Submitted:
03 September 2025
Posted:
04 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacteria Preparation
2.2. Media Preparation
2.3. Starter Culture Preparation
2.4. Bioreactor Procedure
2.5. Polymer Extraction
2.6. Solvent Cast Film Production
2.7. Thermal Analysis
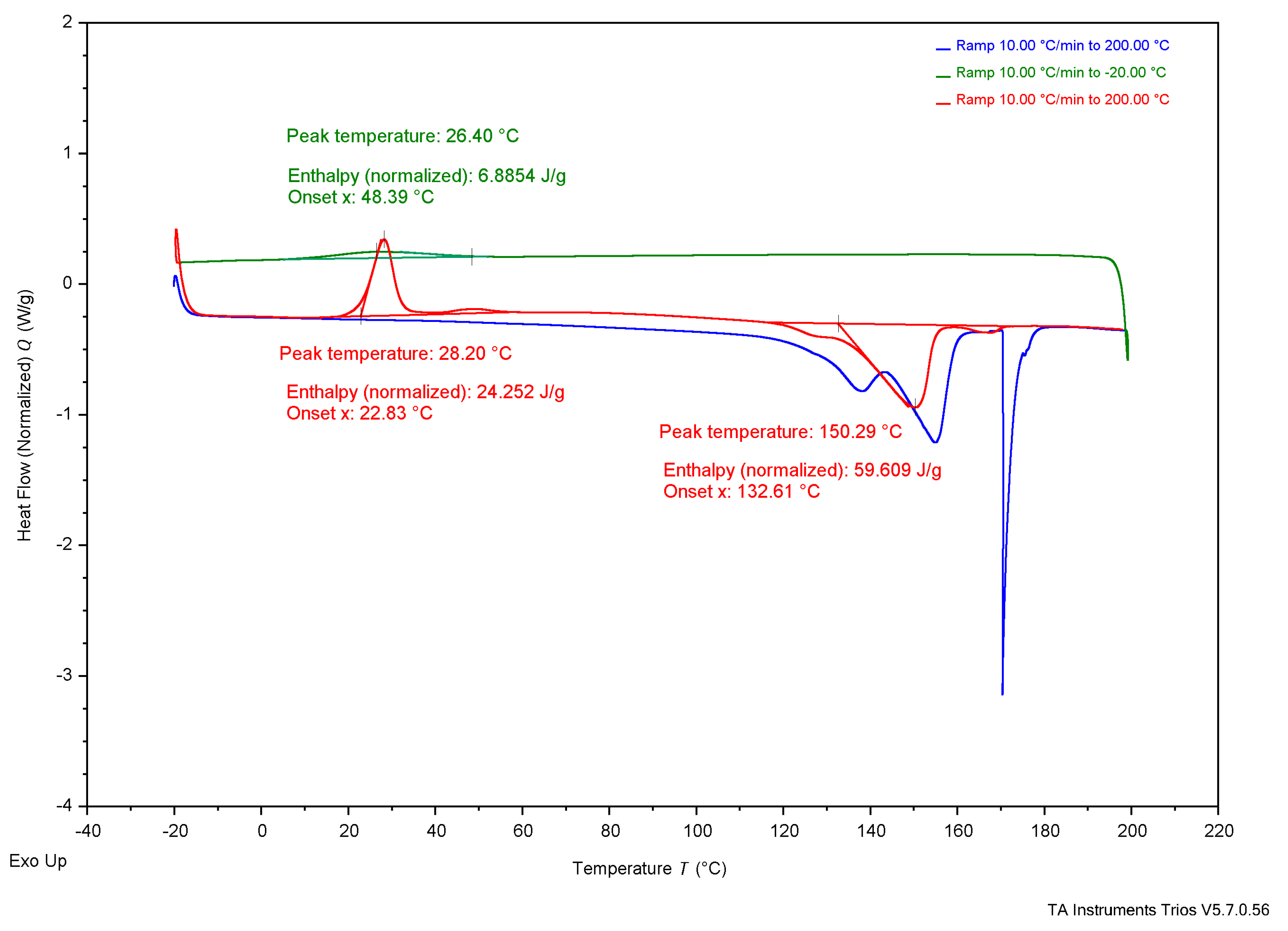
3. Results
3.1. Confirmation of R. eutropha in Culture
3.2. P3HB Yields
3.3. Extraction and Film Casting

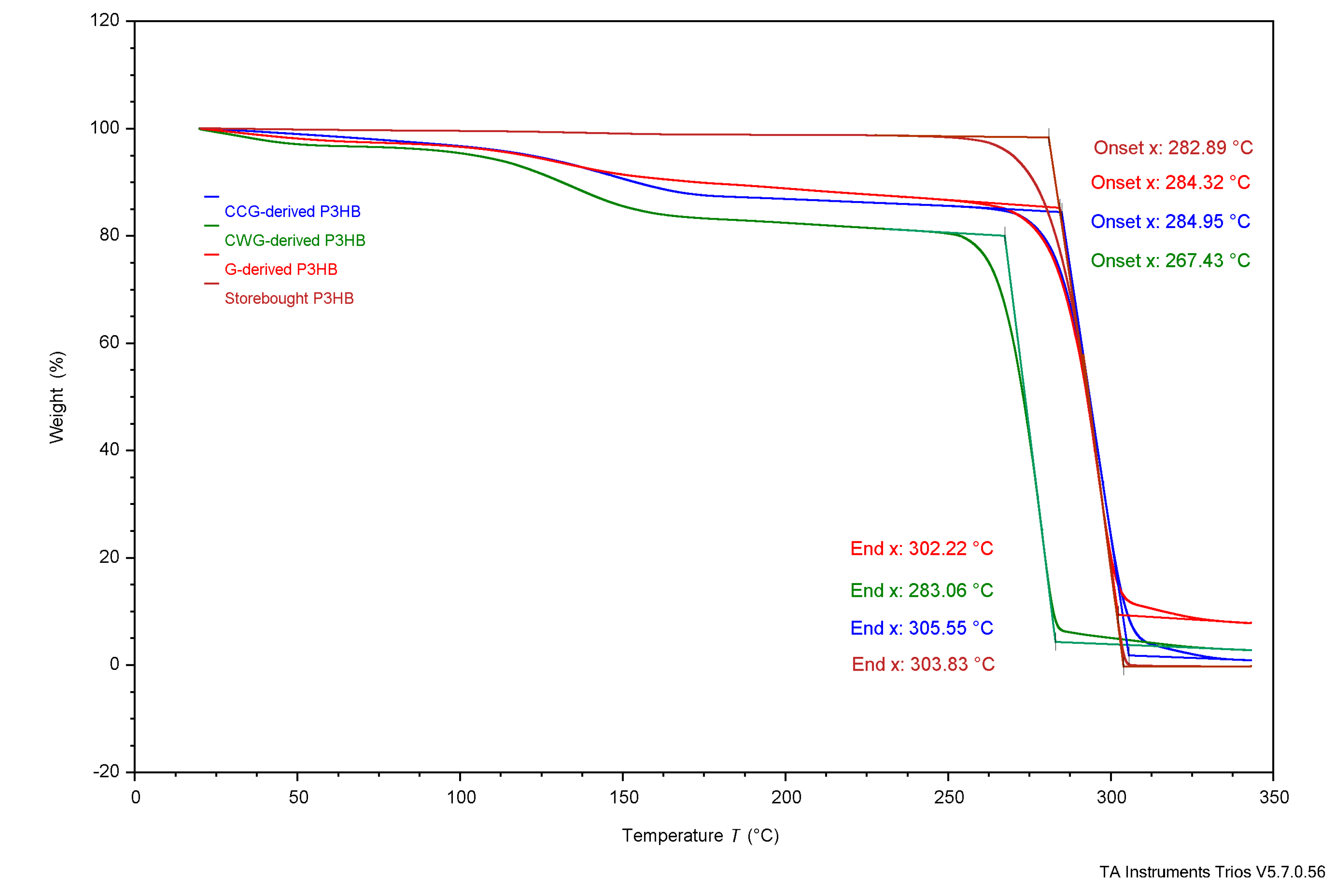
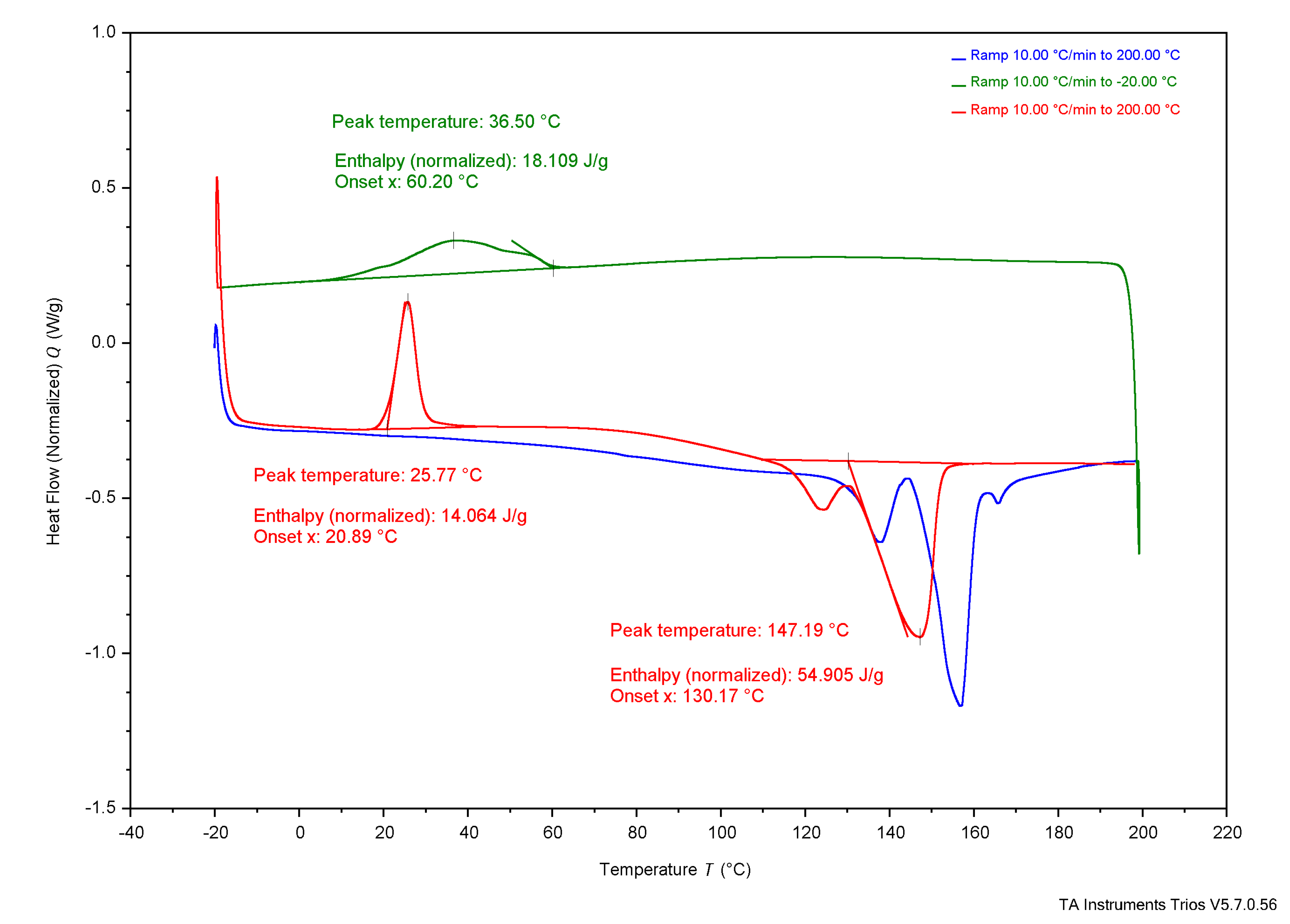
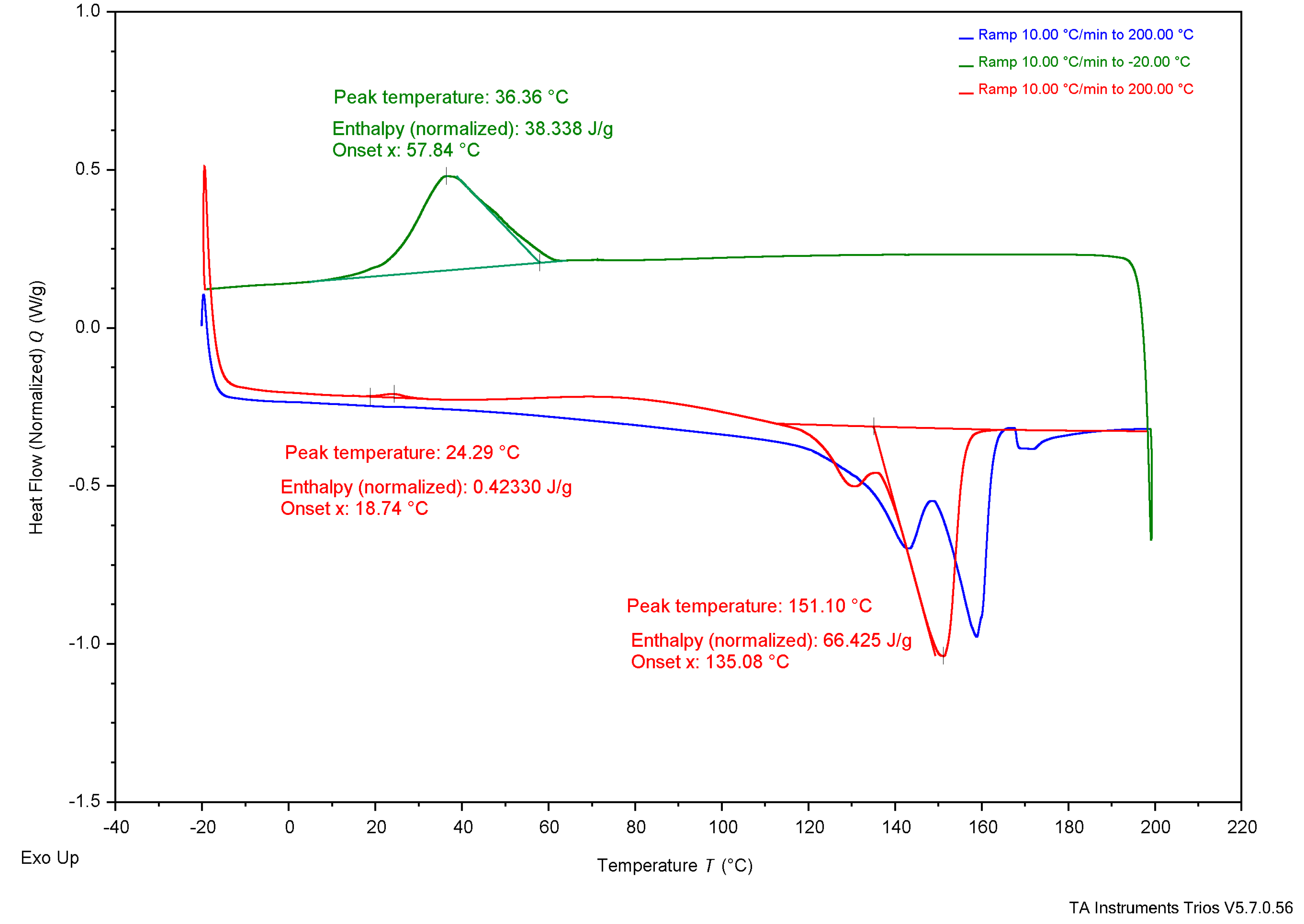
3.4. Thermal Analysis
4. Discussion
4.1. Methodology Success
4.2. P3HB Yields
4.3. Thermal Analysis
4.3.1. Experimental Samples Compared to Literature
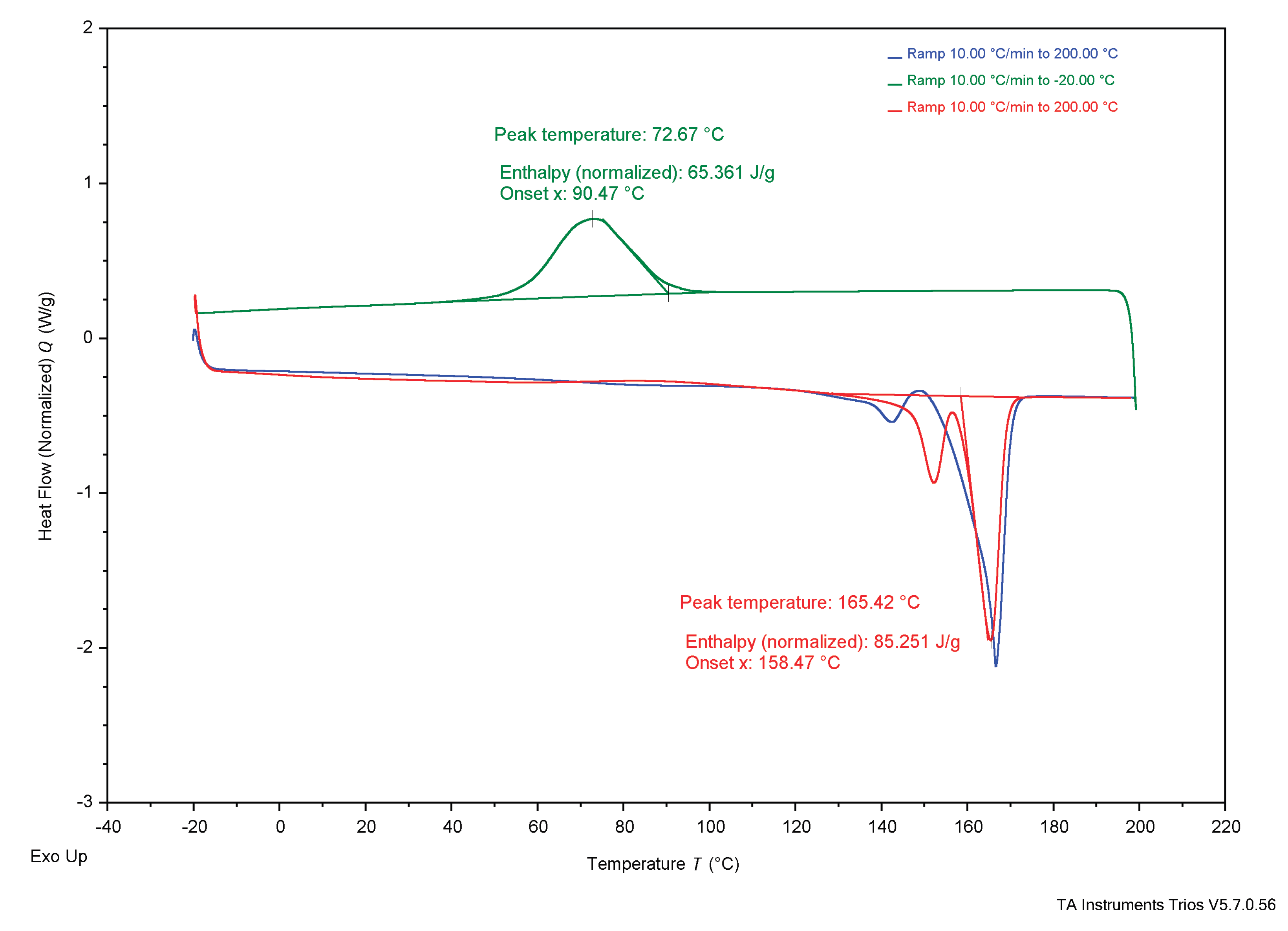
4.3.2. Comparing Experimental Samples
4.4. Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| P3HB | Poly-3-hydroxybutyrate |
| G | Glucose, referring to commercial glucose |
| CCG | Cotton glucose |
| CWG | Cutting waste cotton glucose |
References
- Thunman, H., Berdugo Vilches, T., Seemann, M., Maric, J., Cañete Vela, I., Pissot, S., N.T. Nguyen, H. (2019). Circular use of plastics-transformation of existing petrochemical clusters into thermochemical recycling plants with 100% plastics recovery. Sustainable Materials and Technologies, 22, e00124, ISSN 2214-9937. [CrossRef]
- U.S. Environmental Protection Agency. (2020). Advancing Sustainable Materials Management: Facts and Figures Report. https://www.epa.gov/facts-and-figures-about-materials-waste-and-recycling/national-overview-facts-and-figures-materials.
- Parashar, N., & Hait, S. (2021). Plastics in the time of COVID-19 pandemic: Protector or polluter?. The Science of the total environment, 759, 144274. [CrossRef]
- Barnes, D. K., Galgani, F., Thompson, R. C., & Barlaz, M. (2009). Accumulation and fragmentation of plastic debris in global environments. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 364(1526), 1985–1998. [CrossRef]
- Mannina, G., Presti, D., Montiel-Jarillo, G., & Eugenia Suárez-Ojeda, M. (2019). Bioplastic recovery from wastewater: A new protocol for polyhydroxyalkanoates (PHA) extraction from mixed microbial cultures. Bioresource Technology, 282, 361-369, ISSN 0960-8524. [CrossRef]
- World Economic Forum, Ellen MacArthur Foundation, and McKinsey & Company. (2016). The New Plastics Economy: Rethinking the future of plastics. Retrieved from https://www.ellenmacarthurfoundation.org/the-new-plastics-economy-rethinking-the-future-of-plastics.
- Rhodes, C. J. (2018). Plastic pollution and potential solutions. Science progress, 101(3), 207–260. [CrossRef]
- Bhatia, S. K. , Otari, S. V., Jeon, J. M., Gurav, R., Choi, Y. K., Bhatia, R. K., Pugazhendhi, A., Kumar, V., Rajesh Banu, J., Yoon, J. J., Choi, K. Y., & Yang, Y. H. (2021). Biowaste-to-bioplastic (polyhydroxyalkanoates): Conversion technologies, strategies, challenges, and perspective. Bioresource technology, 326, 124733. doi.org/10.1016/j.biortech.2021.124733.
- Costa, A., Encarnação, T., Tavares, R., Todo Bom, T., & Mateus, A. (2023). Bioplastics: Innovation for Green Transition. Polymers, 15(3), 517.
- Khattab, M. M. , & Dahman, Y. (2019). Production and recovery of poly-3-hydroxybutyrate bioplastics using agro-industrial residues of hemp hurd biomass. Bioprocess and biosystems engineering, 42(7), 1115–1127. [CrossRef]
- Kovalcik, A., Pernicova, I., Obruca, S., Szotkowski, M., Enev, V., Kalina, M., & Marova, I. (2020). Grape winery waste as a promising feedstock for the production of polyhydroxyalkanoates and other value-added products. Food and Bioproducts Processing, vol. 124, pp. 1-10, ISSN 0960-3085. [CrossRef]
- Rysbek, A., Ramankulov, Y., Kurmanbayev, A., Richert, A., & Abeldenov, S. (2022). Comparative Characterization and Identification of Poly-3-hydroxybutyrate Producing Bacteria with Subsequent Optimization of Polymer Yield. Polymers, 14(2), 335. [CrossRef]
- Wang, Y., Chen, R., Cai, J., Liu, Z., Zheng, Y., Wang, H., Li, Q., et al. (2013). Biosynthesis and thermal properties of PHBV produced from levulinic acid by Ralstonia eutropha. Plos One, 8(4), e60318.
- Markets and Markets (2024) Polyhydroxyalkanoate (PHA) Market - Global Forecast to 2028. MarketsandMarkets. Retrieved from https://www.marketsandmarkets.com/Market-Reports/pha-market-395.html.
- Johnson S., Echeverria D., Venditti R., Jameel H., & Yao Y. (2020). Supply Chain of Waste Cotton Recycling and Reuse: A Review. AATCC Journal of Research, 7(1_suppl):19-31. [CrossRef]
- Cotton Incorporated. (2024). Monthly Economic Letter: Cotton Market Fundamentals and Price Outlook. Cotton Incorporated. from https.
- Vera, R. , Zambrano, F., Suarez, A., Pifano, A., Marquez, R., Farrell, M., Ankeny, M., Jameel, H., Gonzalez, R. (2022). Transforming textile wastes into biobased building blocks via enzymatic hydrolysis: A review of key challenges and opportunities. Cleaner and Circular Bioeconomy. 100026. 10.1016/j.clcb.2022.100026.
- Murawski de Mello, A., Vandenberghe, L., Wedderhoff Herrmann, L., Letti, L., Martínez Burgos, W., Scapini, T., Manzoki, M., Zwiercheczewski, P., & Soccol, C. (2023). Strategies and engineering aspects on the scale-up of bioreactors for different bioprocesses. Systems Microbiology and Biomanufacturing. 4. [CrossRef]
- Koller, M., Niebelschütz, H. and Braunegg, G. (2013), Strategies for recovery and purification of poly[(R)-3-hydroxyalkanoates] (PHA) biopolyesters from surrounding biomass. Eng. Life Sci., 13: 549-562. [CrossRef]
- Ramsay, J.A., Berger, E., Voyer, R. et al. Extraction of poly-3-hydroxybutyrate using chlorinated solvents. Biotechnol Tech, 8, 589–594 (1994). [CrossRef]
- Kourmentza, C., Plácido, J., Venetsaneas, N., Burniol-Figols, A., Varrone, C., Gavala, H. N., and Reis, M. A. M. (2017) Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering (Basel), 4.
- Azenta Life Sciences. (2024). Fioravanti4607-Taxonomy.
- Jefferson Institute for Bioprocessing. (2024). Jefferson Institute for Bioprocessing. Available online: https://www.jeffersonbioprocessing.com/.
- De Sousa Junior,. R. R., Cezario, F. E. M., Antonino, L. D., Dos Santos, D. J., & Lackner, M. (2023). Characterization of Poly(3-hydroxybutyrate) (P3HB) from Alternative, Scalable (Waste) Feedstocks. Bioengineering (Basel, Switzerland), 10(12), 1382. [CrossRef]
- García-Cerna, et al. (2022). Evaluation of Poly-3-Hydroxybutyrate (P3HB) Scaffolds Used for Epidermal Cells Growth as Potential Biomatrix. Polymers, 14(19), 4021. [CrossRef]
- Janigová, I., Lacı́k, I., & Chodák, I. (2002). Thermal degradation of plasticized poly(3-hydroxybutyrate) investigated by DSC. Polymer Degradation and Stability, 77, 1, 35-41, ISSN 0141-3910. [CrossRef]
- Lee, S.Y. (1996). Review bacterial polyhydroxyalkanoates. Biotechnol Bioeng, 49, 1–14. [CrossRef]
- Sudesh, K. et al. (2000). Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Progress in Polymer Science, 25, 1503-1555. [CrossRef]
| Growth Medium | Control | G | CCG | CWG |
|---|---|---|---|---|
| Nutrient Broth | 5 mL (100% v/v) | 2.5 mL (50% v/v) | 2.5 mL (50% v/v) | 2.5 mL (50% v/v) |
| MSM 1 (5x concentration) | - | 0.25 mL (5% v/v) | 0.25 mL (5% v/v) | 0.25 mL (5% v/v) |
| MSM 2 (5x concentration) | - | 0.25 mL (5% v/v) | 0.25 mL (5% v/v) | 0.25 mL (5% v/v) |
| Commercial Glucose Solution (7.4% v/w glucose) | - | 0.625 mL (12.5% v/v) |
- | - |
| Cotton Glucose Solution (8.93% v/w glucose) |
- | - | 0.518 mL (10.36% v/v) |
- |
| Cutting Waste Cotton Glucose Solution (9.06% v/w glucose) |
- | - | - | 0.510 mL (10.2% v/v) |
| diH2O | - | 1.375 mL (27.5% v/v) |
1.482 mL (29.64% v/v) |
1.49 mL (29.8% v/v) |
| Growth Medium | G | CCG | CWG |
|---|---|---|---|
| MSM 1 (5x concentration) | 95 mL (9.5% v/v) | 95 mL (9.5% v/v) | 95 mL (9.5% v/v) |
| MSM 2 (5x concentration) | 95 mL (9.5% v/v) | 95 mL (9.5% v/v) | 95 mL (9.5% v/v) |
| Commercial Glucose Solution (7.4% v/w glucose) | 498.75 mL (49.875% v/v) |
- | - |
| Cotton Glucose Solution (8.93% v/w glucose) |
- | 413.3 mL (41.33% v/v) |
- |
| Cutting Waste Cotton Glucose Solution (9.06% v/w glucose) |
- | - | 407.368 mL (40.737% v/v) |
| diH2O | 261.25 mL (26.125% v/v) |
346.7 mL (34.67% v/v) |
352.632 mL (35.263% v/v) |
| Starter Culture Inoculum | 50 mL (5% v/v) | 50 mL (5% v/v) | 50 mL (5% v/v) |
| Glucose Source | Run | Cell Dry Weight (CDW) (g)* |
P3HB Yield (g) |
P3HB Yield (%)** |
|---|---|---|---|---|
| Commercial Glucose (G) | G1 | 5.86 g | 0.10 g | 1.69% |
| G2 | 5.05 g | 0.03 g | 0.59% | |
| G3 | 6.54 g | 0.06 g | 0.92% | |
| G4 | 3.81 g | 0.10 g | 2.62% | |
| G5 | 4.50 g | 0.03 g | 0.67% | |
| Mean ± Std. Dev. | - | - | 1.30 ± 0.86% | |
| Control Cotton Glucose (CCG) | CCG1 | 3.07 g | 1.14 g | 38.5% |
| CCG2 | 6.4 g | 1.09 g | 17.0% | |
| Mean ± Std. Dev. | - | - | 27.8 ± 15.2% | |
| Cutting Waste Cotton Glucose (CWG) | CWG1 | 3.21 g | 0.9 g | 28.0% |
| CWG2 | 6.05 g | 0.82 g | 13.6% | |
| CWG3 | 9.41 g | 0.78 g | 8.29% | |
| Mean ± Std. Dev. | - | - | 16.6 ± 10.2% |
| Test | Property | G-derived P3HB | CCG- derived P3HB |
CWG- derived P3HB |
Purchased P3HB | Literature Source | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wang [13] |
Mannina [5] | Lee [27] |
Sudesh [28] | Rysbek [12] | ||||||
| TGA | Tonset | 284°C | 285°C | 267°C | 283°C | 263.4°C | 185°C | 200°C | - | - |
| Tend | 302°C | 306°C | 283°C | 304°C | 269.8°C | 267°C | - | - | - | |
| DSC | Tg | - | - | - | - | - | - | 5°C | 4°C | - |
| Tc* | 37°C | 36°C | 26°C | 73°C | - | - | - | 50-60°C | - | |
| Tcc* | 26°C | 24°C | 28°C | - | - | - | - | - | - | |
| Tm* | 147°C | 151°C | 150°C | 165°C | 172.05°C | - | 175°C | 180°C | 170°C | |
| Property | G-derived P3HB | CCG- derived P3HB |
CWG- derived P3HB |
Purchased P3HB | Wang [13] |
|---|---|---|---|---|---|
| ΔHc J/g | 18 J/g | 38 J/g | 6.9 J/g | 65 J/g | - |
| Xc (%) | 12.3% | 26.0% | 4.7% | 44.5% | - |
| ΔHcc J/g | 14 J/g | 0.42 J/g | 24 J/g | - | - |
| Xcc (%) | 9.6% | 0.3% | 16.4% | - | - |
| ΔHm J/g | 55 J/g | 66 J/g | 60 J/g | 85 J/g | 89.7 J/g |
| Xm (%) | 37.7% | 45.2% | 41.1% | 58.2% | 61.4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





