Submitted:
05 September 2025
Posted:
05 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials and Treatments
2.2. Cloning of RCA Gene
2.3. Bioinformatics Analysis of RCA Gene
2.4. Measurement of H2O2 and O2•− Contents and Expression Analysis of KcRCAl and KcRCAs in K. Candel under Combined Stress Condition
2.5. Arabidopsis Transformation Assay
2.6. Flooding and Salinity Tolerance Assay of Transgenic Arabidopsis
2.7. Determination of Physiological Indexes in Transgenic Arabidopsis under Combined Flooding and Salinity Tolerance
2.8. Statistical Analysis of Data
3. Results
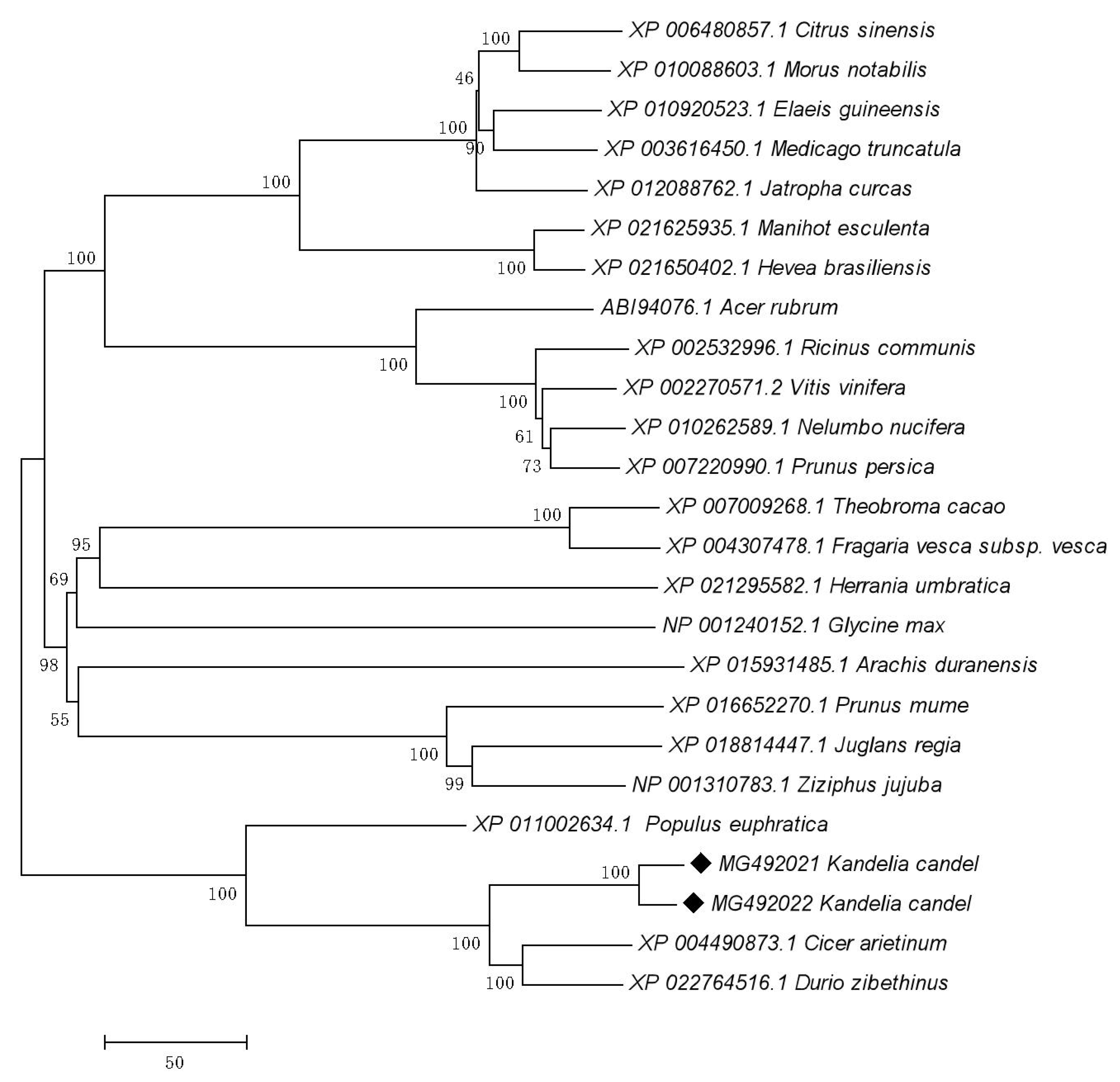
3.1. Isolation and Characterization of Two Distinct RCA Gene Isoforms in Kandelia Candel Leaves
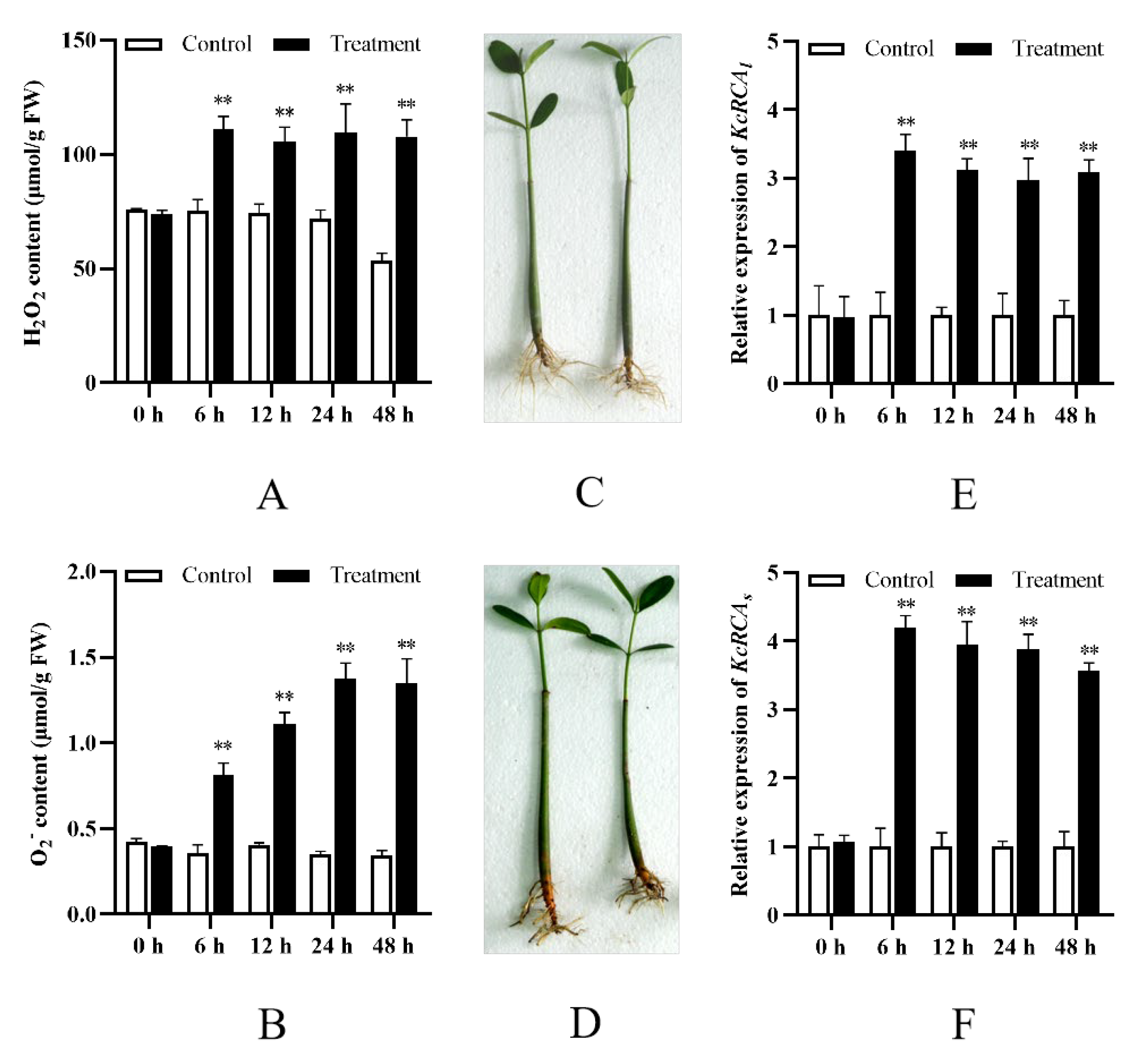
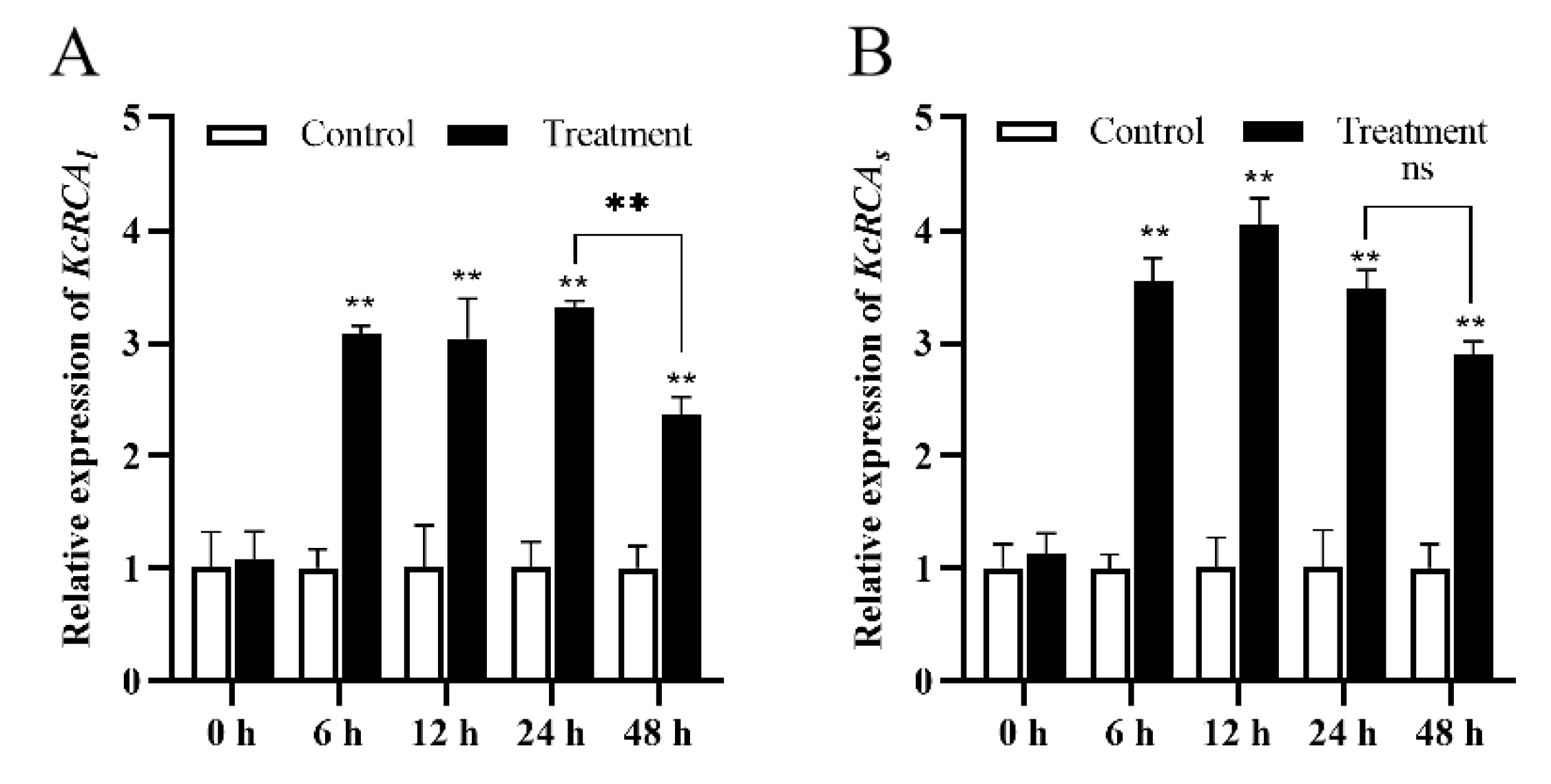
3.2. Analysis of H2O2 and O2•− Contents and KcRCA Gene Expression Levels in Kandelia candel Leaves Under Combined Stress of Flooding and Salinity
3.3. Establishment and Molecular Confirmation of Arabidopsis Transgenic Plants
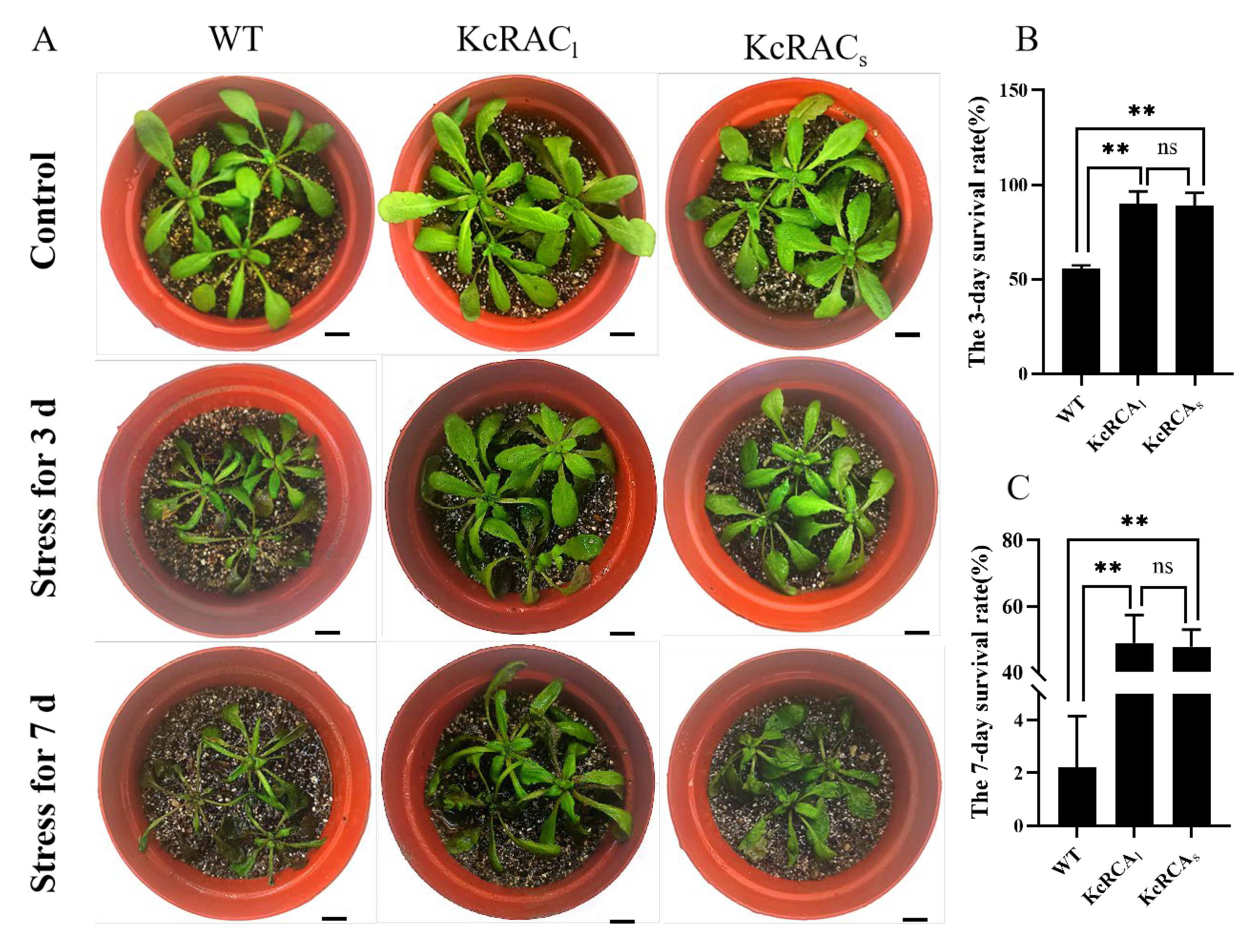
3.4. Transgenic Arabidopsis Plants Exhibited Enhanced Tolerance to Flooding and Salinity Stress
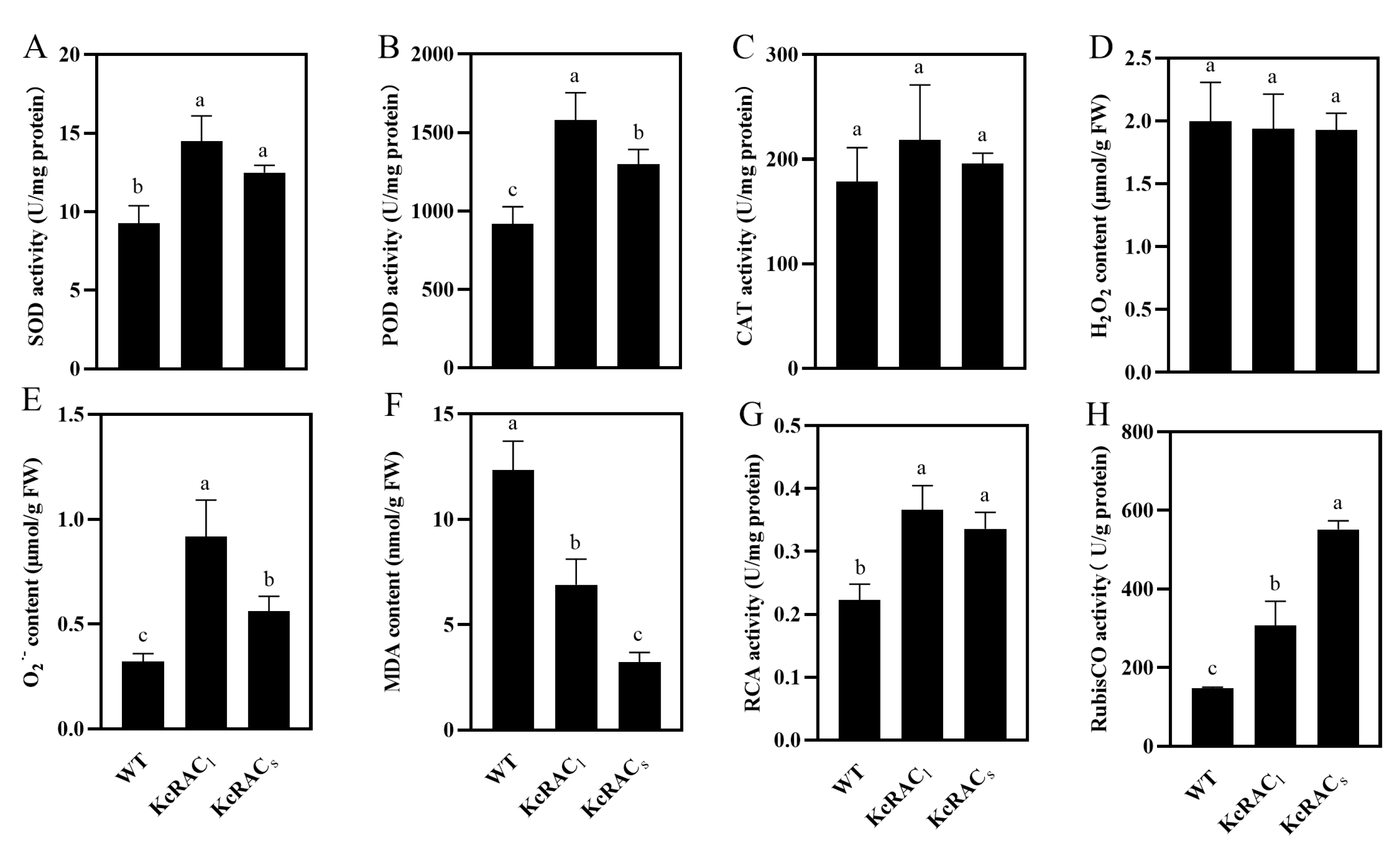
3.5. Physiological Analysis of Transgenic Arabidopsis Under the Combined Flooding and Salinity Stress
4. Discussion
4.1. Successfully Cloned Two Variable Splice Variants of RCA Gene from a Mangrove Plant
4.2. Multifunctional Regulation of RCA in the Flooding and Salinity Tolerance Physiology of Kandelia Candel
4.3. Molecular Characteristics and Functional Roles of Alternatively Spliced Isoforms of RCA in Kandelia Candel
4.4. Biotechnological Potential of RCA Gene Derived from Kandelia Candel for Enhancing Flooding and Salinity Tolerance in Crops
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Gui, G.; Zhang, Q.; Hu, W.; Liu, F. Application of multiomics analysis to plant flooding response. Front. Plant Sci. 2024, 15, 1389379. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Alam, I.; Khoso, M.A.; Ali, Q.; Liu, F. Waterlogging stress in plants: Unraveling the mechanisms and impacts on growth, development, and productivity. Environ. Exp. Bot. 2024, 224, 105824. [Google Scholar] [CrossRef]
- Ngumbi, E.N. Could flooding undermine progress in building climate-resilient crops? Trends Plant Sci. 2025, 30, 85–94. [Google Scholar] [CrossRef]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteome Res. 2012, 11, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Jhan, L.H.; Yang, C.Y.; Huang, C.M.; Lai, M.C.; Huang, Y.H.; Baiya, S.; Kao, C.F. Integrative pathway and network analysis provide insights on flooding-tolerance genes in soybean. Sci. Rep. 2023, 13, 1980. [Google Scholar] [CrossRef]
- Li, G.J.; Wei, N.; Hou, H. Uncovering the secrets of how plants adapt to water stress. Plant Cell Environ. 2025, 10. [Google Scholar] [CrossRef]
- Chakraborty, K.; Ray, S.; Vijayan, J.; Molla, K.A.; Nagar, R.; Jena, P.; Mondal, S.; Panda, B.B.; Shaw, B.P.; Swain, P.; Chattopadhyay, K.; Sarkar, R.K. Preformed aerenchyma determines the differential tolerance response under partial submergence imposed by fresh and saline water flooding in rice. Physiol. Plant. 2021, 173, 1597–1615. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.S.; Li, Y.F.; Cai, Y.H.; Xie, Y.H.; Deng, Z.M.; Li, F.; Hou, Z.Y. Differential strategies to tolerate flooding in polygonum hydropiper plants originating from low- and high-elevation habitats. Front. Plant Sci. 2019, 9, 1970. [Google Scholar] [CrossRef]
- Silveira, J.A.G.; Carvalho, F.E.L. Proteomics, photosynthesis and salt resistance in crops: An integrative view. J. Proteomics. 2016, 143, 24–35. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, Z.; Shen, X.; Liu, D.; Pedersen, O. Flooding-adaptive root and shoot traits in rice. Funct. Plant Biol. 2024, 51, FP23226. [Google Scholar] [CrossRef]
- Qi, M.; Liu, X.; Li, Y.; Song, H.; Yin, Z.; Zhang, F.; He, Q.; Xu, Z.; Zhou, G. Photosynthetic resistance and resilience under drought, flooding and rewatering in maize plants. Photosynth Res. 2021, 148, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Wang, L.; Tan, F.; Lu, S.; Lv, X.; Zaynab, M.; Cheng, C.L.; Abubakar, Y. S; Chen, S; Chen, W. Phosphoproteomics unveils stable energy supply as key to flooding tolerance in Kandelia candel. J. Proteomics. 2018, 176, 1–12. [Google Scholar] [CrossRef]
- Guo, Z.; Ma, D.; Li, J.; Wei, M.; Zhang, L.; Zhou, L.; Zhou, X.; He, S.; Wang, L.; Shen, Y.; Li, Q.Q.; Zheng, H.L. Genome-wide identification and characterization of aquaporins in mangrove plant Kandelia obovata and its role in response to the intertidal environment. Plant Cell Environ. 2022, 45, 1698–1718. [Google Scholar] [CrossRef]
- Wang, L.; Liang, W.; Xing, J.; Tan, F.; Chen, Y.; Huang, L.; Cheng, C.L.; Chen, W. Dynamics of chloroplast proteome in salt-stressed mangrove Kandelia candel (L.) Druce. J. Proteome Res. 2013, 12, 5124–36. [Google Scholar] [CrossRef]
- Nowicka, B.; Ciura, J.; Szymańska, R.; Kruk, J. Improving photosynthesis, plant productivity and abiotic stress tolerance-current trends and future perspectives. J. Plant Physiol. 2018, 231, 415–433. [Google Scholar] [CrossRef]
- Portis, Jr.A.R. Rubisco activase-Rubisco's catalytic chaperone. Photosynth Res. 2003, 75, 11–27. [Google Scholar] [CrossRef]
- Sharkey, T.D. The discovery of rubisco. J. Exp. Bot. 2023, 74, 510–519. [Google Scholar] [CrossRef]
- Portis, Jr.A.R.; Li, C.; Wang, D.; Salvucci, M.E. Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 2008, 59, 1597–604. [Google Scholar] [CrossRef]
- Hayer-Hartl, M.; Hartl, F.U. Chaperone machineries of Rubisco-the most abundant enzyme. Trends Biochem. Sci. 2020, 45, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Bhat, J.Y.; Thieulin-Pardo, G.; Hartl, F.U.; Hayer-Hartl, M. Rubisco activases: AAA+ Chaperones adapted to enzyme repair. Front. Mol. Biosci. 2017, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Werneke, J.M.; Chatfield, J.M.; Ogren, W.L. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell. 1989, 1, 815–25. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Stessman, D.J.; Wright, D.A.; Spalding, M.H.; Huber, S.C.; Ort, D.R. Arabidopsis plants expressing only the redox-regulated Rca-α isoform have constrained photosynthesis and plant growth. Plant J. 2020, 103, 2250–2262. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, X.; Zhou, Z.; Feng, X.; Yang, W.; Jiang, D. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiol. plantarum. 2010, 139, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.M.; Cavanagh, A.P.; Kim, S.Y.; Wright, D.A.; Edquilang, R.G.; Shreeves, K.S.; Perdomo, J.A.; Spalding, M.H.; Ort, D.R.; Bernacchi, C.J.; Huber, S.C. Removal of redox-sensitive Rubisco activase does not alter Rubisco regulation in soybean. Photosynth. Res. 2022, 154, 169–182. [Google Scholar] [CrossRef]
- Leitao, L.; Maoret, J.J.; Biolley, J.P. Changes in PEP carboxylase, rubisco and rubisco activase mRNA levels from maize (Zea mays) exposed to a chronic ozone stress. Biol. Res. 2007, 40, 137–53. [Google Scholar] [CrossRef]
- Khairy, A.I.H.; Oh, M.J.; Lee, S.M.; Kim, D.S.; Roh, K.S. Nitric oxide overcomes Cd and Cu toxicity in in vitro-grown tobacco plants through increasing contents and activities of Rubisco and Rubisco activase. Biochim. Open. 2016, 2, 41–51. [Google Scholar] [CrossRef]
- Bi, H.; Liu, P.; Jiang, Z.; Ai, X. Overexpression of the rubisco activase gene improves growth and low temperature and weak light tolerance in Cucumis sativus. Physiol. Plant. 2017, 161, 224–234. [Google Scholar] [CrossRef]
- Li, X.; Yue, H.; Chu, Y.; Jia, Y. Comparative transcriptomes reveal molecular mechanisms of apple blossoms of different tolerance genotypes to chilling injury. Open Life Sci. 2023, 18, 20220613. [Google Scholar] [CrossRef]
- Carmo-Silva, A.E.; Salvucci, M.E. The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol. 2013, 161, 1645–55. [Google Scholar] [CrossRef]
- Nazari, M.; Kordrostami, M.; Ghasemi-Soloklui, A.A.; Eaton-Rye, J.J.; Pashkovskiy, P.; Kuznetsov, V.; Allakhverdiev, S.I. Enhancing photosynthesis and plant productivity through genetic modification. Cells. 2024, 13, 1319. [Google Scholar] [CrossRef]
- Wijewardene, I.; Shen, G.; Zhang, H. Enhancing crop yield by using Rubisco activase to improve photosynthesis under elevated temperatures. Stress Biol. 2021, 1, 2. [Google Scholar] [CrossRef]
- Sparrow-Muñoz, I.; Chen, T.C.; Burgess, S.J. Recent developments in the engineering of Rubisco activase for enhanced crop yield. Biochem. Soc. Trans. 2023, 51, 627–637. [Google Scholar] [CrossRef]
- Gunn, L.H.; Martin, Avila, E.; Birch, R.; Whitney, S.M. The dependency of red Rubisco on its cognate activase for enhancing plant photosynthesis and growth. Proc. Natl. Acad. Sci. USA. 2020, 117, 25890–25896. [CrossRef]
- Jurczyk, B.; Hura, K.; Trzemecka, A.; Rapacz, M. Evidence for alternative splicing mechanisms in meadow fescue (Festuca pratensis) and perennial ryegrass (Lolium perenne) Rubisco activase gene. J. Plant Physiol. 2015, 176, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Wang, L.; Chen, S.; Lv, X.; Lu, S.; Cheng, C.L.; Tan, F.; Chen, W. Protein acetylation as a mechanism for Kandelia candel's adaption to daily flooding. Tree Physiol. 2018, 38, 895–910. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Liang, M.; Tan, F.; Liang, W.; Chen, Y.; Lin, Y.; Huang, L.; Xing, J.; Chen, W. Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-term stress. PLoS One. 2014, 9, e83141. [Google Scholar] [CrossRef]
- Wang, L.; Pan, D.; Lv, X.; Cheng, C.L.; Li, J.; Liang, W.; Xing, J.; Chen, W. A multilevel investigation to discover why Kandelia candel thrives in high salinity. Plant Cell Environ. 2016, 39, 2486–2497. [Google Scholar] [CrossRef]
- Jambunathan, N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol. Biol. 2010, 639, 292–8. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–43. [Google Scholar] [CrossRef]
- Park, S.I.; Kim, J.J.; Shin, S.Y.; Kim, Y.S.; Yoon, H.S. ASR Enhances environmental stress tolerance and improves grain yield by modulating stomatal closure in rice. Front. Plant Sci. 2020, 10, 1752. [Google Scholar] [CrossRef]
- Renziehausen T, Frings S, Schmidt-Schippers R. 'Against all floods': plant adaptation to flooding stress and combined abiotic stresses. Plant J. 2024, 117, 1836–1855. [CrossRef]
- Liu S, Yang S, Liu H, Hu Q, Liu X, Wang J, Wang J, Xin W, Chen Q. Physiological and transcriptomic analysis of the mangrove species Kandelia obovata in response to flooding stress. Mar. Pollut. Bull. 2023, 196:115598. [CrossRef]
- Karthick, P.V.; Senthil, A.; Djanaguiraman, M.; Anitha, K.; Kuttimani, R.; Boominathan, P.; Karthikeyan, R.; Raveendran, M. Improving crop yield through increasing carbon gain and reducing carbon loss. Plants. 2024, 13, 1317. [Google Scholar] [CrossRef] [PubMed]
- Prywes, N.; Phillips, N. R.; Tuck, O. T.; Valentin-Alvarado, L.E.; Savage, D.F. Rubisco function, evolution, and engineering. Annu. Rev. Biochem. 2023, 92, 385–410. [Google Scholar] [CrossRef] [PubMed]
- Waheeda, K.; Kitchel, H.; Wang, Q.; Chiu, P.L. Molecular mechanism of Rubisco activase: dynamic assembly and Rubisco remodeling. Front. Mol. Biosci. 2023, 10, 1125922. [Google Scholar] [CrossRef]
- Nagarajan, R.; Gill, K.S. Evolution of Rubisco activase gene in plants. Plant Mol. Biol. 2018, 96, 69–87. [Google Scholar] [CrossRef]
- Yin, Z.; Meng, F.; Song, H.; Wang, X.; Xu, X.; Yu, D. Expression quantitative trait loci analysis of two genes encoding Rubisco activase in soybean. Plant Physiol. 2010, 152, 1625–1637. [Google Scholar] [CrossRef]
- Chen, Y; Wang, X.M.; Zhou, L.; He, Y.; Wang, D.; Qi, Y.H.; Jiang, D.A. Rubisco activase is also a multiple responder to abiotic stresses in rice. PLoS One. 2015, 10, e0140934.
- Jiang, Z.; Sun, X.; Ai, X.; Wang, M.; Bi, H.; Wang, H. Responses of Rubisco and Rubisco activase in cucumber seedlings to low temperature and weak light. Ying Yong Sheng Tai Xue Bao. 2010, 21, 2045–2050. (in Chinese). [Google Scholar]
- Somkuwar, R.G.; Dhole, A.M. Somkuwar, R.G.; Dhole, A.M. Understanding the photosynthesis in relation to climate change in grapevines. Theor. Biosci. 2025, 10.1007/s12064-025-00435-w. Advance online publication. [CrossRef] [PubMed]
- Amaral, J.; Lobo, A.K.M.; Carmo-Silva, E. Regulation of Rubisco activity in crops. New Phytol, 2024, 241, 35–51. [Google Scholar] [CrossRef]
- Ma, L.; Liu, C.; Qu, C.; Yin, S.; Liu, J.; Gao, F.; Hong, F. Rubisco activase mRNA expression in spinach: modulation by nanoanatase treatment. Biol. Trace Elem. Res. 2008, 122, 168–78. [Google Scholar] [CrossRef]
- Aliakbari, M.; Cohen, S.P.; Lindlöf, A.; Shamloo-Dashtpagerdi, R. Rubisco activase A (RcaA) is a central node in overlapping gene network of drought and salinity in Barley (Hordeum vulgare L.) and may contribute to combined stress tolerance. Plant Physiol. Bioch. 2021, 161, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Suárez, M.; Ayala-Ochoa, A.; Lozano-Franco, J.; García-Torres, I.; Díaz-Quiñonez, A.; Ortíz-Navarrete, V.F.; Sánchez-de-Jiménez, E. Rubisco activase chaperone activity is regulated by a post-translational mechanism in maize leaves. J. Exp. Bot. 2004, 55, 2533–2539. [Google Scholar] [CrossRef]
- Salvucci, M. E.; Van de Loo F., J.; Stecher, D. Two isoforms of Rubisco activase in cotton, the products of separate genes not alternative splicing. Planta. 2003, 216, 736–744. [Google Scholar] [CrossRef]
- Carmo-Silva, A. E.; Salvucci, M.E. The activity of Rubisco's molecular chaperone, Rubisco activase, in leaf extracts. Photosynth Res. 2011, 108, 143–155. [Google Scholar] [CrossRef]
- Feng, Y.; Wu, H.; Liu, H.; He, Y.; Yin, Z. Effects of OsRCA overexpression on Rubisco activation state and photosynthesis in maize. Plants. 2023, 12, 1614. [Google Scholar] [CrossRef] [PubMed]
- Wijewardene, I.; Mishra, N.; Sun, L.; Smith, J.; Zhu, X.; Payton, P.; Shen, G.; Zhang, H. Improving drought-, salinity-, and heat-tolerance in transgenic plants by co-overexpressing Arabidopsis vacuolar pyrophosphatase gene AVP1 and Larrea Rubisco activase gene RCA. Plant Sci. 2020, 296, 110499. [Google Scholar] [CrossRef]
- Suganami, M.; Suzuki, Y.; Tazoe, Y.; Yamori, W.; Makino, A. Co-overproducing Rubisco and Rubisco activase enhances photosynthesis in the optimal temperature range in rice. Plant physiol. 2021, 185, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Masumoto, C.; Fukayama, H.; Makino, A. Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature. Plant J. 2012, 71, 871–880. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




