,167). In other tissues, CrRLK1Ls interact with ROPs/GEFs to regulate ROS production (32,62,108). This signaling may extend to pollen, as BUPS bind ROP1,3,5,9, and GEF1,12, while LLG3 binds ROP1, all of which are expressed in PTs (35,168).
LRX8,9,10,11 have a partially redundant role in CWI maintenance during PT growth (169). LRX8 interacts with RALF4, positioning the peptide so that its cationic residues are exposed to the medium (22). This conformation enhances RALF4’s affinity for the negatively charged de-esterified pectin (DEP) in the cell wall (28). Two RALF4/LRX8 dimers are capable of forming a disulfide bridge between the LRXs, resulting in a heterotetrameric complex (22). Each of the dimers may bind to a different pectin polymer, acting as a crosslink and therefore strengthening the cell wall mesh. Remarkably, the RALF4/LRX8 complexes disposition along the PT flanks forms a reticulated pattern (28). Since RALF19 shares high identity with RALF4 and binds to LRX8 (22), it probably participates in this CWI mechanism.
Similar to ANXs and BUPs, LRX8-11 modulate the cell wall’s pectin and callose composition (169,170). These proteins also regulate the Ca2+ influx dynamics, mainly at the PT tip (54,171). Curiously, the LRR domain of LRXs interacts with components of the PT’s plasma membrane, and FER’s extracellular domains are capable of binding LRX1-5 (39,40,171). These findings raise the possibility of a functional interaction between CrRLK1Ls and LRXs in the pollen tube, which may be central to RALF signaling. It is important to note that a simultaneous interaction between RALF4, LRX, and CrRLK1Ls was not observed (22), but it remains to be tested whether pectin is necessary for it.
The cytoplasmic proteins MRI, AUN1, and AUN2 are part of RALF’s PT growth regulation. The kinase MRI is activated by RALF4,19 and has a positive role in maintaining CWI (38). Through BUPS1, the RALF/BUPS/ANX/LLG complex phosphorylate MRI, which then interacts with and activates the calcium channels MLO1,5,9, thereby modulating Ca2+ influx (54). Moreover, MRI acts upstream of the ROS-producing RboHH and RboHJ (114). In contrast, the phosphatases AUN1 and AUN2 negatively regulate the CWI maintenance induced by RALF4,19/ANX1,2, by LRX8-11, and by RBOHH,J (113).
In order to reach the ovule, PTs must penetrate the pistil. Along this path, the expanding tube crosses environments with different resistances. At the PT tip, mechanical stress induces ROP1 activation and RALF4 secretion, both dependent on BUPS1. ROP1 is responsible for transducing the signal that leads to rapid exocytosis, reinforcing the cell wall, and thus preventing PT rupture (172). Notably, ANX1 also regulates exocytosis at the PT tip (167). The basis of this mechanical stress perception remains unresolved. An important clue arises from FER’s involvement in the mechanical stress response during cotyledon’s pavement cell morphogenesis, where it binds to pectin and activates ROPs/GEFs (81,173). BUPS1 is also capable of binding to both pectin and ROPs/GEFs (33,168). Therefore, BUPS1 may functions as a mechanical stress sensor in pollen tubes through its pectin-binding ability. Given the secretion of RALF4 induced by the stress stimulus (172), it is also possible that this response is regulated by these peptides.
Figure 12B summarizes the role of RALFs during PT growth.
PT rupture and prevention of multiple fertilization
For fertilization to occur, the two sperm cells located inside the pollen tube must reach the ovule. Once there, the PT penetrates one of two synergids through a complex exchange of signals that one of two synergids through a complex exchange of signals that leads to PT rupture and the release of gametes. This highly coordinated process relies on changes in the synergids’ Ca2+ and ROS profiles (162). FER, its coreceptor LRE, and the Ca²⁺ channel NTA act together in the synergids to control sperm delivery (174). To this end, the pollen-produced RALF4, RALF19, and RALF37 bind to FER/LRE, generating an NTA-mediated Ca²⁺ profile essential for PT perception. It is noteworthy that, in this signaling, the calcium-binding CALMODULIN 7 negatively regulates NTA (23).
RALF peptides also coordinate PT rupture to enable fertilization. RALF34 is produced by the ovule, where it accumulates. When the PT reaches the ovule, the pre-existing RALF34 binds to the pollen’s BUPS/ANX/LLG complexes, dislodging RALF4,19 and triggering PT rupture (20,41). Notably, reproduction is not compromised in the ralf34-1 knockout mutant, suggesting other RALFs with overlapping functions (41). Moreover, in the synergids, the FER/LRE complex regulates ROS production, a response necessary for PT rupture (175). In addition, FER, LRE, HERK1, and ANJ form functionally redundant complexes essential for PT rupture (176). Whether and how RALFs modulate FER/LRE-mediated ROS production and FER/LRE/HERK1/ANJ signaling leading to PT rupture remains to be investigated.
Multiple pollen tube arrival at the ovule (polytubey) is blocked to guarantee a single fertilization event. However, if the first PT fails, the polytubey block is lifted, allowing another tube to reach the ovule (25,162). RALF peptides are involved in this mechanism, which takes place at the ovary’s septum, a tissue between the transmitting tract and the ovule. RALF6,7,16,36,37 (pRALFs) are secreted by the flanks of PTs that penetrated the septum and bind to the FER/HERK1/ANJ complex, present in the female tissue cells. This ligation initiates the blockage, which remains active until PT rupture, when the pRALFs quickly disappear (25). In the case of a successful fertilization, the female tissue ceases to produce PT attractants (25,162). In case of failure, other pollen tubes can penetrate the septum due to the polytubey block being lifted (25).
Figure 12.
RALF peptides participate in fertilization.
Figure 12.
RALF peptides participate in fertilization.
A) The stigma-produced RALF23 and RALF33 bind to the FER/ANJ/LLG1 complex, inducing ROS production via ROP2 and thereby preventing pollen grain (PG) hydration. The RALF23,33/FER/ANJ complex also triggers AHA2 phosphorylation at the inhibitory residue S899, maintaining H⁺ in the cytosol, which results in the protonation and closure of PIPs. Upon pollen arrival, the pollen-produced peptide PCP-Bγ dislodges RALF23,33 from the receptor complex, inhibiting ROS production and promoting AHA2 phosphorylation at the activation residues T881 and T947. This release PIP protonation, opens the water channels, and allows PG hydration. Furthermore, RALF peptides produced by the stigma (sRALFs) bind receptor complexes composed of FER, CVY1, ANJ, and HERK1, blocking pollen tube (PT) penetration. LRX3,4,5 are also involved in this blockage. In turn, the pollen-produced RALFs (pRALFs) dislodge sRALFs from these complexes, lifting the blockage and allowing PT penetration. pRALFs from species distantly related to A. thaliana are unable to lift the blockage.
B) RALF4 and RALF19 maintain cell wall integrity (CWI) during pollen tube growth. At the PT tip, the RALF/BUPS/ANX/LLG complexes phosphorylate MRI, which activates the MLO1,5,9 calcium channels, coordinating the ion influx. These complexes also induce ROS production by RboHH and RboHJ, potentially through MRI and ROPs/GEFs. In addition, LRX8-11 are essential for CWI maintenance. At the PT flanks, RALF4 binds LRX8, and the complex interacts with de-esterified pectin (DEP), rigidifying the CW. AUN1 and AUN2 negatively regulate the RALF4,19, CrRLK1Ls, and LRX8-11 responses. These RALF receptors are also involved in the exocytosis of cell wall material, and during mechanical stress, BUPS1 mediates this process through ROP1. Lastly, CrRLK1Ls and LRXs can bind to each other, and this interaction may be relevant to the RALF signaling pathway.
C) RALF4, RALF19, and RALF37 are produced by the pollen and bind to FER/LRE, activating the calcium channel NTA and creating a Ca2+ profile in the synergids essential for PT perception. By contrast, RALF34 is produced by the synergids and dislodges RALF4,19 from the BUPS/ANX/LLG complexes in the PT, leading to tube rupture. ROS production induced by FER/LRE and the formation of the FER/LRE/HERK1/ANJ complex are also essential for PT rupture and are potentially regulated by pRALFs. At the ovary’s septum, the pollen-produced RALF6,36,37 bind to the FER/HERK1/ANJ complex, blocking the arrival of multiple PTs at the ovule (polytubey). After PT rupture, these pRALFs disappear and the blockage is lifted.
The extensin domain of LRXs has been omitted for better visualization.
Remarkably, FER is part of another mechanism that prevents multiple PTs from arriving at the ovule. Pollen tube arrival induces nitric oxide (NO) production in a FER- and DEP-dependent manner. The NO gas then inhibits the action of the pollen attractant LURE1 peptides (177). Given the intimate relationship between RALFs, FER, DEPs and the control of multiple fertilization, these peptides might regulate this mechanism.
Figure 12C summarizes the role of RALF peptides in PT rupture and in the prevention of multiple PT arrival at the ovule.
XI. Diversity
Although first isolated from tobacco leaves (
Nicotiana tabacum) and synthesized from tomato genes (
Solanum lycopersicum) (10), RALF peptides are extensively studied in the model plant
Arabidopsis thaliana. Nonetheless, important research has been conducted in other species, providing evidence of functional conservation while also revealing novel roles for these peptides. For example, the production of edible fruits and the capacity to fix nitrogen are absent in Arabidopsis but involve RALFs in other species. These peptides are also proving to be good targets for plant breeding, as they regulate immune responses in many, if not all, plant species. The following findings highlight the versatility and relevance of this peptide family for plant development and survival, as well as their potential for enhancing traits of economic interest. A phylogenetic tree with all the RALFs cited in this review can be found in
Figure 13, aiming to help the reader establish comparisons among the peptides.
RALFs are present throughout land plants, and their canonical growth-inhibiting effect is apparently the rule. A Nicotiana attenuata NaRALF gene-silenced mutant (irNaRALF) showed longer primary roots (178), while overexpression mutants of the rice (Oryza sativa) OsRALF26 gene show smaller roots but, curiously, only in the seedling stage (179). The quinoa (Chenopodium quinoa) CqRALF15 interacts with CqFER and its treatment inhibits root growth (180). Likewise, treatment of Russian dandelion (Taraxacum koksaghyz) with TkRALFL1 inhibits root growth due to diminished cell expansion. Surprisingly, lower concentrations of the peptide caused stronger root inhibition than higher concentrations. Moreover, Tkralfl1 knockout plants showed roots with larger volumes that frequently developed a taproot morphology, pointing to a RALF influence on the organ's architecture (181). Treatment of sugarcane (Saccharum spp.) cell suspension with SacRALF1 inhibited microcalli growth, and the SacRALF1,2,3,4 genes are highly expressed during leaf expansion (182). In the opposite direction, tomato plants (S. lycopersicum) lacking SlRALF2 are smaller than wild-type plants, pointing to a novel role for a RALF peptide in growth induction (183).
RALFs from
A. thaliana are directly involved in root hair development (
Figure 6). The tracheophytes’ RHs and the bryophytes’ protonemata are both tip-growing cells, and the moss
Physcomitrium patens PpRALF1 and PpRALF2 have overlapping functions in promoting protonemal tip growth (184). Curiously, PpRALF1 shows a uniform distribution on the plasma membrane, resembling AtRALF22's cellular distribution, while PpRALF2 accumulates at the growing tip, similar to AtRALF1 (12,29,184). However, these peptide pairs do not seem to be closely related (
Figure 13). Nonetheless, the fact that they all promote tip-growth suggests a conserved function throughout hundreds of millions of years of evolution.
The irNaRALF line has shorter and often burst root hairs. This phenotype arises from an imbalance in pH dynamics at the tip of the mutant RHs, where slower oscillations with higher pH peaks take place. Unexpectedly, within days, wild-type plants acidified the medium while irNaRALF plants didn’t. This apparent acidification role of NaRALF is of particular interest, as it contrasts with the canonical alkalinization effect that gives the peptide family its name. In nature, N. attenuata plants are commonly found in basic pH soils. Strikingly, while wild-type and irNaRALF grew similarly in acidic soils, in basic soils the silenced mutant showed significantly less growth, suggesting that NaRALF may modulate the apoplast pH to increase plant fitness (178). Nevertheless, the alkalinization effect seen in most AtRALFs is also found in poplar. The PtdRALF1 and PtdRALF2 peptides, extracted from leaves, alkalize cell suspensions of the Populus tremula × P. tremuloides hybrid (185).
The rubber tree (Hevea brasiliensis) possesses lactiferous cells, whose cytoplasm is called latex, from which rubber is extracted (186). The HbRALF3,16,19,22 genes are expressed in these cells, and the corresponding peptides interact with HbFER1. The receptor was shown to positively regulate rubber biosynthesis, while HbRALF3 and HbRALF19 acidify the latex (187). The Russian dandelion (T. koksaghyz) roots also accumulate natural rubber, as well as inulin. Plants lacking TkRALFL1 produced higher rubber and inulin yields, although with lower content per unit dry weight. Curiously, TkRALFL5,6,10 are highly expressed in the latex (181).
RALF peptides are central to
A. thaliana fertilization (
Figure 12), and their role in reproduction extends to species in both eudicots and monocots. The tomato SlRALF6, present in pollen, was first identified as a potential interactor of a pollen-specific SlLRX protein, and its treatment has a negative effect on pollen tube growth, similar to that of AtRALF4 and AtRALF19 (41,188). The pear (
Pyrus bretschneideri) PbrRALF2 controls PT growth by binding to PbrCrRLK1L13, an AtBUPS homologue. To this end, PbrRALF2 interaction induces CrRLK1L13 phosphorylation and association with PbrMPK18, triggering ROS production (162,189).
The maize (Zea mays) ZmRALF1,2,3,5 genes are also expressed in pollen. Interestingly, ZmRALF2 and ZmRALF3 are secreted and accumulate in the cell wall, while ZmRALF1 and ZmRALF5 are only detected in cytoplasmic compartments. Consistently, plants mutants lacking ZmRALF2,3 show severe PT burst defects, while mutants lacking ZmRALF1,5 do not. Furthermore, ZmRALF2 and ZmRALF3 bind to ZmFERL1,4,7,9, to ZmLLG1,2, and to the maize LRX homologs POLLEN EXTENSION-LIKE 2 (ZmPEX2) (190).
Among the 41 rice (O. sativa) RALF genes, OsRALF17 and OsRALF19 have the highest expression in PTs. The two corresponding peptides have a partially redundant role in promoting pollen germination and PT growth at low concentrations (0.1-0.5 nM), but exhibit the opposite effect at higher doses (191). These two peptides also exert a dose-dependent inhibition of PT burst (192). For their function, OsRALF17 and OsRALF19 interact with MALE-GENE TRANSFER DEFECTIVE 2 (OsMTD2), an AtANX homologue, and with RUPTURED POLLEN TUBE (OsRUPO), an AtBUPSs homologue, inducing ROS production and receptor internalization (191,192). As seen for their Arabidopsis homologues, OsMTD2 and OsRUPO interact with each other (41,192). Notably, during PT growth, OsRUPO autophosphorylates and interacts with the HIGH-AFFINITY POTASSIUM 1 (OsHAK1) transporter, modulating pollen’s K⁺ uptake (193).
The oil-tea tree (Camellia oleifera) genome harbors 50 CoRALFs, with a subgroup being exclusively expressed in pollen. Among these is CoRALF50, whose expression reduction through synthetic antisense oligonucleotides (as-ODNs) results in longer pollen tubes. In contrast, treatment with the mature CoRALF50 peptide inhibits PT growth and reduces ROS levels, along with a decreased pectin and callose deposition. Furthermore, RNA-seq analysis revealed that the peptide presence induces genes involved in carbohydrate metabolism. The oil-tea tree exhibits self-incompatibility, and CoRALF50 expression differs between self- and cross-pollination. Remarkably, the as-ODN approach resulted in pollen with abnormal phenotypes resembling those of self-incompatible PTs (194).
A role for RALF peptides in fruit development was discovered in Solanum chacoense. ScRALF1,2,3,4,5 are intensely expressed during the tomato maturation process, and ScRALF3 interference lines have smaller fruits with markedly fewer seeds (195,196). ScRALF3 is an auxin-induced gene, expressed in the sporophytic tissue of the ovule, where it is up-regulated after fertilization (195). ScRALF3 is also transcribed during pollen development, in the anthers' tapetum and microspores (197). IrScRALF3 mutants have ovules with abnormal cellular organization and a high percentage of pollen with impaired maturation and viability. Therefore, the absence of ScRALF3 seems to disrupt sporophyte–gametophyte communication during both male and female gamete development, suggesting a central role for RALFs in S. chacoense fruit development (195,197).
Food loss due to plant diseases is a major concern for humanity. Research on plant-pathogen interactions in important crop species revealed conserved and novel roles for RALF peptides in plant immunity. The rice OsRALF26 transcription and translation are induced during Xanthomonas oryzae infection, and OsRALF26 overexpression lines show increased resistance to the bacteria. Consistently, OsRALF26 interacts with FER-LIKE RECEPTOR 1 (OsFLR1), and the peptide treatment triggers ROS burst, upregulates PR genes, and causes callose deposition in an OsFLR1-dependent manner (179). Moreover, the expression of OsRALFL7 and OsRALFL8 is strongly upregulated by treatments with fungi-derived chitin or bacteria-derived peptidoglycan, suggesting a role for these genes in the immune response (198).
Soybean (Glycine max) possesses 24 GmRALF (199), all of which are differentially regulated upon P. syringae pv. glycinea (Psg) and/or Phytophthora sojae infection (200). The majority of these genes are induced, while about one-third are repressed. Further analysis of PTI regulation in N. benthamiana leaves showed that transient expression of 21 GmRALFs directly affects the PTI response (200). Among these, GmRALF1 and GmRALF18 stand out for their ability to suppress immunity (200,201). Accordingly, Gmralf1 knockout mutants exhibit enhanced resistance to Psg and P. sojae infections, with minor growth penalty (200). Curiously, haplotype analysis identified signatures of selection for haplotypes of GmRALF1 and GmRALF18, suggesting their involvement in regulating agronomic traits (201).
The molecular mechanism underlying the impact of the two GmRALFs on immunity is being uncovered and involves the RALF/CrRLK1L/LLG complexes observed in Arabidopsis (20,36). GmRALF1 and GmRALF18 bind to GmLLG1 and GmLLG2, which, in turn, interact with the FER homologue LESION MIMIC MUTANT 1 (GmLMM1) (201). This receptor negatively regulates the PTI response by reducing flg22-mediated GmFLS2/BAK1 complex formation (202). Similarly, GmLLG1 and GmLLG2 redundantly impair the PTI response (201). The receptor/coreceptor role conservation extends to LLGs’ chaperone function, since, in the Gmllg1/llg2 double mutant, GmLMM1 shows reduced membrane localization (201), as seen in Arabidopsis (36).
The turnip mosaic virus (TuMV), an important member of the genus Potyvirus, infects hundreds of plant species. The TuMV genome encodes 11 functional proteins, like the SECOND 6-KDA (6K2) (203). Upon TuMV infection of Nicotiana benthamiana, NbRALF1 expression is induced, which provides resistance to the virus. At least part of the RALF-mediated host immunity relies on its upregulation of immune response genes, including JA- and SA-related genes (204). Strikingly, this NbRALF1-mediated resistance is dependent on the whole peptide’s intracellular localization (204), including its N-terminal, canonically processed to generate the secreted mature RALF (5,14). In agreement, the NbRALF1 N-terminal binds to FER’s intracellular domain, which, through its kinase activity, contributes to the host immunity response. Opposing this response, the viral 6K2 protein interacts with NbRALF1 in the cell’s interior and targets the peptide for degradation via the 26S proteasome. Silencing NbRALF1 leads to increased Tobamovirus capsici and Potexvirus ecspotati accumulation, indicating a broader antiviral role for the peptide (204). This remarkable discovery represents a milestone in RALF understanding, providing evidence of a previously undescribed intracellular role for the peptide, which remains to be demonstrated in other species such as Arabidopsis. Notably, NbRALF1 was also shown to negatively regulate PTI and resistance to Pst DC3000 ΔhopQ1, S. sclerotiorum, and P. capsici (200).
Both sweet orange (Citrus sinensis) and the related Rutaceae species Chinese box orange (Atalantia buxifolia) possess 13 RALF genes. Each of these can be paired with a corresponding RALF from the other species due to their high sequence identity, suggesting conserved functions, although in silico promoter analyses indicate that some of these pairs are regulated by different cis-elements. Many of these RALFs are differentially expressed under biotic stresses, and CsRALF11 induces the greater ROS production in sweet orange leaves. Interestingly, treatment with either CsRALF11 or AbRALF11 triggers a higher ROS burst in A. buxifolia than in C. sinensis leaves, suggesting that A. buxifolia is more sensitive to the peptide-induced immunity. This was further supported by the higher expression of the PTI marker genes FRK1, GST1, NPR2, and PDF1.2, as well as by MAPK phosphorylation, in Chinese box orange treated with these peptide (205).
The root-knot nematode (Meloidogyne incognita) produces MiRALFs, which are secreted as effectors of pathogenicity. MiRALF1 and MiRALF3 act in an AtFER-dependent manner to facilitate the infection of A. thaliana (9). MiRALF1 and AtRALF23 can also bind to the soybean GmLMM1, and mutants lacking this receptor are resistant to the nematode (206). Curiously, unlike MiRALF1 and AtRALF23, AtRALF1 and MiRALF3 cannot bind to GmLMM1, suggesting binding specificity (206). Moreover, the rice OsFLR1 is important for M. incognita pathogenicity (9). Together, these results suggest a conserved infection strategy employed by the root-knot nematode in Arabidopsis, soybean, and rice, involving the MiRALFs and the host's FER homologues.
Plant infection by Fusarium fungi involves RALF peptides. In soybean, F. oxysporum invasion induces the upregulation of five GmRALFs and the downregulation of nine. Of the repressed genes, the peptide products of GmRALF4, GmRALF10, and GmRALF24 are able to bind GmFER (207). The related species F. graminearum possesses a FgRALF expressed during wheat (Triticum aestivum) infection (155). FgRALF inhibits root growth and alkalizes the host's rhizosphere, which is known to affect fungal pathogenicity (155,208). FgRALF acts by binding to TaFER and suppressing the PTI response to promote fungal pathogenicity (155). F. oxysporum also possesses a RALF-like gene (FoRALF), expressed during infection (155,156). Treatment of tomato plants (S. lycopersicum) with FoRALF alkalizes the root medium, inhibits its growth, and negatively regulates plant immunity. Accordingly, fungal mutants lacking FoRALF had reduced virulence and elicited a strong host immune response (156). Thus, the Fusarium RALF-like peptides are used as effectors of pathogenicity by the fungus.
Lignin acts as a barrier against pathogens, but it also limits plant growth (209). Consistent with this tradeoff, the tomato SlRALF2 and SlFER negatively regulate lignin content and F. oxysporum resistance in roots. This receptor/ligand pair does so by diminishing the presence of SlMYB63 and DIRIGENT PROTEIN 19 (SlDIR19). The transcription factor SlMYB63 binds to the SlDIR19 promoter and induces its expression. In turn, SlDIR19 positively regulates lignin deposition and F. oxysporum resistance at the expense of root growth. Opposing this signaling, SlFER interacts with and phosphorylates SlMYB63, inducing its degradation, and SlRALF2 treatment enhances this response. Thus, SlRALF2 and SlFER orchestrate the growth/defense balance by modulating lignin levels (183).
SlRALF2 is also involved in resistance against late blight, caused by Phytophthora infestans, one of the most devastating tomato diseases. Overexpression of SlRALF2 enhanced plant resistance to the oomycete, upregulating SlPRs and positively regulating ROS levels by reducing POD and SOD activities while increasing CAT activity. Interestingly, transcriptome analysis showed induction of zeatin and carotenoid biosynthesis genes in the SlRALF2-overexpressing plants, as well as genes involved in MAPK signaling (210).
The role of RALF peptides in immunity extends to other species. The majority of the 61 BnRALFs from oilseed rape (Brassica napus) are differentially expressed in the presence of PAMPs or DAMPs. Of those, BnRALF10 is upregulated by infection with Sclerotinia sclerotiorum. In this response, the peptide interacts with BnFER, inducing ROS production and resistance to the pathogenic fungus. BnRALF10 treatment also leads to hundreds of differently expressed proteins, with an enrichment of those involved in stress responses (211). In contrast, FaRALF33L from strawberry (Fragaria × ananassa) shows the opposite effect. FaRALF33L is upregulated in the fruit’s red-ripe stage during infection by the fungus Colletotrichum acutatum. The silencing of FaRALF33L leads to a delay in fungal colonization in red fruits, while overexpressing the gene results in more disease symptoms in white fruits, suggesting that FaRALF33L plays a negative role in the response against C. acutatum (212).
Although many plant-microorganism interactions are detrimental to the host, some are beneficial. An important example is the symbiosis between rhizobia and leguminous plants, which results in nitrogen fixation (213). In the common bean (Phaseolus vulgaris), PvRALF1, PvRALF6, and PvFER1 are upregulated during nodule organogenesis of roots inoculated with Rhizobium tropici. Experiments with silencing and overexpression lines of each of these genes point to a positive effect of their proteins on nodulation in the absence of nitrate, but a negative effect in its presence. Moreover, PvRALF1 and PvRALF6 interact with PvFER1, suggesting that these ligand/receptor pairs regulate nodule formation in P. vulgaris (214). Similarly, MtRALF1 from Medicago truncatula is upregulated by bacterial Nod factors, which mediate nodulation. Upon inoculation with Sinorhizobium meliloti, plants overexpressing MtRALFL1 showed a reduction in the number of nodules and an increase in nodules with abnormal development when compared to wild-type (215). Furthermore, upon inoculation with the nitrogen-fixing bacterium Bradyrhizobium japonicum, 16 GmRALFs were differentially expressed (199). Thus, RALF peptides are involved in regulating nodulation in different species of the subfamily Faboideae.
Several GmRALFs, especially GmRALF22, are upregulated under low phosphate (LP) conditions. The promoters of these genes possess known binding sites for PHR1, and GmPHR1 is able to bind the GmRALF22 promoter and induce its expression in vivo. When GmRALF22 is delivered to the roots via Bacillus subtilis, it enhances the vegetative growth of soybean plants under LP, increasing the soluble Pi content in both roots and leaves. GmRALF22 also induces the transcription of PI TRANSPORTER (GmPT2,7,11). Furthermore, RNA sequencing of plants with and without GmRALF22 delivery shows DEGs involved in JA and SA signaling (216). It is noteworthy that the phosphate starvation response in Arabidopsis involves the PHR1-induced expression of RALFs, which modulate immunity to enrich the colonization of bacteria that facilitate phosphate uptake (153). Thus, GmRALF22-triggered response might also alter soybean microbiome.
The moss P. patens PpRALF2 and PpRALF3 are involved in stress responses. Mutants lacking PpRALF2 or PpRALF3 are more resistant to the bacteria Pectobacterium carotovorum and to the fungi Fusarium solani infections. Both mutants are also more tolerant to salt and oxidative stresses. Curiously, their protoplasts showed increased cell wall regeneration compared to wild-type, which may explain part of the observed resistances (217). The involvement of RALFs in bryophyte stress responses suggests that these regulatory mechanisms may have arisen hundreds of millions of years ago.
The role of RALFs in salt stress, well-investigated in
A. thaliana (
Figure 9), has also been shown in a couple of other tracheophytes. The quinoa
CqRALF15 is induced by NaCl, and, similar to its paralog AtRALF22 (133), Arabidopsis plants overexpressing
CqRALF15 are hypersensitive to salt stress (180). Furthermore, the allotetraploid Mexican cotton (
Gossypium hirsutum) possesses an impressive number of 104
GhRALFs genes. Of these,
GhRALF14,
GhRALF27 and
GhRALF41 are expressed after drought and salt stress. Consistently, silencing each of these genes resulted in enhanced salt tolerance in seedlings (218).
The markedly high number of GhRALFs is a consequence of whole-genome duplications (WGD) (218). As a result, gene clusters were formed, such as GhRALF4L and GhRALF30L, composed of 5 and 23 RALFs, respectively, with the latter further expanding due to tandem duplication events. Although these genes were named after their protein homology to AtRALF4 and AtRALF30, members of the GhRALF30L cluster are remarkably shorter and, with mature peptides containing only 19 amino acids. Genes from both clusters are mainly expressed in anthers and pollen grains, and under high-temperature (HT) stress, GhRALF4L and GhRALF30L are downregulated, but less so in an HT-tolerant line. Through CRISPR-generated mutants with cumulative editing of genes within each cluster, it was shown that GhRALF4L and GhRALF30L promote anther dehiscence and pollen viability under HT stress in a dose-dependent manner. Plants with simultaneous mutations in both gene clusters exhibit more severe male sterility under HT stress, which extends to microspore development. Moreover, G. hirsutum possesses 80 CrRLK1L genes, and mutants of GhFERs, GhANXs, GhBUPSs, and GhCAPs show severe male sterility phenotypes under HT. Both GhRALF30L and GhRALF4L physically interact with the ectodomains of these receptors. Under HT stress, GhRALF4L induces GhANXD1 and GhBUPSA2 condensation at the plasma membrane, while GhRALF30L promotes the condensation of GhFERA1 and GhCAPA1. The analysis of ROS accumulation and expression of ROS-scavenging genes in GhRALF and GhCrRLK1L mutants suggests that the GhRALF4L/30L-mediated pathways are essential to prevent oxidative stress under HT. A similar pattern was observed for the expression of heat shock protein (HSP) genes (219). Therefore, under high temperature stress, GhRALF4L,30L peptides modulate GhCrRLK1L membrane organization and signaling to promote ROS scavenging and HSP responses, providing thermotolerance to male reproductive organs in cotton.
The first direct involvement of RALF peptides in drought tolerance was described in rice and involves the FER homologue MALECTIN/MALECTIN-LIKE DOMAIN-CONTAINING RECEPTOR-LIKE KINASE 63 (OsMRLK63), which is mainly expressed in leaf blades. OsMRLK63 transcription is induced by different stresses, and rice plants overexpressing OsMRLK63 are more tolerant to drought. At least part of this tolerance is achieved by OsMRLK63’s direct phosphorylation of OsRbohA, inducing ROS production (220). Accordingly, OsRbohA was previously shown to positively regulate rice drought tolerance (221). OsRALF45 and OsRALF46 are upregulated by drought and ROS, and the encoded peptides are secreted into the apoplast, where they bind OsMRLK63 to induce ROS production in an OsRbohA-dependent manner. Curiously, combined application of OsRALF45 and OsRALF46 results in improved drought tolerance compared to treatments with each peptide alone, suggesting a synergistic action (220).
The Tartary buckwheat (Fagopyrum tataricum) possesses nine FtRALFs, the majority of which are differentially expressed under NaCl, cadmium (Cd), antimony (Sb), cold, and heat stresses. Among these, FtRALF6 exhibited the most robust transcriptional response, being induced in leaves and roots under all cited stresses. Pre-treatment with FtRALF6 mitigated root growth inhibition caused by salt, heavy metal, or temperature stress, without affecting growth of Tartary buckwheat under control conditions. Accordingly, stress phenotypes such as impaired biosynthesis of photosynthetic pigments, reduced dehydrogenase activity, and oxidative stress were alleviated by the presence of FtRALF6 (222). Thus, this peptide functions as a broad-spectrum abiotic stress resistance gene.
A genome-wide investigation of peanut (Arachis hypogaea) revealed the presence of 24 AhRALF genes. Among these, seven were downregulated under Al stress, including AhRALF1. The encoded peptide is secreted, and its treatment inhibits Arabidopsis primary root growth. Further characterization of AhRALF1 through VIGS interference resulted in smaller peanut plants with higher basal activities of enzymes related to oxidative stress compared with the wild type. In contrast, under Al stress, the SOD, POD, and CAT activities were lower in interference lines than in wild-type plants, as was the MDA content. Together, these results suggest an involvement of AhRALF1, and potentially other AhRALFs, in the Al stress response (223).
Figure 13.
Phylogenetic tree of all RALFs cited in this review.
Figure 13.
Phylogenetic tree of all RALFs cited in this review.
The predicted mature RALFs sequences were aligned using MUSCLE. The phylogenetic tree was generated using the UPGMA method with 1000 bootstrap replicates in MEGA12 software. The figure was generated with the iTOL online tool. The size of the grey circles indicates relative bootstrap values.
References
- Kende, H.; Zeevaart, J. The Five “Classical” Plant Hormones. Plant Cell. 1997, 9, 1197–1210. [Google Scholar] [CrossRef]
- Pearce, G.; Strydom, D.; Johnson, S.; Ryan, C.A. A Polypeptide from Tomato Leaves Induces Wound-Inducible Proteinase Inhibitor Proteins. Science 1991, 253, 895–897. [Google Scholar] [CrossRef]
- Gancheva, M.S.; Malovichko, Y.V.; Poliushkevich, L.O.; Dodueva, I.E.; Lutova, L.A. Plant Peptide Hormones. Russ. J. Plant Physiol. 2019, 66, 171–189. [Google Scholar] [CrossRef]
- Campbell, L.; Turner, S.R. A Comprehensive Analysis of RALF Proteins in Green Plants Suggests There Are Two Distinct Functional Groups. Front. Plant Sci. 2017, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Abarca, A.; Franck, C.M.; Zipfel, C. Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 2021, 187, 996–1010. [Google Scholar] [CrossRef]
- Blackburn, M.R.; Haruta, M.; Moura, D.S. Twenty Years of Progress in Physiological and Biochemical Investigation of RALF Peptides. Plant Physiol. 2020, 182, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Shi, F. Evolution of the RALF Gene Family in Plants: Gene Duplication and Selection Patterns. Evol. Bioinform. 2012, 8, 271–292. [Google Scholar] [CrossRef]
- Thynne, E.; Saur, I.M.L.; Simbaqueba, J.; Ogilvie, H.A.; Gonzalez-Cendales, Y.; Mead, O.; Taranto, A.; Catanzariti, A.; McDonald, M.C.; Schwessinger, B.; et al. Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 2016, 18, 811–824. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, H.; Zhu, S.; Xing, J.; Li, X.; Zhu, Z.; Zheng, J.; Wang, L.; Wang, B.; Chen, J.; et al. Nematode-Encoded RALF Peptide Mimics Facilitate Parasitism of Plants through the FERONIA Receptor Kinase. Mol. Plant 2020, 13, 1434–1454. [Google Scholar] [CrossRef]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. 2001, 98, 12843–12847. [Google Scholar] [CrossRef]
- Wang, L.; Yang, T.; Wang, B.; Lin, Q.; Zhu, S.; Li, C.; Ma, Y.; Tang, J.; Xing, J.; Li, X.; et al. RALF1-FERONIA complex affects splicing dynamics to modulate stress responses and growth in plants. Sci. Adv. 2020, 6, eaaz1622. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Estévez, J.M.; Liao, H.; Zhu, Y.; Yang, T.; Li, C.; Wang, Y.; Li, L.; Liu, X.; Pacheco, J.M.; et al. The RALF1–FERONIA Complex Phosphorylates eIF4E1 to Promote Protein Synthesis and Polar Root Hair Growth. Mol. Plant 2020, 13, 698–716. [Google Scholar] [CrossRef]
- Chen, J.; Xu, F.; Qiang, X.; Liu, H.; Wang, L.; Jiang, L.; Li, C.; Wang, B.; Luan, S.; Wu, D.; et al. Regulated cleavage and translocation of FERONIA control immunity in Arabidopsis roots. Nat. Plants 2024, 10, 1761–1774. [Google Scholar] [CrossRef]
- Matos, J.L.; Fiori, C.S.; Silva-Filho, M.C.; Moura, D.S. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 2008, 582, 3343–3347. [Google Scholar] [CrossRef]
- Srivastava, R.; Liu, J.; Guo, H.; Yin, Y.; Howell, S.H. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 2009, 59, 930–939. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.-C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.-K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. 2018, 115, 13123–13128. [Google Scholar] [CrossRef]
- Xiao, Y.; Stegmann, M.; Han, Z.; DeFalco, T.A.; Parys, K.; Xu, L.; Belkhadir, Y.; Zipfel, C.; Chai, J. Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 2019, 572, 270–274. [Google Scholar] [CrossRef]
- Dressano, K.; O Ceciliato, P.H.; Silva, A.L.; Guerrero-Abad, J.C.; Bergonci, T.; Ortiz-Morea, F.A.; Bürger, M.; Silva-Filho, M.C.; Moura, D.S. BAK1 is involved in AtRALF1-induced inhibition of root cell expansion. PLOS Genet. 2017, 13, e1007053. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Zhao, Y.; Liu, M.-C.; Zhou, L.-Z.; Wang, L.; Zhong, S.; Hou, S.; Jiang, J.; Liu, T.; Huang, Q.; et al. LLG2/3 Are Co-receptors in BUPS/ANX-RALF Signaling to Regulate Arabidopsis Pollen Tube Integrity. Curr. Biol. 2019, 29, 3256–3265.e5. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Moussu, S.; Broyart, C.; Santos-Fernandez, G.; Augustin, S.; Wehrle, S.; Grossniklaus, U.; Santiago, J. Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc. Natl. Acad. Sci. 2020, 117, 7494–7503. [Google Scholar] [CrossRef]
- Gao, Q.; Wang, C.; Xi, Y.; Shao, Q.; Li, L.; Luan, S. A receptor–channel trio conducts Ca2+ signalling for pollen tube reception. Nature 2022, 607, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Lan, Z.; Song, Z.; Wang, Z.; Li, L.; Liu, Y.; Zhi, S.; Wang, R.; Wang, J.; Li, Q.; Bleckmann, A.; et al. Antagonistic RALF peptides control an intergeneric hybridization barrier on Brassicaceae stigmas. Cell 2023, 186, 4773–4787.e12. [Google Scholar] [CrossRef]
- Zhong, S.; Li, L.; Wang, Z.; Ge, Z.; Li, Q.; Bleckmann, A.; Wang, J.; Song, Z.; Shi, Y.; Liu, T.; et al. RALF peptide signaling controls the polytubey block in Arabidopsis. Science 2022, 375, 290–296. [Google Scholar] [CrossRef]
- Bhalla, H.; Sudarsanam, K.; Srivastava, A.; Sankaranarayanan, S. Structural insights into the recognition of RALF peptides by FERONIA receptor kinase during Brassicaceae pollination. Plant Mol. Biol. 2025, 115, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-C.J.; Yeh, F.-L.J.; Yvon, R.; Simpson, K.; Jordan, S.; Chambers, J.; Wu, H.-M.; Cheung, A.Y. Extracellular pectin-RALF phase separation mediates FERONIA global signaling function. Cell 2023, 187, 312–330.e22. [Google Scholar] [CrossRef]
- Moussu, S.; Lee, H.K.; Haas, K.T.; Broyart, C.; Rathgeb, U.; De Bellis, D.; Levasseur, T.; Schoenaers, S.; Fernandez, G.S.; Grossniklaus, U.; et al. Plant cell wall patterning and expansion mediated by protein-peptide-polysaccharide interaction. Science 2023, 382, 719–725. [Google Scholar] [CrossRef]
- Schoenaers, S.; Lee, H.K.; Gonneau, M.; Faucher, E.; Levasseur, T.; Akary, E.; Claeijs, N.; Moussu, S.; Broyart, C.; Balcerowicz, D.; et al. Rapid alkalinization factor 22 has a structural and signalling role in root hair cell wall assembly. Nat. Plants 2024, 10, 494–511. [Google Scholar] [CrossRef]
- Lindner, H.; Müller, L.M.; Boisson-Dernier, A.; Grossniklaus, U. CrRLK1L receptor-like kinases: not just another brick in the wall. Current Opinion in Plant Biology 2012, 15, 659–669. [Google Scholar] [CrossRef]
- Gonneau, M.; Desprez, T.; Martin, M.; Doblas, V.G.; Bacete, L.; Miart, F.; Sormani, R.; Hématy, K.; Renou, J.; Landrein, B.; et al. Receptor Kinase THESEUS1 Is a Rapid Alkalinization Factor 34 Receptor in Arabidopsis. Curr. Biol. 2018, 28, 2452–2458.e4. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, L.; Xiao, Y.; Vyshedsky, D.; Peng, C.; Sun, X.; Liu, Z.; Cheng, L.; Zhang, H.; Han, Z.; et al. Pollen PCP-B peptides unlock a stigma peptide–receptor kinase gating mechanism for pollination. Science 2021, 372, 171–175. [Google Scholar] [CrossRef]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.-C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca2+ Signaling. Curr. Biol. 2018, 28, 666–675.e5. [Google Scholar] [CrossRef]
- Kong, Y.; Chen, J.; Jiang, L.; Chen, H.; Shen, Y.; Wang, L.; Yan, Y.; Zhou, H.; Zheng, H.; Yu, F.; et al. Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation. Plant Commun. 2023, 4, 100559. [Google Scholar] [CrossRef]
- Feng, H.; Liu, C.; Fu, R.; Zhang, M.; Li, H.; Shen, L.; Wei, Q.; Sun, X.; Xu, L.; Ni, B.; et al. LORELEI-LIKE GPI-ANCHORED PROTEINS 2/3 Regulate Pollen Tube Growth as Chaperones and Coreceptors for ANXUR/BUPS Receptor Kinases in Arabidopsis. Mol. Plant 2019, 12, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yeh, F.-L.; Cheung, A.Y.; Duan, Q.; Kita, D.; Liu, M.-C.; Maman, J.; Luu, E.J.; Wu, B.W.; Gates, L.; et al. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 2015, 4. [Google Scholar] [CrossRef]
- Zhou, K. Glycosylphosphatidylinositol-Anchored Proteins in Arabidopsis and One of Their Common Roles in Signaling Transduction. Front. Plant Sci. 2019, 10, 1022. [Google Scholar] [CrossRef] [PubMed]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P.; et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef]
- Dünser, K.; Gupta, S.; Herger, A.; I Feraru, M.; Ringli, C.; Kleine-Vehn, J. Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Herger, A.; Gupta, S.; Kadler, G.; Franck, C.M.; Boisson-Dernier, A.; Ringli, C. Overlapping functions and protein-protein interactions of LRR-extensins in Arabidopsis. PLOS Genet. 2020, 16, e1008847. [Google Scholar] [CrossRef]
- Ge, Z.; Bergonci, T.; Zhao, Y.; Zou, Y.; Du, S.; Liu, M.-C.; Luo, X.; Ruan, H.; García-Valencia, L.E.; Zhong, S.; et al. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 2017, 358, 1596–1600. [Google Scholar] [CrossRef]
- Campos, W.F.; Dressano, K.; Ceciliato, P.H.; Guerrero-Abad, J.C.; Silva, A.L.; Fiori, C.S.; Canto, A.M.D.; Bergonci, T.; Claus, L.A.; Silva-Filho, M.C.; et al. Arabidopsis thaliana rapid alkalinization factor 1–mediated root growth inhibition is dependent on calmodulin-like protein 38. J. Biol. Chem. 2018, 293, 2159–2171. [Google Scholar] [CrossRef]
- Li, L.; Chen, H.; Alotaibi, S.S.; Pěnčík, A.; Adamowski, M.; Novák, O.; Friml, J. RALF1 peptide triggers biphasic root growth inhibition upstream of auxin biosynthesis. Proc. Natl. Acad. Sci. 2022, 119. [Google Scholar] [CrossRef]
- Liu, Z.; Chu, X.; Ren, W.; Cheng, L.; Liu, C.; Wang, C.; Gao, S.; Dai, S.; Li, C. PCP-B peptides and CrRLK1L receptor kinases control pollination via pH gating of aquaporins in Arabidopsis. Dev. Cell 2025, 60, 1336–1347.e5. [Google Scholar] [CrossRef]
- Gjetting, S.K.; Mahmood, K.; Shabala, L.; Kristensen, A.; Shabala, S.; Palmgren, M.; et al. Evidence for multiple receptors mediating RALF-triggered Ca2+ signaling and proton pump inhibition. The Plant Journal 2020, 104, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Hager, A.; Menzel, H.; Krauss, A. Versuche und Hypothese zur Prim rwirkung des Auxins beim Streckungswachstum. Planta 1971, 100, 47–75. [Google Scholar] [CrossRef]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef] [PubMed]
- Fuglsang, A.T.; Kristensen, A.; Cuin, T.A.; Schulze, W.X.; Persson, J.; Thuesen, K.H.; Ytting, C.K.; Oehlenschlæger, C.B.; Mahmood, K.; Sondergaard, T.E.; et al. Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J. 2014, 80, 951–964. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef] [PubMed]
- Day, I.S.; Reddy, V.S.; Ali, G.S.; Reddy, A. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.-G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef]
- Haruta, M.; Monshausen, G.; Gilroy, S.; Sussman, M.R. A Cytoplasmic Ca2+ Functional Assay for Identifying and Purifying Endogenous Cell Signaling Peptides in Arabidopsis Seedlings: Identification of AtRALF1 Peptide. Biochemistry 2008, 47, 6311–6321. [Google Scholar] [CrossRef] [PubMed]
- Frederick, R.O.; Haruta, M.; Tonelli, M.; Lee, W.; Cornilescu, G.; Cornilescu, C.C.; Sussman, M.R.; Markley, J.L. Function and solution structure of the Arabidopsis thaliana RALF8 peptide. Protein Sci. 2019, 28, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Wang, C.; Xi, Y.; Shao, Q.; Hou, C.; Li, L.; Luan, S. RALF signaling pathway activates MLO calcium channels to maintain pollen tube integrity. Cell Res. 2023, 33, 71–79. [Google Scholar] [CrossRef]
- Morato Do Canto, A.; Ceciliato, P.H.O.; Ribeiro, B.; Ortiz Morea, F.A.; Franco Garcia, A.A.; Silva-Filho, M.C.; et al. Biological activity of nine recombinant AtRALF peptides: Implications for their perception and function in Arabidopsis. Plant Physiology and Biochemistry 2014, 75, 45–54. [Google Scholar] [CrossRef]
- Yu, Y.; Assmann, S.M. Inter-relationships between the heterotrimeric Gβ subunit AGB1, the receptor-like kinase FERONIA, and RALF1 in salinity response. Plant Cell & Environment 2018, 41, 2475–2489. [Google Scholar]
- He, Y.; Chen, S.; Chen, X.; Xu, Y.; Liang, Y.; Cai, X. RALF22 promotes plant immunity and amplifies the Pep3 immune signal. J. Integr. Plant Biol. 2023, 65, 2519–2534. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhao, L.; Qi, F.; Htwe, N.M.P.S.; Li, Q.; Zhang, D.; Lin, F.; Shang-Guan, K.; Liang, Y. The receptor-like cytoplasmic kinase RIPK regulates broad-spectrum ROS signaling in multiple layers of plant immune system. Mol. Plant 2021, 14, 1652–1667. [Google Scholar] [CrossRef]
- Berken, A.; Thomas, C.; Wittinghofer, A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 2005, 436, 1176–1180. [Google Scholar] [CrossRef]
- Smokvarska, M.; Bayle, V.; Maneta-Peyret, L.; Fouillen, L.; Poitout, A.; Dongois, A.; Fiche, J.-B.; Gronnier, J.; Garcia, J.; Höfte, H.; et al. The receptor kinase FERONIA regulates phosphatidylserine localization at the cell surface to modulate ROP signaling. Sci. Adv. 2023, 9. [Google Scholar] [CrossRef]
- Song, Y.; Wilson, A.J.; Zhang, X.-C.; Thoms, D.; Sohrabi, R.; Song, S.; Geissmann, Q.; Liu, Y.; Walgren, L.; He, S.Y.; et al. FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat. Plants 2021, 7, 644–654. [Google Scholar] [CrossRef]
- Bergonci, T.; Ribeiro, B.; Ceciliato, P.H.; Guerrero-Abad, J.C.; Silva-Filho, M.C.; Moura, D.S. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 2014, 65, 2219–2230. [Google Scholar] [CrossRef]
- Friml, J. Auxin transport — shaping the plant. Current Opinion in Plant Biology 2003, 6, 7–12. [Google Scholar] [CrossRef]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis Root Development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis: A Simple Two-Step Pathway Converts Tryptophan to Indole-3-Acetic Acid in Plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Li, X.; Chen, J.; Chen, W.; Li, B.; Li, C.; Wang, L.; Li, J.; Zhao, X.; Lin, J.; et al. Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. 2016, 113, E8326–E8334. [Google Scholar] [CrossRef] [PubMed]
- Barbez, E.; Dünser, K.; Gaidora, A.; Lendl, T.; Busch, W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2017, 114, E4884–E4893. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Zhang, Z.; Liu, Y.; Tao, L.; Liu, H. FERONIA regulates auxin-mediated lateral root development and primary root gravitropism. FEBS Lett. 2018, 593, 97–106. [Google Scholar] [CrossRef]
- Li, E.; Wang, G.; Zhang, Y.; Kong, Z.; Li, S. FERONIA mediates root nutating growth. Plant J. 2020, 104, 1105–1116. [Google Scholar] [CrossRef]
- Li, C.; Chen, J.; Li, X.; Zhang, X.; Liu, Y.; Zhu, S.; et al. FERONIA is involved in phototropin 1 -mediated blue light phototropic growth in Arabidopsis. JIPB 2022, 64, 1901–1915. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, R.; Cui, Y.; Chen, W.; Li, B.; Zhang, X.; Bu, Y.; Cao, Y.; Xing, J.; Jewaria, P.K.; et al. The RALF1-FERONIA interaction modulates endocytosis to mediate control of root growth in Arabidopsis. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146. [Google Scholar] [CrossRef]
- Kim, T.-W.; Wang, Z.-Y. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu. Rev. Plant Biol. 2010, 61, 681–704. [Google Scholar] [CrossRef]
- Chaudhary, A.; Hsiao, Y.-C.; Yeh, F.-L.J.; Župunski, M.; Zhang, H.; Aizezi, Y.; Malkovskiy, A.; Grossmann, G.; Wu, H.-M.; Cheung, A.Y.; et al. FERONIA signaling maintains cell wall integrity during brassinosteroid-induced cell expansion in Arabidopsis. Mol. Plant 2025, 18, 603–618. [Google Scholar] [CrossRef]
- Sun, Y.; Han, Z.; Tang, J.; Hu, Z.; Chai, C.; Zhou, B.; Chai, J. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013, 23, 1326–1329. [Google Scholar] [CrossRef]
- Park, C.H.; Kim, T.-W.; Son, S.-H.; Hwang, J.-Y.; Lee, S.C.; Chang, S.C.; Kim, S.-H.; Kim, S.W.; Kim, S.-K. Brassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana. Phytochemistry 2010, 71, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Bergonci, T.; Silva-Filho, M.C.; Moura, D.S. Antagonistic relationship between AtRALF1and brassinosteroid regulates cellexpansion-related genes. Plant Signal. Behav. 2014, 9, e976146–e976146. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Structure and growth of plant cell walls. Nat. Rev. Mol. Cell Biol. 2023, 25, 340–358. [Google Scholar] [CrossRef]
- Lin, W.; Tang, W.; Pan, X.; Huang, A.; Gao, X.; Anderson, C.T.; Yang, Z. Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling. Curr. Biol. 2022, 32, 497–507.e4. [Google Scholar] [CrossRef] [PubMed]
- Biermann, D.; von Arx, M.; Munzert-Eberlein, K.S.; Xhelilaj, K.; Séré, D.; Stegmann, M.; Vert, G.; Wolf, S.; Engelsdorf, T.; Zipfel, C.; et al. A RALF-brassinosteroid signaling circuit regulates Arabidopsis hypocotyl cell shape. Curr. Biol. 2025, 35, 5002–5017.e5. [Google Scholar] [CrossRef] [PubMed]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, X.; Qiang, X.; Li, X.; Li, X.; Zhu, S.; et al. EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol. 2018, 16, e2006340. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. New Modes of Translational Control in Development, Behavior, and Disease. Mol. Cell 2007, 28, 721–729. [Google Scholar] [CrossRef]
- Zhu, S.; Li, Y.; Wu, Y.; Shen, Y.; Wang, Y.; Yan, Y.; Chen, W.; Fu, Q.; Wang, Y.; Yu, X.; et al. The FERONIA-YUELAO module participates in translational control by modulating the abundance of tRNA fragments in Arabidopsis. Dev. Cell 2023, 58, 2930–2946.e9. [Google Scholar] [CrossRef]
- Lalande, S.; Merret, R.; Salinas-Giegé, T.; Drouard, L. Arabidopsis tRNA-derived fragments as potential modulators of translation. RNA Biol. 2020, 17, 1137–1148. [Google Scholar] [CrossRef]
- Stegmann, M. EBP1: A crucial growth regulator downstream of receptor kinases across kingdoms. PLOS Biol. 2018, 16, e3000056. [Google Scholar] [CrossRef]
- Staiger, D.; Zecca, L.; Kirk, D.A.W.; Apel, K.; Eckstein, L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003, 33, 361–371. [Google Scholar] [CrossRef]
- Steffen, A.; Elgner, M.; Staiger, D. Regulation of Flowering Time by the RNA-Binding Proteins AtGRP7 and AtGRP8. Plant Cell Physiol. 2019, 60, 2040–2050. [Google Scholar] [CrossRef]
- Clark, G.B.; Morgan, R.O.; Fernandez, M.-P.; Salmi, M.L.; Roux, S.J. Breakthroughs spotlighting roles for extracellular nucleotides and apyrases in stress responses and growth and development. Plant Sci. 2014, 225, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Guérin, A.; Herger, A.; Hou, X.; Schaufelberger, M.; Roulard, R.; Diet, A.; Roffler, S.; Lefebvre, V.; Wicker, T.; et al. Growth-inhibiting effects of the unconventional plant APYRASE 7 of Arabidopsis thaliana influences the LRX/RALF/FER growth regulatory module. PLOS Genet. 2024, 20, e1011087. [Google Scholar] [CrossRef] [PubMed]
- Rößling, A.K.; Dünser, K.; Liu, C.; Lauw, S.; Rodriguez-Franco, M.; Kalmbach, L.; et al. Pectin methylesterase activity is required for RALF1 peptide signalling output [Internet]. 2024 [cited 2024 Aug 11]. Available from: https://elifesciences.org/reviewed-preprints/96943v1.
- Manmohit Kalia, P.K. Pectin Methylesterases: A Review. J Bioprocess Biotech [Internet]. 2015 [cited 2024 Aug 11];05(05). Available from: https://www.omicsonline.org/open-access/pectin-methylesterases-a-review-2155-9821-1000227.php?aid=52733.
- Xu, F.; Chen, J.; Li, Y.; Ouyang, S.; Yu, M.; Wang, Y.; Fang, X.; He, K.; Yu, F. The soil emergence-related transcription factor PIF3 controls root penetration by interacting with the receptor kinase FER. Dev. Cell 2024, 59, 434–447.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Shi, H.; Xue, C.; Wei, N.; Guo, H.; Deng, X.W. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. 2014, 111, 3913–3920. [Google Scholar] [CrossRef]
- Fang, X.; Liu, B.; Shao, Q.; Huang, X.; Li, J.; Luan, S.; He, K. AtPiezo Plays an Important Role in Root Cap Mechanotransduction. Int. J. Mol. Sci. 2021, 22, 467. [Google Scholar] [CrossRef]
- Shen, Y.; Xie, Q.; Wang, T.; Wang, X.; Xu, F.; Yan, Z.; Li, X.; Ouyang, S.; Chen, J.; Wang, Y.; et al. RALF33–FERONIA signaling orchestrates postwounding root-tip regeneration via TPR4–ERF115 dynamics. Plant Cell 2025, 37. [Google Scholar] [CrossRef]
- Canher, B.; Lanssens, F.; Zhang, A.; Bisht, A.; Mazumdar, S.; Heyman, J.; Wolf, S.; Melnyk, C.W.; De Veylder, L. The regeneration factors ERF114 and ERF115 regulate auxin-mediated lateral root development in response to mechanical cues. Mol. Plant 2022, 15, 1543–1557. [Google Scholar] [CrossRef]
- Heyman, J.; Cools, T.; Canher, B.; Shavialenka, S.; Traas, J.; Vercauteren, I.; Van den Daele, H.; Persiau, G.; De Jaeger, G.; Sugimoto, K.; et al. The heterodimeric transcription factor complex ERF115–PAT1 grants regeneration competence. Nat. Plants 2016, 2, 16165. [Google Scholar] [CrossRef]
- Murphy, E.; Vu, L.D.; Broeck, L.V.D.; Lin, Z.; Ramakrishna, P.; van de Cotte, B.; Gaudinier, A.; Goh, T.; Slane, D.; Beeckman, T.; et al. RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. J. Exp. Bot. 2016, 67, 4863–4875. [Google Scholar] [CrossRef]
- Grierson, C.; Nielsen, E.; Ketelaarc, T.; Schiefelbein, J. Root Hairs. The Arabidopsis Book 2014, 12, e0172. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of Genes Involved in the Response of Arabidopsis to Simultaneous Biotic and Abiotic Stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Hocq, L.; Pelloux, J.; Lefebvre, V. Connecting Homogalacturonan-Type Pectin Remodeling to Acid Growth. Trends Plant Sci. 2017, 22, 20–29. [Google Scholar] [CrossRef]
- Morcillo, R.J.L.; Leal-López, J.; Férez-Gómez, A.; López-Serrano, L.; Baroja-Fernández, E.; Gámez-Arcas, S.; Tortosa, G.; E López, L.; Estevez, J.M.; Doblas, V.G.; et al. RAPID ALKALINIZATION FACTOR 22 is a key modulator of the root hair growth responses to fungal ethylene emissions in Arabidopsis. Plant Physiol. 2024, 196, 2890–2904. [Google Scholar] [CrossRef]
- Molendijk, A.J.; Bischoff, F.; Rajendrakumar, C.S.V.; Friml, J.; Braun, M.; Gilroy, S.; Palme, K. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 2001, 20, 2779–2788. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. 2010, 107, 17821–17826. [Google Scholar] [CrossRef]
- Datta, S.; Prescott, H.; Dolan, L. Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat. Plants 2015, 1, 15138. [Google Scholar] [CrossRef]
- Yi, K.; Menand, B.; Bell, E.; Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010, 42, 264–267. [Google Scholar] [CrossRef]
- Masucci, J.D.; Schiefelbein, J.W. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996, 8, 1505–1517. [Google Scholar]
- Huang, G.; Li, E.; Ge, F.; Li, S.; Wang, Q.; Zhang, C.; Zhang, Y. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol. 2013, 200, 1089–1101. [Google Scholar] [CrossRef]
- Franck, C.M.; Westermann, J.; Bürssner, S.; Lentz, R.; Lituiev, D.S.; Boisson-Dernier, A. The Protein Phosphatases ATUNIS1 and ATUNIS2 Regulate Cell Wall Integrity in Tip-Growing Cells. Plant Cell 2018, 30, 1906–1923. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Franck, C.M.; Lituiev, D.S.; Grossniklaus, U. Receptor-like cytoplasmic kinase MARIS functions downstream of Cr RLK1L-dependent signaling during tip growth. Proc Natl Acad Sci USA 2015, 112, 12211–12216. [Google Scholar] [CrossRef]
- Donaldson, J.; Poratshliom, N.; Cohen, L. Clathrin-independent endocytosis: A unique platform for cell signaling and PM remodeling. Cell. Signal. 2009, 21, 1–6. [Google Scholar] [CrossRef]
- Borner, G.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; MacAskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of Detergent-Resistant Membranes in Arabidopsis. Evidence for Plasma Membrane Lipid Rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, H.; Xu, F.; Yu, M.; Coego, A.; Rodriguez, L.; Lu, Y.; Xie, Q.; Fu, Q.; Chen, J.; et al. CAR modulates plasma membrane nano-organization and immune signaling downstream of RALF1-FERONIA signaling pathway. New Phytol. 2023, 237, 2148–2162. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Holehouse, A.S.; Strader, L.C. Biological Phase Separation and Biomolecular Condensates in Plants. Annu. Rev. Plant Biol. 2021, 72, 17–46. [Google Scholar] [CrossRef]
- Diaz, M.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Rodriguez, L.; Fernandez, D.; Antoni, R.; Yunta, C.; Belda-Palazon, B.; Gonzalez-Guzman, M.; Peirats-Llobet, M.; et al. Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling. Proc. Natl. Acad. Sci. 2015, 113, 201512779–405. [Google Scholar] [CrossRef]
- Kay, J.G.; Fairn, G.D. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun. Signal. 2019, 17, 1–8. [Google Scholar] [CrossRef]
- Platre, M.P.; Bayle, V.; Armengot, L.; Bareille, J.; Marquès-Bueno, M.d.M.; Creff, A.; Maneta-Peyret, L.; Fiche, J.-B.; Nollmann, M.; Miège, C.; et al. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 2019, 364, 57–62. [Google Scholar] [CrossRef]
- Smokvarska, M.; Francis, C.; Platre, M.P.; Fiche, J.-B.; Alcon, C.; Dumont, X.; Nacry, P.; Bayle, V.; Nollmann, M.; Maurel, C.; et al. A Plasma Membrane Nanodomain Ensures Signal Specificity during Osmotic Signaling in Plants. Curr. Biol. 2020, 30, 4654–4664.e4. [Google Scholar] [CrossRef]
- Leung, J.; Giraudat, J. ABSCISIC ACID SIGNAL TRANSDUCTION. Annu. Rev. Plant Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Chen, J.; Yu, F.; Liu, Y.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodriguez, P.L.; Liu, X.; et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. 2016, 113, E5519–E5527. [Google Scholar] [CrossRef]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. 2012, 109, 14693–14698. [Google Scholar] [CrossRef]
- Fu, Q.; Li, H.; Wang, B.; Chen, W.; Wu, D.; Gao, C.; Yu, F.; Shen, W. The RALF1 peptide-FERONIA complex phosphorylates the endosomal sorting protein FREE1 to attenuate abscisic acid signaling. Plant Physiol. 2024, 197. [Google Scholar] [CrossRef]
- Gao, C.; Luo, M.; Zhao, Q.; Yang, R.; Cui, Y.; Zeng, Y.; Xia, J.; Jiang, L. A Unique Plant ESCRT Component, FREE1, Regulates Multivesicular Body Protein Sorting and Plant Growth. Curr. Biol. 2014, 24, 2556–2563. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Zhao, Q.; Li, T.; Wei, J.; Li, B.; et al. The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat Plants. 2019, 5, 512–524. [Google Scholar] [CrossRef]
- Mohanasundaram, B.; Pandey, S. Moving beyond the arabidopsis-centric view of G-protein signaling in plants. Trends Plant Sci. 2023, 28, 1406–1421. [Google Scholar] [CrossRef]
- Yu, Y.; Chakravorty, D.; Assmann, S.M. The G Protein β-Subunit, AGB1, Interacts with FERONIA in RALF1-Regulated Stomatal Movement. Plant Physiol. 2018, 176, 2426–2440. [Google Scholar] [CrossRef]
- Chakravorty, D.; Yu, Y.; Assmann, S.M. A kinase-dead version of FERONIA receptor-like kinase has dose-dependent impacts on rosette morphology and RALF 1-mediated stomatal movements. FEBS Letters 2018, 592, 3429–3437. [Google Scholar] [CrossRef]
- Zhao, C.; Jiang, W.; Zayed, O.; Liu, X.; Tang, K.; Nie, W.; Li, Y.; Xie, S.; Li, Y.; Long, T.; et al. The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 2020, 8, nwaa149. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of Reactive Oxygen Species and Antioxidant Defense in Plants under Salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Liu, L.; Li, Y.; Ogden, M.; Somssich, M.; Liu, Y.; Zhang, Y.; Ran, M.; Persson, S.; et al. FERONIA adjusts CC1 phosphorylation to control microtubule array behavior in response to salt stress. Sci. Adv. 2024, 10, eadq8717. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, W.; Li, Y.; Nie, H.; Cui, L.; Li, R.; Tan, L.; Peng, L.; Li, C.; Luo, J.; et al. FERONIA coordinates plant growth and salt tolerance via the phosphorylation of phyB. Nat. Plants 2023, 9, 645–660. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; van Zelm, E.; Huo, W.; Lamers, J.; Testerink, C. Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development 2022, 149. [Google Scholar] [CrossRef]
- Sato, T.; Maekawa, S.; Yasuda, S.; Yamaguchi, J. Carbon and nitrogen metabolism regulated by the ubiquitin-proteasome system. Plant Signal. Behav. 2011, 6, 1465–1468. [Google Scholar] [CrossRef]
- Xu, G.; Chen, W.; Song, L.; Chen, Q.; Zhang, H.; Liao, H.; Zhao, G.; Lin, F.; Zhou, H.; Yu, F. FERONIA phosphorylates E3 ubiquitin ligase ATL6 to modulate the stability of 14-3-3 proteins in response to the carbon/nitrogen ratio. J. Exp. Bot. 2019, 70, 6375–6388. [Google Scholar] [CrossRef]
- Burkart, G.M.; Brandizzi, F. A Tour of TOR Complex Signaling in Plants. Trends Biochem. Sci. 2021, 46, 417–428. [Google Scholar] [CrossRef]
- Song, L.; Xu, G.; Li, T.; Zhou, H.; Lin, Q.; Chen, J.; Wang, L.; Wu, D.; Li, X.; Wang, L.; et al. The RALF1-FERONIA complex interacts with and activates TOR signaling in response to low nutrients. Mol. Plant 2022, 15, 1120–1136. [Google Scholar] [CrossRef]
- Wang, P.; Clark, N.M.; Nolan, T.M.; Song, G.; Whitham, O.G.; Liao, C.-Y.; Montes-Serey, C.; Bassham, D.C.; Walley, J.W.; Yin, Y.; et al. FERONIA functions through Target of Rapamycin (TOR) to negatively regulate autophagy. Front. Plant Sci. 2022, 13, 961096. [Google Scholar] [CrossRef]
- Hou, S.; Tsuda, K. Salicylic acid and jasmonic acid crosstalk in plant immunity. Kanyuka K, Hammond-Kosack K, editors. Essays in Biochemistry 2022, 66, 647–656. [Google Scholar]
- Guo, H.; Nolan, T.M.; Song, G.; Schnable, P.S.; Walley, J.W.; Yin, Y.; Guo, H.; Nolan, T.M.; Song, G.; Liu, S.; et al. FERONIA Receptor Kinase Contributes to Plant Immunity by Suppressing Jasmonic Acid Signaling in Arabidopsis thaliana. Curr. Biol. 2018, 28, 3316–3324.e6. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, J.-M. Plant Immunity Triggered by Microbial Molecular Signatures. Mol. Plant 2010, 3, 783–793. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef]
- Gronnier, J.; Franck, C.M.; Stegmann, M.; A DeFalco, T.; Abarca, A.; von Arx, M.; Dünser, K.; Lin, W.; Yang, Z.; Kleine-Vehn, J.; et al. Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. eLife 2022, 11. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Roy, S.; Kritsas, K.; Grobei, M.A.; Jaciubek, M.; Schroeder, J.I.; Grossniklaus, U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 2009, 136, 3279–3288. [Google Scholar] [CrossRef] [PubMed]
- Mang, H.; Feng, B.; Hu, Z.; Boisson-Dernier, A.; Franck, C.M.; Meng, X.; Huang, Y.; Zhou, J.; Xu, G.; Wang, T.; et al. Differential Regulation of Two-Tiered Plant Immunity and Sexual Reproduction by ANXUR Receptor-Like Kinases. Plant Cell 2017, 29, 3140–3156. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Bourdais, G.; Pan, H.; Robatzek, S.; Tang, D. Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl. Acad. Sci. 2017, 114, 5749–5754. [Google Scholar] [CrossRef]
- Tang, J.; Wu, D.; Li, X.; Wang, L.; Xu, L.; Zhang, Y.; Xu, F.; Liu, H.; Xie, Q.; Dai, S.; et al. Plant immunity suppression via PHR1-RALF-FERONIA shapes the root microbiome to alleviate phosphate starvation. EMBO J. 2022, 41, e109102. [Google Scholar] [CrossRef]
- García-Gómez, P.; Bahaji, A.; Gámez-Arcas, S.; Muñoz, F.J.; Sánchez-López, Á.M.; Almagro, G.; Baroja-Fernández, E.; Ameztoy, K.; De Diego, N.; Ugena, L.; et al. Volatiles from the fungal phytopathogen Penicillium aurantiogriseum modulate root metabolism and architecture through proteome resetting. Plant, Cell Environ. 2020, 43, 2551–2570. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Yuan, B.; Chen, X.; Zhao, H.; Ali, Q.; Zheng, M.; Tan, Z.; Yao, H.; Zheng, S.; et al. Fusarium graminearum rapid alkalinization factor peptide negatively regulates plant immunity and cell growth via the FERONIA receptor kinase. Plant Biotechnol. J. 2024, 22, 1800–1811. [Google Scholar] [CrossRef]
- Masachis, S.; Segorbe, D.; Turrà, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef]
- Liao, Y.; Wen, X.; Zheng, J.; Li, X.; Deng, X.; Tan, X.; Liang, X.; Yi, X.; Liao, H. RALF-like peptide improves the colonization of endophytic Colletotrichum tofieldiae through interacting with plant receptor-like kinase. Plant Pathol. 2023, 72, 1649–1661. [Google Scholar] [CrossRef]
- Pin, P.A.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant, Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Le, C.; Wang, Y.; Li, Z.; Jiang, D.; Wang, Y.; He, Y. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013, 4, 1947. [Google Scholar] [CrossRef]
- Michaels, S.D.; Himelblau, E.; Kim, S.Y.; Schomburg, F.M.; Amasino, R.M. Integration of Flowering Signals in Winter-Annual Arabidopsis. Plant Physiol. 2005, 137, 149–156. [Google Scholar] [CrossRef]
- Wang, L.; Yang, T.; Lin, Q.; Wang, B.; Li, X.; Luan, S.; Yu, F. Receptor kinase FERONIA regulates flowering time in Arabidopsis. BMC Plant Biol. 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Johnson, M.A.; Harper, J.F.; Palanivelu, R. A Fruitful Journey: Pollen Tube Navigation from Germination to Fertilization. Annu. Rev. Plant Biol. 2019, 70, 809–837. [Google Scholar] [CrossRef]
- Törnroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; et al. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Chebli, Y.; Kaneda, M.; Zerzour, R.; Geitmann, A. The Cell Wall of the Arabidopsis Pollen Tube—Spatial Distribution, Recycling, and Network Formation of Polysaccharides. Plant Physiol. 2012, 160, 1940–1955. [Google Scholar] [CrossRef]
- Wudick, M.M.; Feijó, J.A. At the Intersection: Merging Ca 2+ and ROS Signaling Pathways in Pollen. Mol. Plant 2014, 7, 1595–1597. [Google Scholar] [CrossRef] [PubMed]
- Seitz, P.L.; Qu, L.-J.; Dresselhaus, T.; Zhou, L.-Z. Spatial organization and trafficking dynamics of ANX/BUPS-RALF-LLG signaling complexes during pollen tube growth. Plant Reprod. 2025, 38, 1–10. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Lituiev, D.S.; Nestorova, A.; Franck, C.M.; Thirugnanarajah, S.; Grossniklaus, U. ANXUR Receptor-Like Kinases Coordinate Cell Wall Integrity with Growth at the Pollen Tube Tip Via NADPH Oxidases. Nasrallah JB, editor. PLoS Biol. 2013, 11, e1001719. [Google Scholar] [CrossRef]
- Zhu, L.; Chu, L.; Liang, Y.; Zhang, X.; Chen, L.; Ye, D. The Arabidopsis CrRLK1L protein kinases BUPS1 and BUPS2 are required for normal growth of pollen tubes in the pistil. Plant J. 2018, 95, 474–486. [Google Scholar] [CrossRef]
- Sede, A.R.; Borassi, C.; Wengier, D.L.; Mecchia, M.A.; Estevez, J.M.; Muschietti, J.P. Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett. 2018, 592, 233–243. [Google Scholar] [CrossRef]
- Wang, X.; Wang, K.; Yin, G.; Liu, X.; Liu, M.; Cao, N.; Duan, Y.; Gao, H.; Wang, W.; Ge, W.; et al. Pollen-Expressed Leucine-Rich Repeat Extensins Are Essential for Pollen Germination and Growth. Plant Physiol. 2017, 176, 1993–2006. [Google Scholar] [CrossRef]
- Fabrice, T.N.; Vogler, H.; Draeger, C.; Munglani, G.; Gupta, S.; Herger, A.G.; Knox, P.; Grossniklaus, U.; Ringli, C. LRX Proteins Play a Crucial Role in Pollen Grain and Pollen Tube Cell Wall Development. Plant Physiol. 2017, 176, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lu, J.; Zhang, Y.; Guo, J.; Lin, W.; Van Norman, J.M.; Qin, Y.; Zhu, X.; Yang, Z. Membrane receptor-mediated mechano-transduction maintains cell integrity during pollen tube growth within the pistil. Dev. Cell 2021, 56, 1030–1042.e6. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Lin, W.; Zhou, X.; Guo, J.; Dang, X.; Li, B.; Lin, D.; Yang, Z. Mechano-transduction via the pectin-FERONIA complex activates ROP6 GTPase signaling in Arabidopsis pavement cell morphogenesis. Curr. Biol. 2022, 32, 508–517.e3. [Google Scholar] [CrossRef]
- Ngo, Q.A.; Vogler, H.; Lituiev, D.S.; Nestorova, A.; Grossniklaus, U. A Calcium Dialog Mediated by the FERONIA Signal Transduction Pathway Controls Plant Sperm Delivery. Dev. Cell 2014, 29, 491–500. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Johnson, E.A.; Aggarwal, M.; Gates, L.; Wu, H.-M.; Cheung, A.Y. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 2014, 5, 3129. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Trigo, S.; Blanco-Touriñán, N.; A DeFalco, T.; Wells, E.S.; E Gray, J.; Zipfel, C.; Smith, L.M. CrRLK1L receptor-like kinases HERK1 and ANJEA are female determinants of pollen tube reception. Embo Rep. 2019, 21, e48466. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Liu, M.-C.J.; Kita, D.; Jordan, S.S.; Yeh, F.-L.J.; Yvon, R.; Carpenter, H.; Federico, A.N.; Garcia-Valencia, L.E.; Eyles, S.J.; et al. FERONIA controls pectin- and nitric oxide-mediated male–female interaction. Nature 2020, 579, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kurten, E.L.; Monshausen, G.; Hummel, G.M.; Gilroy, S.; Baldwin, I.T. NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 2007, 52, 877–890. [Google Scholar] [CrossRef]
- Kwon, O.; Moon, H.; Jeong, A.; Yeom, G.; Park, C. Rice small secreted peptide, OsRALF26, recognized by FERONIA-like receptor 1 induces immunity in rice and Arabidopsis. Plant J. 2024, 118, 1528–1549. [Google Scholar] [CrossRef]
- Jiang, W.; Li, C.; Li, L.; Li, Y.; Wang, Z.; Yu, F.; Yi, F.; Zhang, J.; Zhu, J.-K.; Zhang, H.; et al. Genome-Wide Analysis of CqCrRLK1L and CqRALF Gene Families in Chenopodium quinoa and Their Roles in Salt Stress Response. Front. Plant Sci. 2022, 13, 918594. [Google Scholar] [CrossRef]
- Wieghaus, A.; Prüfer, D.; Gronover, C.S. Loss of function mutation of the Rapid Alkalinization Factor (RALF1)-like peptide in the dandelion Taraxacum koksaghyz entails a high-biomass taproot phenotype. PLOS ONE 2019, 14, e0217454. [Google Scholar] [CrossRef]
- Mingossi, F.B.; Matos, J.L.; Rizzato, A.P.; Medeiros, A.H.; Falco, M.C.; Silva-Filho, M.C.; Moura, D.S. SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol. Biol. 2010, 73, 271–281. [Google Scholar] [CrossRef]
- Fan, Y.; Bai, J.; Wu, S.; Zhang, M.; Li, J.; Lin, R.; et al. The RALF2-FERONIA-MYB63 module orchestrates growth and defense in tomato roots. New Phytologist 2024, 243, 1123–1136. [Google Scholar] [CrossRef]
- Ginanjar, E.F.; Teh, O.; Fujita, T. Characterisation of rapid alkalinisation factors in Physcomitrium patens reveals functional conservation in tip growth. New Phytol. 2021, 233, 2442–2457. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Constabel, C.P. Rapid Alkalinization Factors in Poplar Cell Cultures. Peptide Isolation, cDNA Cloning, and Differential Expression in Leaves and Methyl Jasmonate-Treated Cells. Plant Physiol. 2003, 131, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Gidrol, X.; Chrestin, H.; Mounoury, G.; D'Auzac, J. Early Activation by Ethylene of the Tonoplast H+-Pumping ATPase in the Latex from Hevea brasiliensis. Plant Physiol. 1988, 86, 899–903. [Google Scholar] [CrossRef]
- Sui, J.; Xiao, X.; Yang, J.; Fan, Y.; Zhu, S.; Zhu, J.; Zhou, B.; Yu, F.; Tang, C. The rubber tree RALF peptide hormone and its receptor protein kinase FER implicates in rubber production. Plant Sci. 2022, 326, 111510. [Google Scholar] [CrossRef]
- Covey, P.A.; Subbaiah, C.C.; Parsons, R.L.; Pearce, G.; Lay, F.T.; Anderson, M.A.; Ryan, C.A.; Bedinger, P.A. A Pollen-Specific RALF from Tomato That Regulates Pollen Tube Elongation. Plant Physiol. 2010, 153, 703–715. [Google Scholar] [CrossRef]
- Kou, X.; Sun, J.; Wang, P.; Wang, D.; Cao, P.; Lin, J.; Chang, Y.; Zhang, S.; Wu, J. PbrRALF2-elicited reactive oxygen species signaling is mediated by the PbrCrRLK1L13-PbrMPK18 module in pear pollen tubes. Hortic. Res. 2021, 8, 222. [Google Scholar] [CrossRef]
- Zhou, L.-Z.; Wang, L.; Chen, X.; Ge, Z.; Mergner, J.; Li, X.; Küster, B.; Längst, G.; Qu, L.-J.; Dresselhaus, T. The RALF signaling pathway regulates cell wall integrity during pollen tube growth in maize. Plant Cell 2023, 36, 1673–1696. [Google Scholar] [CrossRef]
- Kim, E.; Kim, J.; Hong, W.; Kim, E.Y.; Kim, M.; Lee, S.K.; Min, C.W.; Kim, S.T.; Park, S.K.; Jung, K.; et al. Rice pollen-specific OsRALF17 and OsRALF19 are essential for pollen tube growth. J. Integr. Plant Biol. 2023, 65, 2218–2236. [Google Scholar] [CrossRef]
- Kim, J.-H.; Son, Y.-J.; Kim, E.-J.; Jung, K.-H.; Kim, Y.-J. Pollen tube-expressed RUPO forms a complex with OsMTD2 and OsRALF17 and OsRALF19 peptides in rice (Oryza sativa). J. Plant Physiol. 2025, 305, 154421. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, C.; Kuang, B.; Wei, L.; Yan, L.; Wang, T. Receptor-Like Kinase RUPO Interacts with Potassium Transporters to Regulate Pollen Tube Growth and Integrity in Rice. PLOS Genet. 2016, 12, e1006085. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhou, J.; Gu, Y.; Zeng, Y.; Lu, K.; Peng, S.; Tan, X. Genome-wide identification of the RALF gene family in Camellia oleifera Abel. and the potentiality of CoRALF50 in pollen tube growth. Plant Sci. 2025, 359, 112603. [Google Scholar] [CrossRef]
- Chevalier, E.; Loubert-Hudon, A.; Matton, D.P. ScRALF3, a secreted RALF-like peptide involved in cell–cell communication between the sporophyte and the female gametophyte in a solanaceous species. Plant J. 2012, 73, 1019–1033. [Google Scholar] [CrossRef]
- Germain, H.; Matton, D.P.; Chevalier, É.; Caron, S. Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta 2004, 220, 447–454. [Google Scholar] [CrossRef]
- Loubert-Hudon, A.; Mazin, B.D.; Chevalier, É.; Matton, D.P. The ScRALF3 secreted peptide is involved in sporophyte to gametophyte signalling and affects pollen mitosis I. Plant Biol. 2020, 22, 13–20. [Google Scholar] [CrossRef]
- Wang, P.; Yao, S.; Kosami, K.; Guo, T.; Li, J.; Zhang, Y.; Fukao, Y.; Kaneko-Kawano, T.; Zhang, H.; She, Y.; et al. Identification of endogenous small peptides involved in rice immunity through transcriptomics- and proteomics-based screening. Plant Biotechnol. J. 2019, 18, 415–428. [Google Scholar] [CrossRef]
- Jia, Y.; Li, Y. Genome-Wide Identification and Comparative Analysis of RALF Gene Family in Legume and Non-Legume Species. Int. J. Mol. Sci. 2023, 24, 8842. [Google Scholar] [CrossRef]
- Wang, W.; He, Z.; Wang, D.; Xu, G.; Yang, J.; Feng, X.; Liang, X.; Dou, D.; Chen, R. Functional analysis of RALF peptides in soybean immunity and disease resistance. Phytopathol. Res. 2025, 7, 1–14. [Google Scholar] [CrossRef]
- Wang, W.; Chen, R.; Wang, D.; Zhang, W.; Zhang, Q.; Zhang, Y.; Xu, G.; Yang, J.; Cheng, Z.; Yang, S.; et al. A pair of GPI-anchored proteins regulate soybean immunity and disease resistance in coordination with GmLMM1 and GmRALFs. Plant Biotechnol. J. 2025, 23, 4353–4366. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liang, X.; Bao, Y.; Yang, S.; Zhang, X.; Yu, H.; Zhang, Q.; Xu, G.; Feng, X.; Dou, D. A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. Embo Rep. 2020, 21, e50442. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Fang, X.; Yu, T.; Chen, J.; Yan, F. Turnip mosaic virus pathogenesis and host resistance mechanisms in Brassica. Hortic. Plant J. 2024, 10, 947–960. [Google Scholar] [CrossRef]
- Rui, P.; Jia, Z.; Fang, X.; Yu, T.; Mao, W.; Lin, J.; et al. A plant viral effector subverts FER - RALF1 module-mediated intracellular immunity. Plant Biotechnology Journal 2025, 23, 2734–2751. [Google Scholar] [CrossRef]
- Shen, W.; Yuan, M.; Chen, L.; Zhang, X. Comparative analysis of rapid alkalinization factor peptide-triggered plant immunity in citrus and closely related species. Plant Physiol. Biochem. 2025, 224, 109941. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, D.; Chen, J.; Wu, D.; Feng, X.; Yu, F. Nematode RALF-Like 1 Targets Soybean Malectin-Like Receptor Kinase to Facilitate Parasitism. Front. Plant Sci. 2021, 12, 775508. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.; Jiang, H.; Shui, Z.; Zhong, Y.; Shang, J.; Yang, H.; Sun, X.; Du, J. Genome-wide characterization of soybean RALF genes and their expression responses to Fusarium oxysporum. Front. Plant Sci. 2022, 13, 1006028. [Google Scholar] [CrossRef]
- Prusky, D.; Yakoby, N. Pathogenic fungi: leading or led by ambient pH? Molecular Plant Pathology 2003, 4, 509–516. [Google Scholar] [CrossRef]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef]
- Wei, H.; Yang, R.; Xue, Z.; Zhu, J.; Zhang, Q.; Luan, Y. Molecular Traits of Rapid Alkalinization Factor Family and Functional Analysis of SlRALF2 in Tomato Resistance to Phytophthora infestans. J. Agric. Food Chem. 2025, 73, 3622–3634. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-H.; Zhang, Z.-R.; Xu, Y.-P.; Chen, S.-Y.; Cai, X.-Z. Genome-Wide Identification of Rapid Alkalinization Factor Family in Brassica napus and Functional Analysis of BnRALF10 in Immunity to Sclerotinia sclerotiorum. Front. Plant Sci. 2022, 13, 877404. [Google Scholar] [CrossRef]
- Merino, M.C.; Guidarelli, M.; Negrini, F.; De Biase, D.; Pession, A.; Baraldi, E. Induced expression of the Fragaria × ananassa Rapid alkalinization factor-33-like gene decreases anthracnose ontogenic resistance of unripe strawberry fruit stages. Mol. Plant Pathol. 2019, 20, 1252–1263. [Google Scholar] [CrossRef]
- van Rhijn, P.; Vanderleyden, J. The Rhizobium-plant symbiosis. Microbiol. Rev. 1995, 59, 124–142. [Google Scholar] [CrossRef]
- Solís-Miranda, J.; Juárez-Verdayes, M.A.; Nava, N.; Rosas, P.; Leija-Salas, A.; Cárdenas, L.; Quinto, C. The Phaseolus vulgaris Receptor-Like Kinase PvFER1 and the Small Peptides PvRALF1 and PvRALF6 Regulate Nodule Number as a Function of Nitrate Availability. Int. J. Mol. Sci. 2023, 24, 5230. [Google Scholar] [CrossRef]
- Combier, J.-P.; Küster, H.; Journet, E.-P.; Hohnjec, N.; Gamas, P.; Niebel, A. Evidence for the Involvement in Nodulation of the Two Small Putative Regulatory Peptide-Encoding Genes MtRALFL1 and MtDVL1. Mol. Plant-Microbe Interactions 2008, 21, 1118–1127. [Google Scholar] [CrossRef]
- Li, F.; Mai, C.; Liu, Y.; Deng, Y.; Wu, L.; Zheng, X.; He, H.; Huang, Y.; Luo, Z.; Wang, J. Soybean PHR1-regulated low phosphorus-responsive GmRALF22 promotes phosphate uptake by stimulating the expression of GmPTs. Plant Sci. 2024, 348, 112211. [Google Scholar] [CrossRef]
- Mamaeva, A.; Lyapina, I.; Knyazev, A.; Golub, N.; Mollaev, T.; Chudinova, E.; Elansky, S.; Babenko, V.V.; Veselovsky, V.A.; Klimina, K.M.; et al. RALF peptides modulate immune response in the moss Physcomitrium patens. Front. Plant Sci. 2023, 14, 1077301. [Google Scholar] [CrossRef]
- Lin, H.; Han, X.; Feng, X.; Chen, X.; Lu, X.; Yuan, Z.; Li, Y.; Ye, W.; Yin, Z. Molecular traits and functional analysis of Rapid Alkalinization Factors (RALFs) in four Gossypium species. Int. J. Biol. Macromol. 2022, 194, 84–99. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Y.; Li, Y.; Ma, H.; Zuo, C.; Yang, J.; Ma, Y.; Kong, J.; Wei, X.; Zhu, L.; et al. Polyploidization-driven formation of the GhRALF30L gene cluster confers basal thermotolerance in cotton male reproductive organs via GhFERA1/GhCAPA1. Sci. Adv. 2025, 11, eady1386. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Shi, P.-T.; Zhang, R.; Zhou, M.-R.; Shalmani, A.; Wang, G.-F.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. Rice kinase OsMRLK63 contributes to drought tolerance by regulating reactive oxygen species production. Plant Physiol. 2023, 194, 2679–2696. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, M.; Wang, Y.; Gao, Y.; Li, R.; Wang, G.; Li, W.; Liu, W.; Chen, K. The plasma membrane NADPH oxidase OsRbohA plays a crucial role in developmental regulation and drought-stress response in rice. Physiol. Plant. 2015, 156, 421–443. [Google Scholar] [CrossRef]
- Sun, W.; Qin, Y.; Cao, Y.; Tong, J.; Shi, L.; Liu, H.; Wang, G.; Zheng, X.; Zou, L.; Jiang, L.; et al. Genome-wide identification and functional characterization of rapid alkalinization factor 6 as a key peptide regulator of abiotic stress tolerance in Tartary buckwheat. Plant Sci. 2025, 362, 112747. [Google Scholar] [CrossRef]
- Qiao, Q.; Ren, Z.; Fu, X.; Qiao, W.; Sheng, F.; Li, S.; Xiao, D.; He, L. Genome-wide exploration and characterization of the RALFs and analysis of its role in peanut (Arachis hypogaea L.). BMC Plant Biol. 2025, 25, 1–21. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).