Submitted:
29 October 2025
Posted:
03 November 2025
You are already at the latest version
Abstract
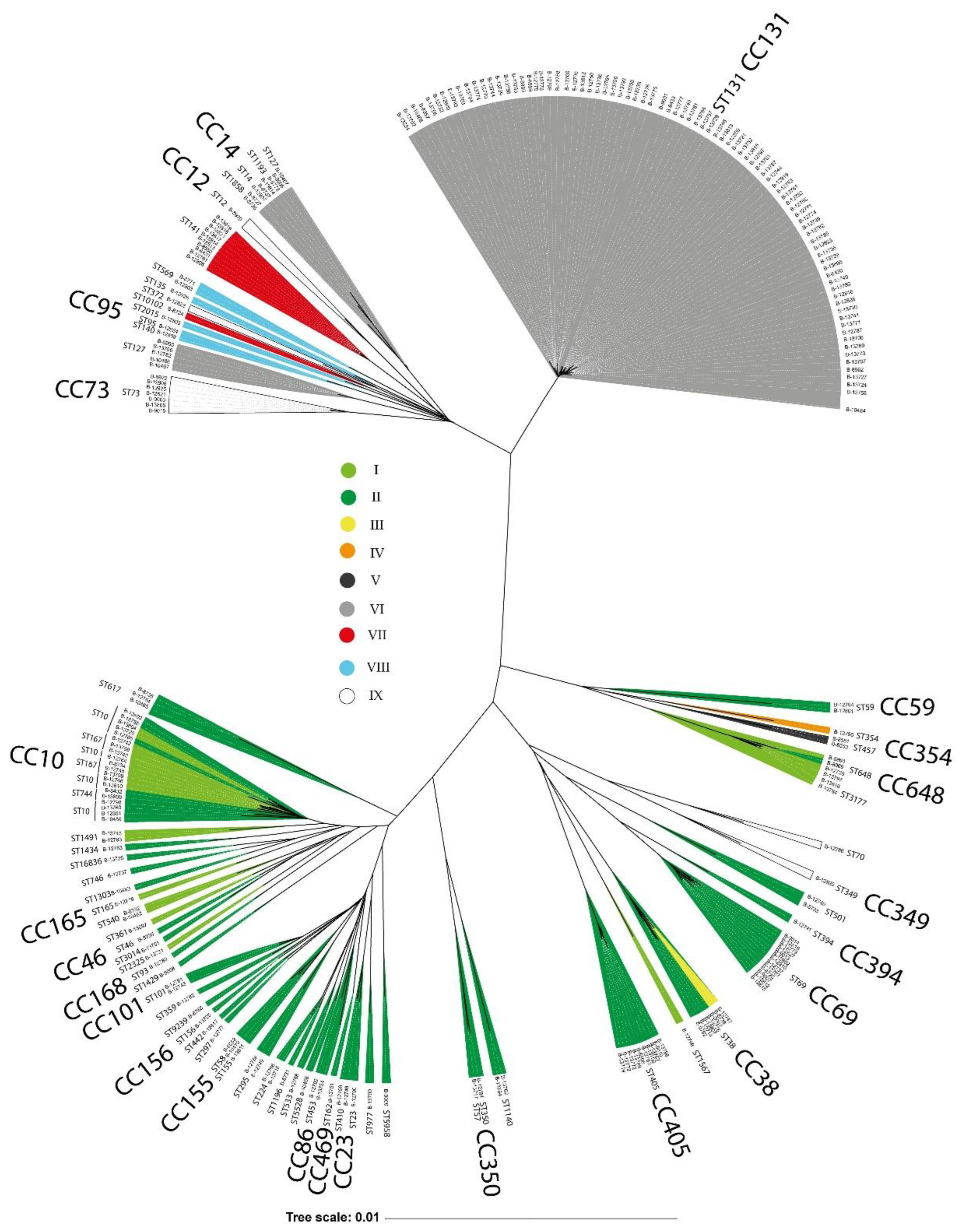
The type I-E and I-F CRISPR-Cas systems were identified in 237 E. coli strains isolated from patients with urinary tract infections (UTIs) between 2004 and 2019. The strains were classified into nine distinct groups (I-IX) based on the presence or absence of cas genes and repeat regions (RRs). Within the type I-E systems, two sequence variants were identified, distinguished by polymorphisms in the casB, cas3, cas7, cas5, and cas6 genes. The direct repeats (DRs) also differed, with I-E-associated RRs ranging from 26–32 bp and I-F-associated RRs being a consistent 28 bp. We identified 762 unique spacers (29–35 bp in length) across the strain collection. The number of spacers per strain varied from 1 to 47, and potential DNA targets were determined for 83 spacers, targeting 56 bacteriophage genomes, 19 plasmids, and 8 cas genes of the I-F type. Multilocus sequence typing (MLST) revealed 68 sequence types and 24 clonal complexes (CCs), with ST131, CC10, CC69, CC405, CC14, CC38, CC73, and CC648 being the most prevalent. Significant correlations were observed between specific phylogroups/CCs, the type of CRISPR-Cas system present, and distinct profiles of virulence and antibiotic resistance genes.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. DNA Isolation and PCR Amplification
2.3. Whole-Genome Sequencing
2.4. Genomic Analysis
2.5. Statistical Methods
3. Results
3.1. General Characteristics of the Strains
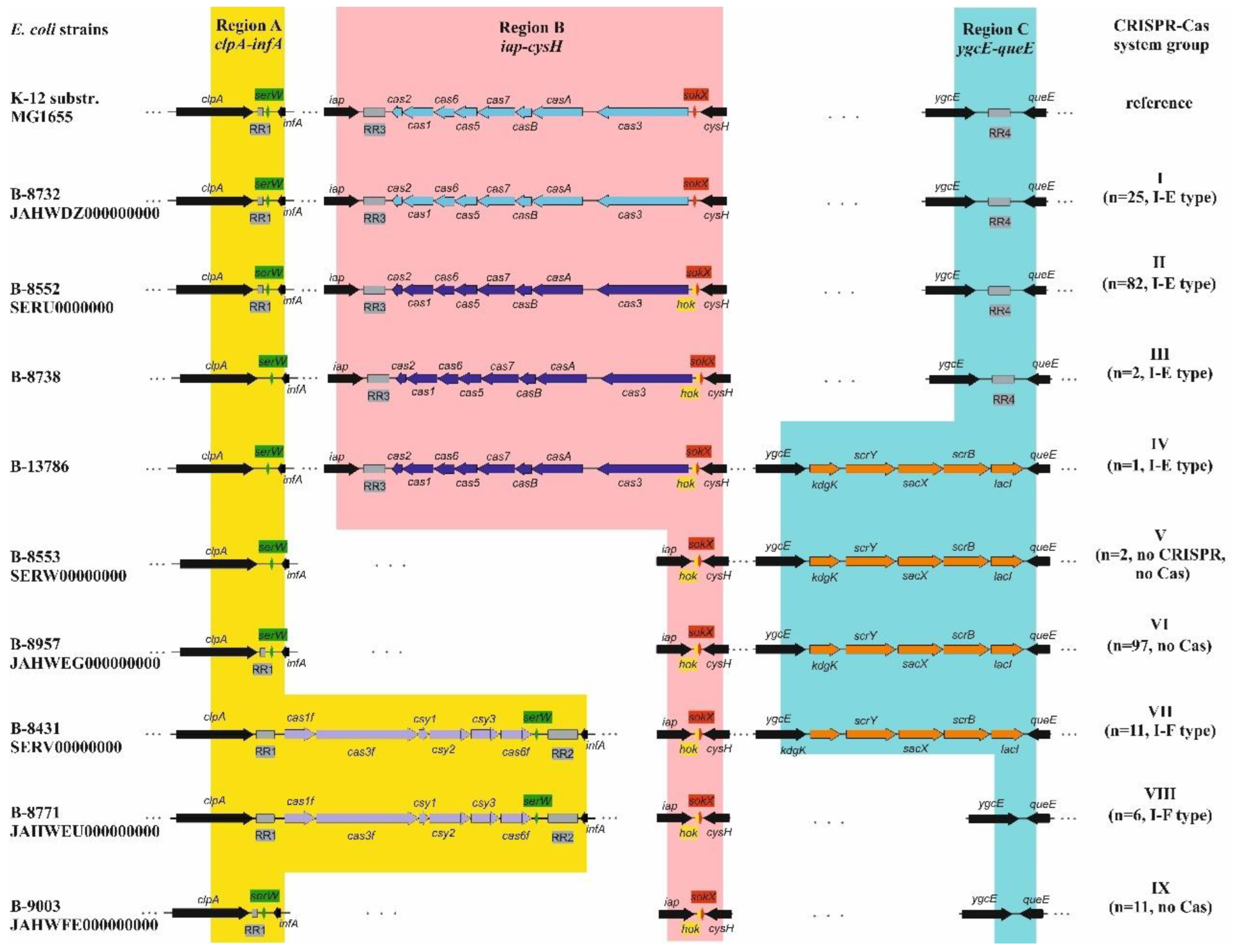
3.2. Identification and Characterization of CRISPR-Cas Systems
3.3. Types of CRISPR-Cas Systems
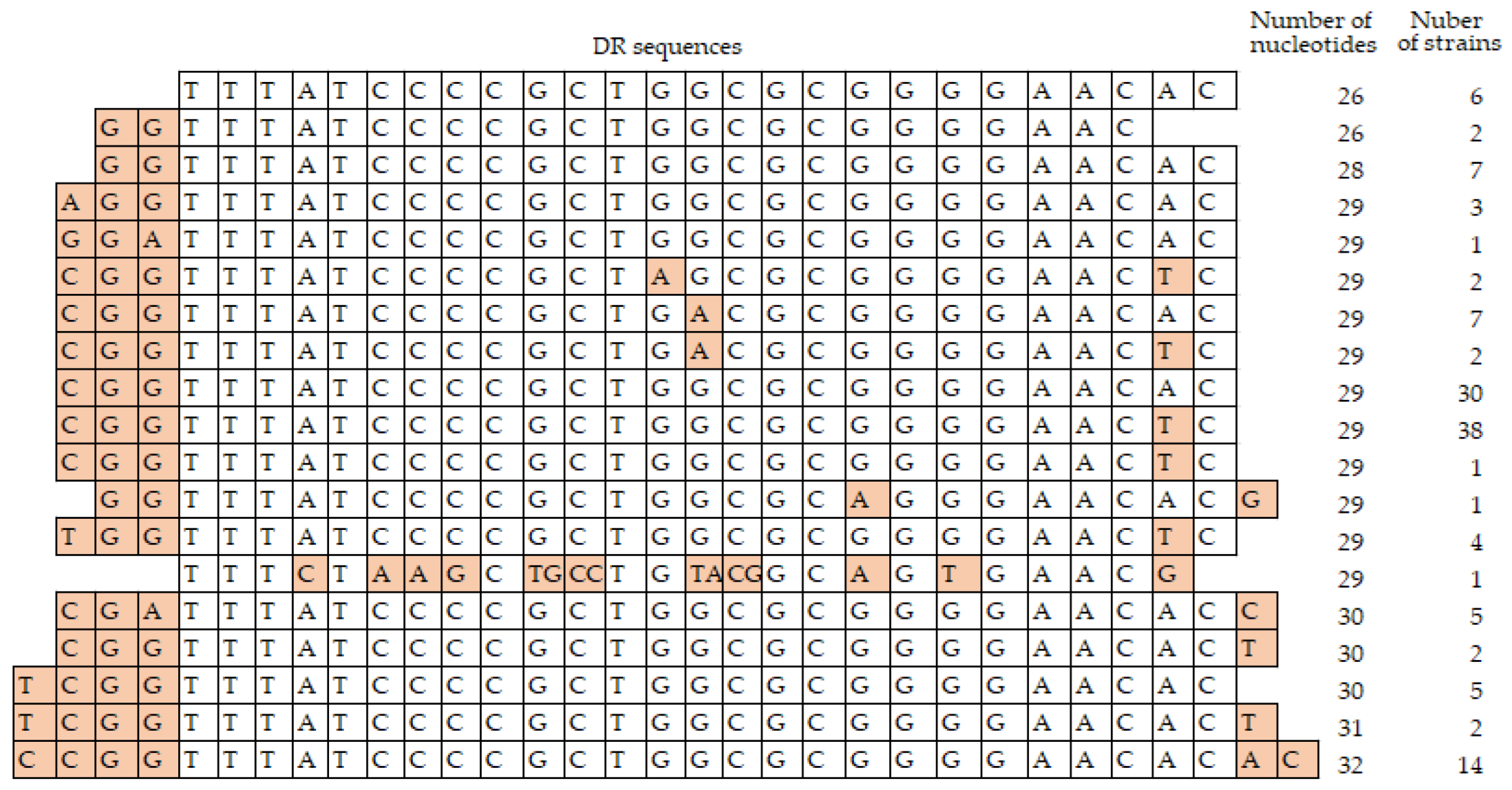
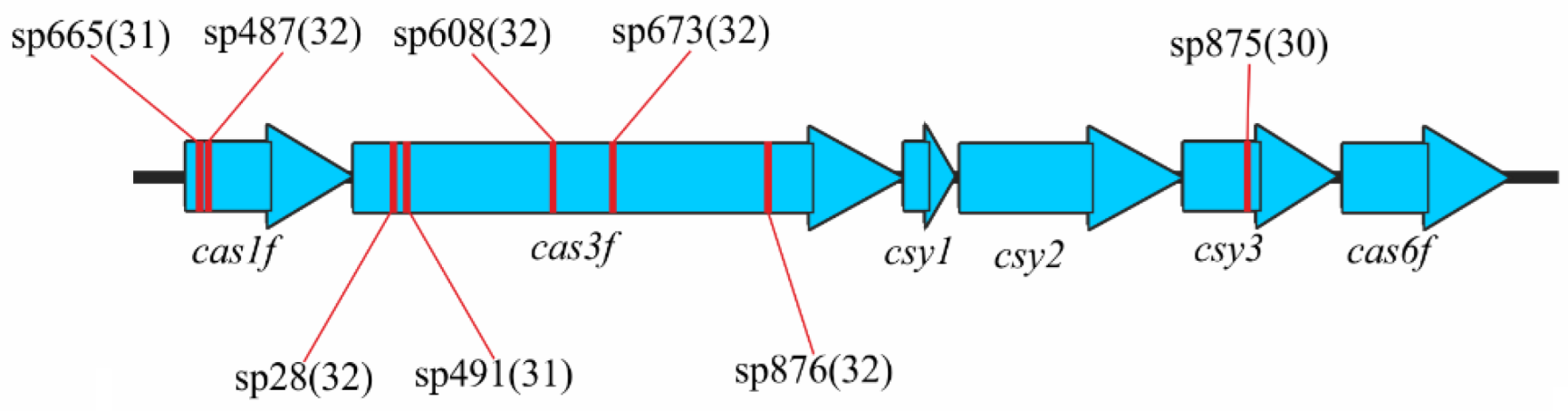
3.4. Analysis of the Repeat Regions, RRs
3.4.1. Direct Repeats, DRs
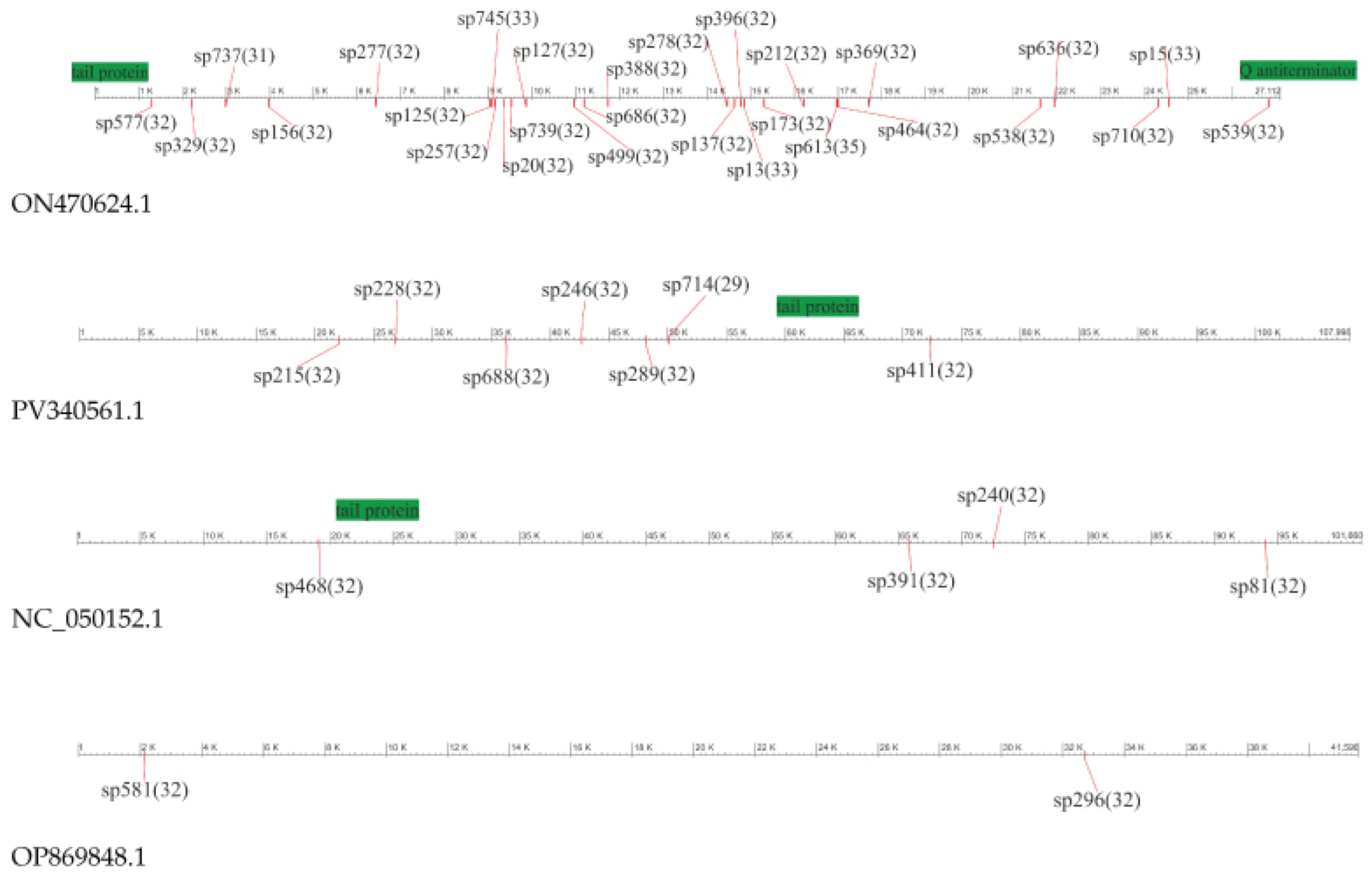
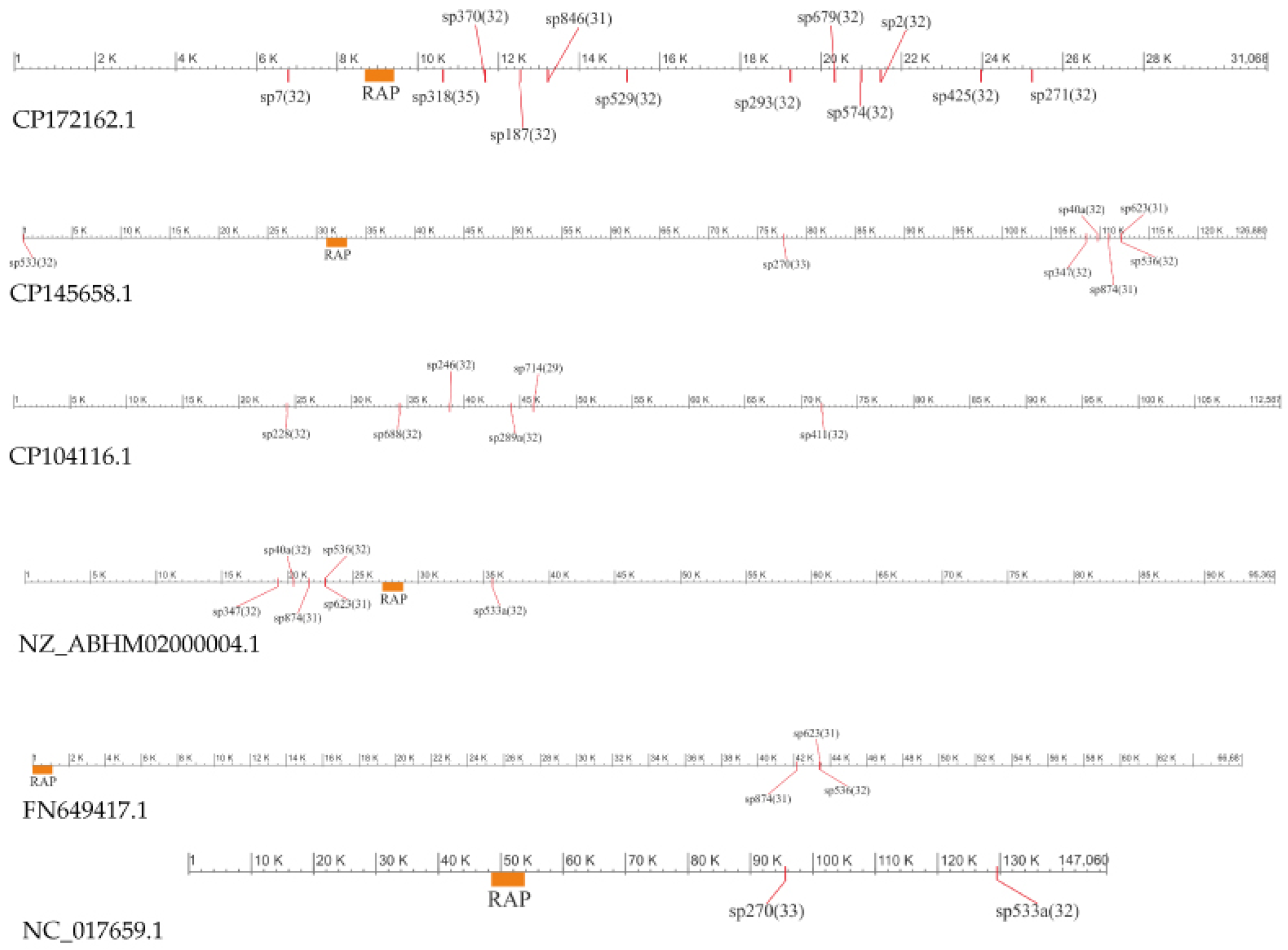
3.4.2. Spacer Sets
3.5. E. coli Phylogenetic Groups and CRISPR-Cas Systems Prevalence
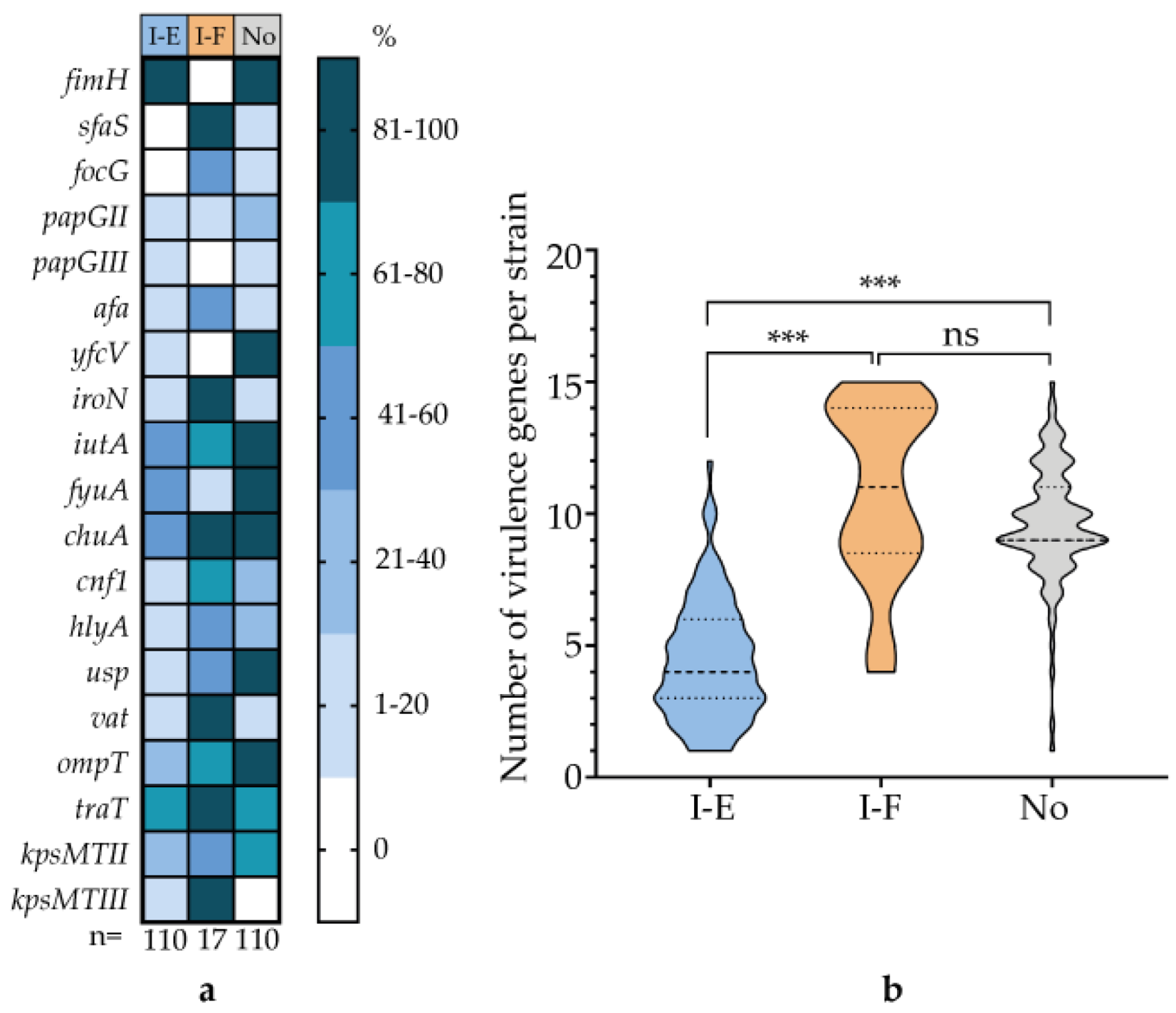
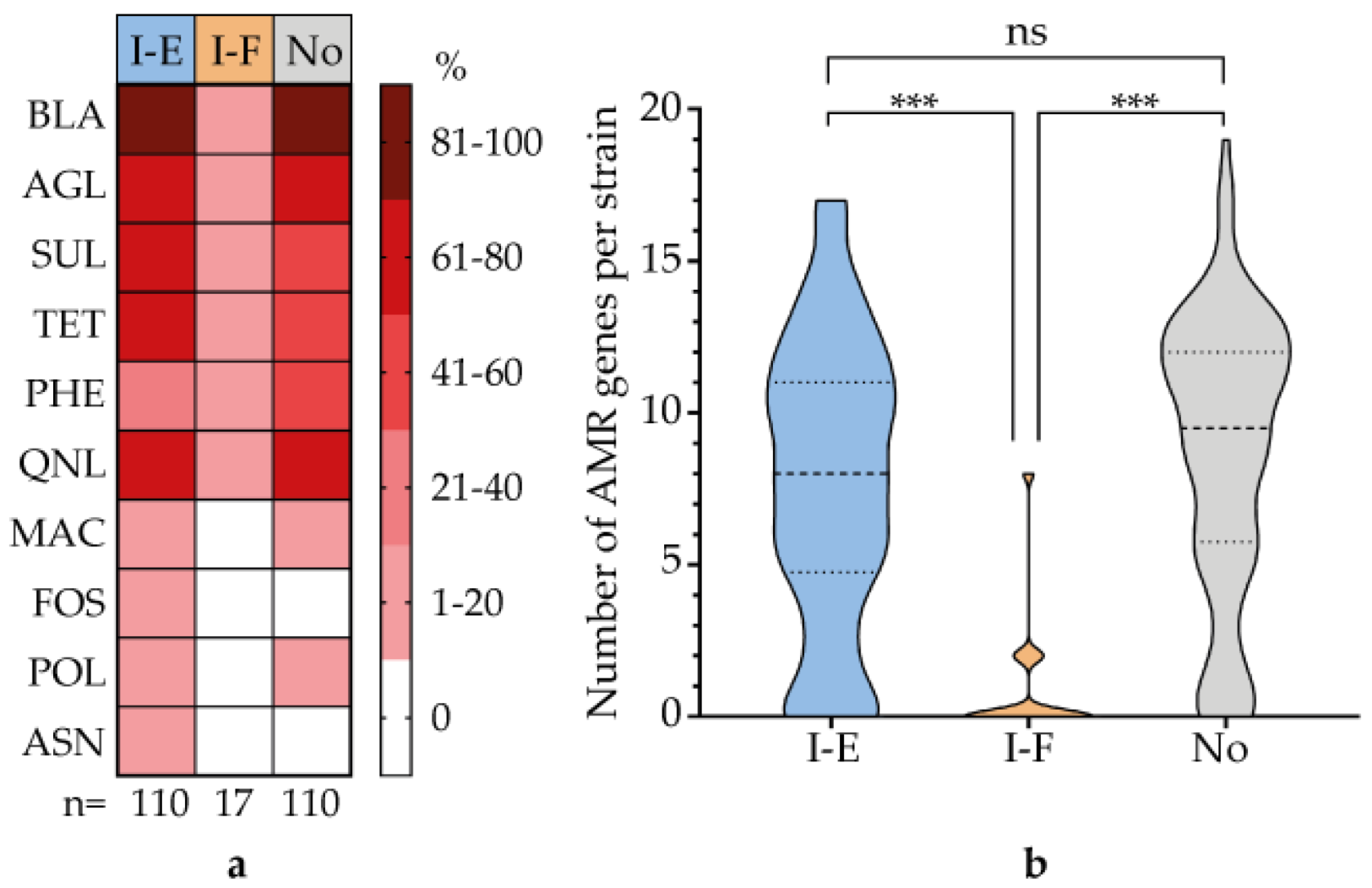
3.6. Correlation of CRISPR-Cas System Type with Virulence Genes and AMR Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Cas | CRISPR-associated proteins |
| RRs | Repeat Regions |
| DRs | Direct Repeats |
| UPEC | Uropathogenic E. coli |
| UTI | Urinary Tract Infection |
| WGS | Whole Genome Sequencing |
| ST | Sequence Type |
| CC | Clonal Complex |
References
- Toval, F.; Köhler, C.-D.; Vogel, U.; Wagenlehner, F.; Mellmann, A.; Fruth, A.; Schmidt, M.A.; Karch, H.; Bielaszewska, M.; Dobrindt, U. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 2014. 52, 407–418. [CrossRef]
- Gagaletsios, L.A.; Kikidou, E.; Galbenis, C.; Bitar, I.; Papagiannitsis, C.C. Exploring virulence characteristics of clinical Escherichia coli isolates from Greece. Microorganisms. 2025. 13(7), 1488.
- Mitić, D.; Bolt, E.L.; Ivančić-Baće, I. CRISPR-Cas adaptation in Escherichia coli. Biosci Rep. 2023, 43(3), BSR20221198.
- Mosterd, C.; Rousseau, G.M.; Moineau, S. A short overview of the CRISPR-Cas adaptation stage. Can J Microbiol. 2021, 67(1), 1-12.
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015. 13, 722–736. [CrossRef]
- Song, S.; Semenova, E.; Severinov, K.; Fernández-García, L.; Benedik, M.J.; Maeda, T.; Wood, T.K. CRISPR-Cas controls cryptic prophages. Int J Mol Sci. 2022, 23(24), 16195. [CrossRef]
- Dudley, E.G. The E. coli CRISPR-Cas conundrum: are they functional immune systems or genomic singularities? EcoSal Plus. 2025. 9, eesp00402020.
- Díez-Villaseñor, C.; Almendros, C.; García-Martínez, J.; Mojica, F.J. Diversity of CRISPR loci in Escherichia coli. Microbiology (Reading). 2010, 156(5), 1351-1361.
- Dion, M.B.; Shah, S.A.; Deng, L.; Thorsen, J.; Stokholm, J.; Krogfelt, K.A.; Schjørring, S.; Horvath, P.; Allard, A.; Nielsen, D.S.; Petit, M.A.; Moineau, S. Escherichia coli CRISPR arrays from early life fecal samples preferentially target prophages. ISME J. 2024, 18(1), wrae005. [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; van der Oost, J.; Koonin, E.V. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011, 9(6), 467-477.
- Touchon, M.; Charpentier, S.; Clermont, O.; Rocha, E.P.; Denamur, E.; Branger, C. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J Bacteriol. 2011, 193(10), 2460-7. [CrossRef]
- Iordache, D.; Baci, G.M.; Căpriță, O.; Farkas, A.; Lup, A.; Butiuc-Keul, A. Correlation between CRISPR loci diversity in three enterobacterial taxa. Int J Mol Sci. 2022, 23(21), 12766.
- Touchon, M.; Rocha, E.P. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010, 5(6), e11126.
- Mikhaylova, Y.; Tyumentseva, M.; Karbyshev, K.; Tyumentsev, A.; Slavokhotova, A.; Smirnova, S.; Akinin, A.; Shelenkov, A.; Akimkin, V. Interrelation between pathoadaptability factors and Crispr-element patterns in the genomes of Escherichia coli isolates collected from healthy puerperant women in Ural region, Russia. Pathogens. 2024, 13(11), 997. [CrossRef]
- Yoshimi, K.; Takeshita, K.; Kodera, N.; Shibumura, S.; Yamauchi, Y.; Omatsu, M.; Umeda, K.; Kunihiro, Y.; Yamamoto, M.; Mashimo, T. Dynamic mechanisms of CRISPR interference by Escherichia coli CRISPR-Cas3. Nat Commun. 2022, 13(1), 4917. [CrossRef]
- Sontheimer, E.J.; Barrangou, R. The bacterial origins of the CRISPR genome-editing revolution. Hum Gene Ther. 2015, 26(7), 413-424.
- Tajkarimi, M.; Wexler, H.M. CRISPR-Cas systems in Bacteroides fragilis, an important pathobiont in the human gut microbiome. Front Microbiol. 2017, 23(8),2234.
- Sheludchenko, M.S.; Huygens, F.; Stratton, H.; Hargreaves, M. CRISPR diversity in E. coli isolates from Australian animals, humans and environmental waters. PLoS One. 2015, 10(5), e0124090.
- Dong, H.; Cui, Y.; Zhang, D. CRISPR/Cas technologies and their applications in Escherichia coli. Front Bioeng Biotechnol. 2021, 9, 762676. [CrossRef]
- Kim, K.; Lee, Y.J. Relationship between CRISPR sequence type and antimicrobial resistance in avian pathogenic Escherichia coli. Vet Microbiol. 2022, 266, 109338.
- Dziuba, A.; Dzierżak, S.; Sodo, A.; Wawszczak-Kasza, M.; Zegadło, K.; Białek, J.; Zych, N.; Kiebzak, W.; Matykiewicz, J.; Głuszek, S.; Adamus-Białek, W. Comparative study of virulence potential, phylogenetic origin, CRISPR-Cas regions and drug resistance of Escherichia coli isolates from urine and other clinical materials. Front Microbiol. 2023, 14, 1289683. [CrossRef]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology. 2015, 161, 980–988.
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020, 75(12), 3491-3500.
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. 2018. 19(1), 307.
- Larsen, M.; Cosentino, S.; Rasmussen, S.; Rundsten, C.; Hasman, H.; Marvig, R.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.; Aarestrup, F.; Lund, O. Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol. 2012, 50(4), 1355-1361. [CrossRef]
- Joyce, S.; Belmont, C.; Scheffler, A.W.; Ravi, K.; Kim, H.; Rubin-Saika, N.; Elises, M.; Soto, A.; Mahesh, P. A.; Chambers, H.; Raphael, E. Trends in uropathogenic Escherichia coli genotype and antimicrobial resistance from 2019 to 2022 in a San Francisco Public Hospital Network. Open Forum Infect Dis. 2025. 12(9), ofaf579.
- Gebremedhin, K.B.; Amogne, W.; Alemayehu, H.; Bopegamage, S.; Eguale, T. The role of uropathogenic Escherichia coli virulence factors in the development of urinary tract infection. J Med Life. 2025. 18(8), 701-709.
- Nasrollahian, S.; Graham, J.P.; Halaji, M. A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli. Front Cell Infect Microbiol. 2024. 14, 1387497. [CrossRef]
- Almendros, C.; Mojica, F.J.; Díez-Villaseñor, C.; Guzmán, N.M.; García-Martínez, J. CRISPR-Cas functional module exchange in Escherichia coli. mBio. 2014. 5(1), e00767-13.
- Iordache, D.; Baci, G.M.; Căpriță, O.; Farkas, A.; Lup, A.; Butiuc-Keul, A. Correlation between CRISPR loci diversity in three enterobacterial taxa. Int J Mol Sci. 2022. 23(21), 12766. [CrossRef]
- García-Gutiérrez, E.; Almendros, C.; Mojica, F.J.M.; Guzmán, N.M.; García-Martínez, J. CRISPR content correlates with the pathogenic potential of Escherichia coli. PLoS ONE. 2015. 10, e0131935.
| Gene | var. B-8552 / var. K-12 | ||||
|---|---|---|---|---|---|
| Gene length, bp | GC-content, % | Gene QC / PI, % | Protein QC / PI, % | ||
| cas3 | 2700 / 2667 | 50 / 45 | 0 / 0 | 87 / 29 | |
| casA | 1563 / 1509 | 50 / 44 | 1 / 94 | 53 / 23 | |
| casB | 537 / 483 | 52 / 46 | 0 / 0 | 0 / 0 | |
| cas7 | 1056 / 1092 | 51 / 44 | 0 / 0 | 82 / 31 | |
| cas5 | 747 / 675 | 55 / 48 | 0 / 0 | 66 / 32 | |
| cas6 | 651 / 600 | 55 / 45 | 0 / 0 | 99 / 29 | |
| cas1 | 924 / 918 | 52 / 51 | 90 / 73 | 98 / 84 | |
| cas2 | 294 / 285 | 48 / 46 | 99 / 75 | 90 / 86 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





