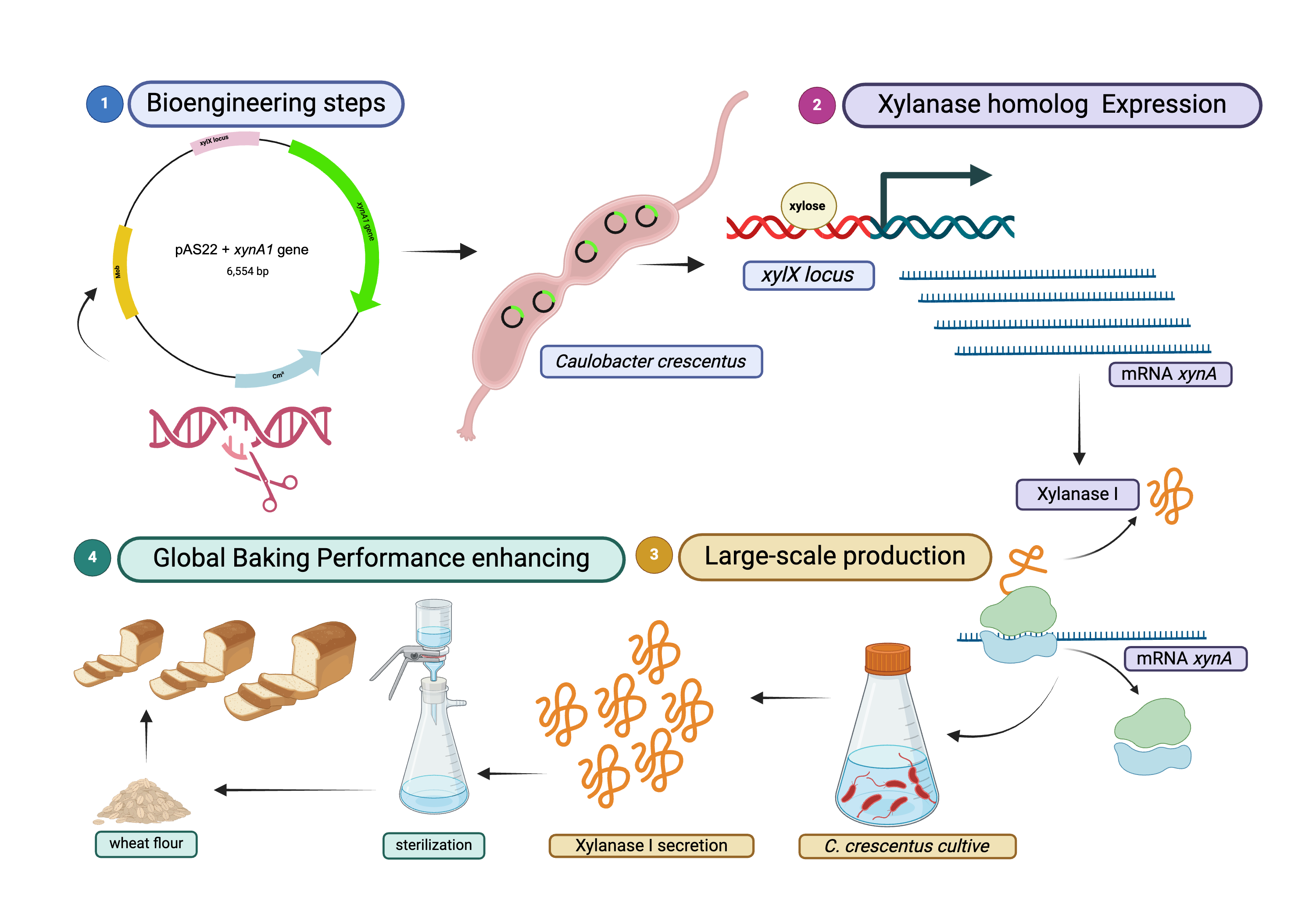
1. Introduction
Xylan is a representative part of the lignocellulosic mass in plants, and its degradation involves the synergistic action of different enzymes, with endo-β,4-D-xylanases (E.C.3.2.1.8) and
β-D-xylosidases (E.C. 3.2.1.37) being the most important [
1]. Endo-β-1,4-D-xylanases cleave the xylan main chain, producing xylo-oligosaccharides that can be converted to xylose by β-D-xylosidases [
2]. In many biotechnological and industrial applications, bacterial xylanases have shown better performance than fungal xylanases [
3,
4,
5]. The significant differences found were low cellulase activity, low cellulase stability, low cellulase activity in the alkaline range, and high cellulase thermostability, among others [
6,
7]. Moreover, superxylanases are very difficult to produce when all these features are combined, implying that most of the available bacterial xylanases have only one or two of these properties, leading industrial processes to depend on expensive and hazardous chemical processes or strategies for designing or engineering superenzymes [
6,
8].
Xylanases have been widely used in the baking industry for decades. They are employed in baking together with α-amylase, maleate amylase, glucose oxidase and proteases. Xylanases breakdown hemicellulose in wheat flour, helping to redistribute water and make the dough softer and easier to knead. During the bread baking process, they generate greater flexibility and elasticity, allowing the dough to grow. The use of xylanases in baking assures an increase in bread volume, better water absorption and greater resistance to fermentation [
9,
10]. In the manufacture of cookies and crackers, xylanase is recommended to make them creamy and lighter and to improve their texture, palatability, and uniformity [
1].
Xylanases are widely used as processing aids in the grain milling industry to produce flour. In this industry, xylanases are often referred to as hemicellulases or pentosanases. Thus, the term hemicellulase refers to the ability of xylanase to hydrolyze insoluble nonstarch compounds found in flour, while the term pentosanase indicates that the substrate for the xylan enzyme is composed of pentose monomers [
11]. Therefore, xylanases can improve the quality of the baking process and bread dough. In
Bacillus sp., for instance, the
xynHB gene encoding xylanase was cloned and inserted into an antibiotic-free vector and integrated into the
Saccharomyces cerevisiae genome. The yeast strain A13 overexpressing xylanase was applied in baking and showed a reduction in the time required for kneading and an increase in bread dough height and diameter [
8].
Recently, a xylanase-producing fungus,
Phoma sp. MF13, was isolated from mangrove sediment, and the xylanase gene (
xynMF13A) was cloned from this fungus and expressed successfully in
Pichia pastoris. The resulting expressed protein was subsequently purified and characterized. The results demonstrated that XynMF13A is a salt-tolerant enzyme with a high degree of salt resistance up to 4 M NaCl. The hydrolysis products generated from corncob xylan by this xylanase include xylobiose, xylotriose, xylotetraose, and xylopentaose. Furthermore, recombinant xylanase (XynMF13A) had a beneficial effect on Chinese steamed bread, a traditional food, by improving its specific volume and elasticity while simultaneously decreasing its hardness and chewiness [
12].
Bacteria of the
Caulobacter
genus are gram-negative and are present in virtually all aquatic environments and in many soil types [
13,
14]. The genome of
Caulobacter crescentus (NA1000 strain) contains eight genes [
15,
16], which are directly involved in biomass degradation and are therefore relevant for biotechnological applications. Among these genes, only one encodes cellulase (
celA1), two encode enzymes with endoxylanase activity (
xynA1-2), and five encode enzymes with β-xylosidase activity (
xynB-xynB5) [
2,
3,
4,
5,
17,
18,
19,
20,
21,
22]. In addition,
C. crescentus has traditionally been considered nonpathogenic and assumed to be nontoxic to humans, making it a promising bioengineering vector for environmental remediation, the food industry, and medical applications [
23].
The
C. crescentus xynA1 gene was successfully overexpressed in
E. coli. The purified recombinant XynA1 was thoroughly characterized, revealing an enzymatic activity of 18.26 U/mL and a specific activity of 2.22 U/mg when xylan from beechwood was employed as the substrate. Kinetic analysis of XynA1 revealed KM and Vmax values of 3.77 mg/mL and 10.20 μM/min, respectively. The enzyme exhibited an optimal pH of 6 and peaked at 50 °C. Impressively, XynA1 has demonstrated remarkable thermal stability for biotechnological applications, retaining 80% of its activity over a span of 4 hours at 50 °C [
3]. Building upon previous work involving the purification and biochemical characterization of
C. crescentus XynA1, this study introduces a novel approach in which a conditional mutant was engineered. This mutant enabled homologous overexpression of the
C. crescentus xynA1 gene within the NA1000 strain, leveraging an induced promoter regulated by xylose. The resulting overexpressed and secreted
C. crescentus XynA1 was incorporated into flour and applied throughout baking processes.
2. Materials and Methods
2.1. Bacterial Strains and Growing Conditions
The strains and plasmids used in the present work are described in
Table 1.
Escherichia coli DH5α and S17 bacterial strains [
24] were used for subcloning and conjugation, respectively. Both strains were grown at 37 °C and maintained at 4 °C in Luria–Bertani (LB) medium [
25].
The recombinant NA1000 strain [
26,
27] containing the pAS22-
xynA1 [
28] construct was grown at 37 °C and maintained at 4 °C in LB media supplemented with chloramphenicol (1 µg/mL). The bacterial strain
C. crescentus NA1000 was grown at 30 °C and maintained at 4 °C in PYE media (0.2% bactopeptone, 0.1% yeast extract, 1.7 mM MgSO
4, 0.5 mM CaCl
2). The BS-
xynA1 mutant strain was isolated at 30 °C and maintained at 4 °C in PYE media supplemented with chloramphenicol (1 µg/mL) and nalidixic acid (20 µg/mL).
To induce the expression of xylanase I (XynA1) and other enzymes, the NA1000 strain and C. crescentus mutant BS-xynA1 were cultivated at 30 °C and 120 rpm in minimal medium (M2) (Na2HPO4 12 mM, NH4Cl 9 mM, KH2PO4 8 mM, MgSO4 1 mM, CaCl2 0.5 mM, FeSO4 10 mM) (Ely, 1991) supplemented with 0.2% (w/v) glucose, 0.3% (w/v) xylose and 0.3% xylose added from different agro-industrial residues at 0.2% (w/v) (CS) corn straw, (CC) corn cob, (HPM) hemicellulose from corn straw, (WS) wheat straw, (PR) polisher residue (WF) wheat flour industry vacuum cleaner residue (RF) rice flour (SB) sugarcane bagasse (RS) rice straw and (SR) soybean residue.
2.2. Cloning of the xynA1 Gene in the pAS22 Expression Vector
The DNA fragment corresponding to the
xynA1 gene (CCNA_02894) was isolated from the pJET1.2-
xynA1 construct [
3] as described below. The
xynA1 gene of interest containing 1,158 base pairs present in the pJET1.2-
xynA1 construct, along with the priming methionine and its stop codon, was digested with the restriction enzyme
XhoI (Thermo Fischer Scientific®). This enzyme cleaves this construct only to promote the linearization of the same construct. Then, a reaction with the enzyme DNA Blunting (Fermentas®) was carried out to obtain a noncohesive end, which allowed the subcloning process in the pAS22 vector to another noncohesive site in subsequent steps. With the gene still partially attached to the pJET1.2 vector at one end but containing a noncohesive end, it was digested with the restriction enzyme
EcoRI. (Thermo Fischer Scientific®). After digestion, the genetic material was resolved by 1% agarose gel electrophoresis in 1× TAE buffer. The 1,793 bp gel band, corresponding to the
xynA1 gene and an untranslated portion of the pJET1.2 blunt plasmid that remained attached after the stop codon of the
xynA1 gene, was cut from the scalpel gel and recovered with a DNA extraction kit (
Pure Link Gel Extraction Kit - Invitrogen®).
The plasmid used for xylanase (XynA1) expression [
3] in
C. crescentus was pAS22. This plasmid has a xylose-inducible promoter, which allows for an increase in the expression of the cloned xylanase gene upstream of the promoter. Construction planning was based on experimental evidence that the
C. crescentus xynA1 gene is not regulated by xylose according to transcriptomic analysis [
29]. Furthermore, the gene was also cloned without the control of its original promoter. The increase in gene expression in this case can easily be accompanied by an increase in xylanase activity, dispensing with evidence for the accumulation of essential protein mass in the case of proteins that do not present measurable enzymatic function. Double plasmid digestion was performed with
EcoRI and
EcoRV restriction enzymes (Thermo Fischer Scientific®). This linearized vector was ligated to the DNA fragment corresponding to the
xynA1 gene (
EcoRI/Blunt) with the aid of T4 DNA Ligase (BioLabs®). Subsequently, the pAS22-
xynA1 construct was used for transformation of the
E. coli strain DH5α. Transformants were selected by growth in media supplemented with chloramphenicol (1 μg/mL), a resistance marker of the pAS22 vector. After the transformants were selected, the plasmid material was extracted by mini plasmid preparation, double-digested with the restriction enzymes
EcoRI/
KpnI and resolved by 1% TAE 1X agarose gel electrophoresis to confirm the gene size.
2.3. Construction of the BS-xynA1 Mutant Strain
The pAS22-xynA1 construct was used to transform the E. coli strain S17, which has conjugative functions. Transformants were selected by growth in media supplemented with chloramphenicol (1 μg/mL), the resistance marker of the pAS22 vector. After selection of the transformant, plasmid material was extracted by mini plasmid preparation, double-digested with the restriction enzymes EcoRI/KpnI and resolved by 1% TAE 1X agarose gel electrophoresis to confirm the recombinant size. E. coli S17 transformant strains containing the pAS22-xynA1 construct were used for the transformation of the C. crescentus NA1000 strain by conjugation. An aliquot of a fresh E. coli strain containing the pAS22-xynA1 vector was mixed with twice the amount of fresh C. crescentus cells of the NA1000 strain in solid PYE media. After 48 hours of growth at 30 °C, samples of the bacterial cultures grown on the plate were streaked on a new solid PYE plate containing chloramphenicol (1 μg/mL), the pAS22 vector selection mark and nalidixic acid (20/μg mL), a selection mark of the NA1000 strain and not the E. coli strain, at 30 °C for 48 hours. Isolated colonies grown on these plates were again streaked onto a new PYE plate containing the same amount of chloramphenicol and nalidixic acid. Microscopy analysis was carried out to confirm that only C. crescentus cells were present in the medium. Aliquots of the mutant cells were then named BS-xynA1 and stored at -20 °C and 80 °C for subsequent characterization. In parallel to the construction of the BS-xynA1 mutant, a control strain of the NA1000 strain was constructed via conjugation of the empty plasmid pAS22.
2.4. Growth, Xylose Consumption and Xylanase Expression
C. crescentus strains Cc-pAS22 and BS-
xynA1 were preinoculated in 10 mL of liquid PYE medium supplemented with chloramphenicol (1 µg/mL) and nalidixic acid (20 µg/mL) and grown for 12 hours at 30 °C and 120 rpm. When the cells reached the stationary phase of growth, they were diluted (OD
λ600 nm= 0.1) in M2 medium supplemented with 0.3% (w/v) xylose and grown at 30 °C with 120 rpm agitation. Every two hours of growth, a 2 mL aliquot of the growing culture was collected. From this volume, 1 mL was used to measure spectrophotometer cell growth, 1 mL was centrifuged at 15,000 xg for 5 min at 4 °C, and the supernatant was reserved for extracellular xylanase activity analysis (optimizing for pH and temperature of xylanase I) in addition to the dosage of xylose consumption by the Orcinol method for pentoses [
30]. Cell precipitates from centrifugation were frozen for further quantification of intracellular xylanase. The frozen cell pellet was lysed with 350 μL of 50 mM phosphate buffer, pH 6.0, under vigorous vortexing until thawing. The samples were kept on ice, and the overall activity levels of xylanolytic enzymes were determined. The assay and dosages were performed in biological and experimental triplicates, respectively.
2.5. XynA1 Expression in Different Agro-Industrial Residues
Preinoculation and inoculation were performed as described in the previous section, but the M2 medium was supplemented with 0.3% (w/v) xylose and 0.2% (w/v) different agro-industrial residues. (CS) Corn Straw; (CC) Corn Cob; (HPM) Hemicellulose from Corn Straw; (WS) Wheat Straw, (PR) Polisher Residue (WF) Wheat Flour Industry Vacuum Cleaner Residue (RF) Rice Flour. (SB) Sugarcane bagasse (RS) rice straw; (SR) soybean residue. All residues were prepared as described previously [
18]. Cultures were incubated at 30 °C with shaking at 120 rpm for 18 hours. Then, the samples were centrifuged at 15,000 × g for 5 min at 4 °C. The supernatants were used to quantify XynA1 activity. The supernatant with the highest xylanase I activity was also characterized by the presence of cellulase, pectinase, α-L-arabinofuranosidase, β-glycosidase, β-xylosidase and α-amylase. Since the bacterium has two genes for xylanases (
xynA1 and
xyn2), two enzymes can be expressed under induced conditions. In these experiments, the NA1000 strain containing only the pAS22 (Cc-pAS22) vector was used as a control.
2.6. Dosages of Different Extracellular Enzymes
Reactions to verify xylanase I and II activity were performed using 1% (w/v) xylan from beechwood (Sigma®) substrate (in 50 mM sodium phosphate buffer pH 6.0 for xylanase I and buffer McIvaine pH 8.0 for xylanase II) followed by incubation at 50 and 60 °C, respectively. Reactions to verify cellulase and α-amylase activity were performed using 1% (w/v) carboxymethylcellulose (CMC) as the substrate in 50 mM sodium citrate buffer (pH 5.5) and 1% starch as the substrate (w/v) in 50 mM sodium citrate buffer (pH 5.0), respectively, followed by incubation at 40 °C. Reducing sugars were measured using 3,5-dinitrosalicylic acid (DNS) [
31]. Xylanolytic and cellulolytic activities were defined in U/mL as the amount of enzyme capable of releasing 1 μmol of xylose per mL of solution per min of reaction (U). The enzymatic activities of β-glycosidase, β-xilosidase, and α-L-arabinosidase were determined using ρ-nitrophenyl-β-D-glucopyranoside (ρNPG) and ρ-nitrophenyl-β-D-xylopyranoside (ρNPX) reagents. and ρ-nitrophenyl-α-L-arabinofuranoside (ρNPA) (Sigma®), respectively, according to the adapted methodology described by Justo et al. (2015), estimating the amount of ρ-nitrophenol (ρNP) released from the respective reagents. The total protein concentration was estimated by the Bradford method [
32], which uses bovine serum albumin (BSA; Bio-Rad®) as a standard.
2.7. Commercially Available Xylanases Versus Xylanase of Mutant BS-xynA1
Xylanolytic activity was also measured in different commercial xylanase mixtures available for application in the bakery industry. Among the variants tested were Spring CA 400 (produced by Granotec from Brail S.A. Nutrition and Biotechnology), Mega Cell 899 and Xylamax 292 (produced by Prozin Industrial and Comercial Ltd.). Enzymatic assays were performed as described in the previous section, with varying incubation temperatures (40, 50, 60, 70, 80, 90 and 100 °C). The same procedure was performed for the extracellular crude extract containing the C. crescentus enzyme XynA1.
2.8. Test for Confirmation of the Absence of Viable Bacteria in Enzyme Extracts
The total enzyme extracts produced were centrifuged at 5,000 × g at 4 °C for 10 min. The centrifuged supernatant containing C. crescentus XynA1 was sterilized with a 0.22 µm polystyrene vacuum filtration system containing sterile, pyrogen-free low protein binding polyether sulfone (Ciencor®). The complete absence of viable bacteria in the extracellular extracts was tested by plating a 100 µl aliquot of extracellular enzymatic crude extract into each of the three PYE media plates at 30 °C and LB at 37 °C followed by incubation for 48 hours for microbial growth investigation. Aliquots of the filtrates and cell-free Xylanase I was used for enzymatic assay testing after centrifugation and filtration to confirm the maintenance of xylanase activity.
2.9. Application of C. crescentus XynA1 cell-Free in Bread Formulation
To ensure food safety, the C. crescentus BS-xynA1 mutant was grown in the absence of chloramphenicol since the cells maintained the pAS22 plasmid and xylanase production under the tested conditions for up to three consecutive days. The extracellular enzymatic extracts enriched with recombinant homologous xylanase I from C. crescentus were filtered with a PES (polyestersulfone) vacuum filtration system of 0.1 μm and 500 mL capacity (431475-Corning®) and cell-free Xylanase I was used in the baking assays. Therefore, it is certain that the final product used in the tests meets food safety standards. The xylanolytic activity of enzymatic extracts used as processing aids in the production of white wheat flour bread has been investigated. This study aimed to evaluate the effects of these enzyme extracts on bread quality using a standardized loaf formulation adhering to the guidelines of Art. 661 of the Argentine Food Code.
The baking trials were meticulously executed in triplicate, employing carefully defined formulations. The core ingredients consisted of white wheat flour (as specified), sugar, salt, fresh yeast (Fleischmann®), vegetable shortening (Coamo®), and water. To explore the impact of enzyme extracts, two additional formulations were developed based on an experimental design in which the enzyme extract concentrations were manipulated. Adjustments in water content were made according to the volume of enzyme solution employed, ensuring consistency with the standard formulation. Refer to
Table 2 for a breakdown of the ingredient quantities in each test formulation.
The dough preparation procedure included distinct phases, each of which contributed to the final bread characteristics: 1. Mixing (Phase I and Phase II): Initial ingredients, including flour, sugar, salt, and water (also incorporating enzymatic solution for tests T2 and T3), were combined in a trough maintained at a temperature of 4 to 6 °C. This mixture was blended at a constant speed (100 rpm) for 3 minutes. In Phase II, the blend welcomed vegetable fat and fresh yeast, and mixing was continued at 190 rpm until the desired dough consistency was achieved; 2. Division: The resulting dough was divided into uniform 500-gram portions with precision, aided by spatulas and scales; 3. Modeling: The modeling phase involved a single pass through a specialized machine designed to roll, fold, stretch, and thereby homogenize the dough. This three-step process ensured an even dough texture and structure, promoting symmetrical bread formation during fermentation and baking; 4. Fermentation and Growth: The molded dough was shaped into standard forms and positioned within a controlled fermentation chamber set at 28 °C. This allowed for an exact fermentation period of 4 hours, promoting optimal dough development; 5. Baking (Final Step): The fully formed, fermented, and grown dough was placed into a preheated oven set at 180 °C. Baking transpired over a 30-minute period. After baking, the loaves were allowed to cool to room temperature. Bread height measurements were acquired using a measuring tape, and cross-sectional observations were made by halving the loaves to assess alveolar structure. Additionally, photographic documentation facilitated a comprehensive comparison between control loaves and loaves subjected to varying enzyme extract concentrations.
2.10. Alveograph Test: Evaluating Flour Viscoelastic Characteristics
The enzymatic impact on the viscoelastic properties of flour is assessed through alveography, a technique that emulates dough behavior during fermentation by replicating the formation of air pockets (alveoli) generated by yeast-released carbon dioxide (CO
2). This assessment involves the analysis of various alveographic parameters to gauge the viscoelastic traits of different dough samples. Employing the Chopin Alveograph model, AlveoPC, crafted by CHOPIN Technologies (Villeneuve-la-Garenne, France) and adhering to the International Association for Cereal Science and Technology guidelines [
33], this test method provides precision to the characterization process.
For the enzyme addition experiment (
Table 3), the enzyme extract volume was determined according to the ratio of enzyme extract per kilogram of flour as utilized in baking test T3. The control run, devoid of enzyme supplementation, served as a baseline. Consistency was maintained in both the control and test groups according to the following parameters: humidity, 14.4%; hydration, 50%; and B concentration, 15% H
2O, in line with the AACC methodology recommendations. The preparation involved diluting sodium chloride in sterile water to afford a 2.5% (w/v) saline solution, which was subsequently combined with the enzyme solution in the enzyme addition test. A total of 250 grams of flour was introduced into the mixer at 24±2 °C. The process begins, and within 20 seconds, saline solution is added. The mixing continued for one minute, encompassing the 20 seconds prior to saline addition. Following this, the mixer halts, allowing for the clearing of its sides using a spatula – a one-minute procedure.
The mixture was mixed for an additional 7 minutes, resulting in a total of 8 minutes. The mixer is then paused to alter the rotation direction of the paddle, leading to the division of the resulting dough into five uniform portions. These portions were placed within an alveograph rest chamber maintained at 25 ± 0.2 °C. Twenty-eight minutes after the initiation of dough mixing, the first portion was placed at the center of the stationary alveograph plate and coated with Vaseline. The lid is positioned and secured, and the plate undergoes two full turns within 20 seconds. Five seconds later, the lid and ring were removed, and the dough was allowed to inflate until a bubble formed. This sequence is repeated for the other four dough portions.
Parameters derived from the alveograms include toughness (P), which represents the peak pressure exerted during mass expansion (measured in millimeters); extensibility (L), which quantifies the length of the curve (measured in millimeters); and mass deformation energy (W), which signifies the mechanical work needed to expand the bubble until rupture, expressed as 10-4 J. The ratio of toughness to extensibility (P/L) offers insight into mass equilibrium, where P signifies the mass's resistance to deformation, and L indicates mass extensibility.
2.11. RNA Extraction, RT-qPCR Assays and Statistical Analysis
The RNA extraction and RT-qPCR assays closely followed the protocols outlined in the study by Corrêa et al. [
21]. These procedures were meticulously replicated in three distinct experimental trials, except for the alveograph tests, which underwent a comprehensive fivefold investigation. To ensure the accuracy of the results, standard errors were employed for result validation.
For the enzymatic treatment test within the bread formulation and the alveographic test, the acquired data were subjected to rigorous scrutiny through analysis of variance (ANOVA). Subsequently, a Tukey post hoc test was conducted to determine significant differences, maintaining a significance level of 99%.
Author Contributions
B.S.: investigation, visualization, and writing - original draft; P.M.C.R, A. F, J.L.C.S.; R.F.G. and AM: methodology; M.K.: methodology and resources; RCGS: Software, project administration, funding acquisition, supervision, writing, review, and editing. All authors have read and agreed to the published version of the manuscript.
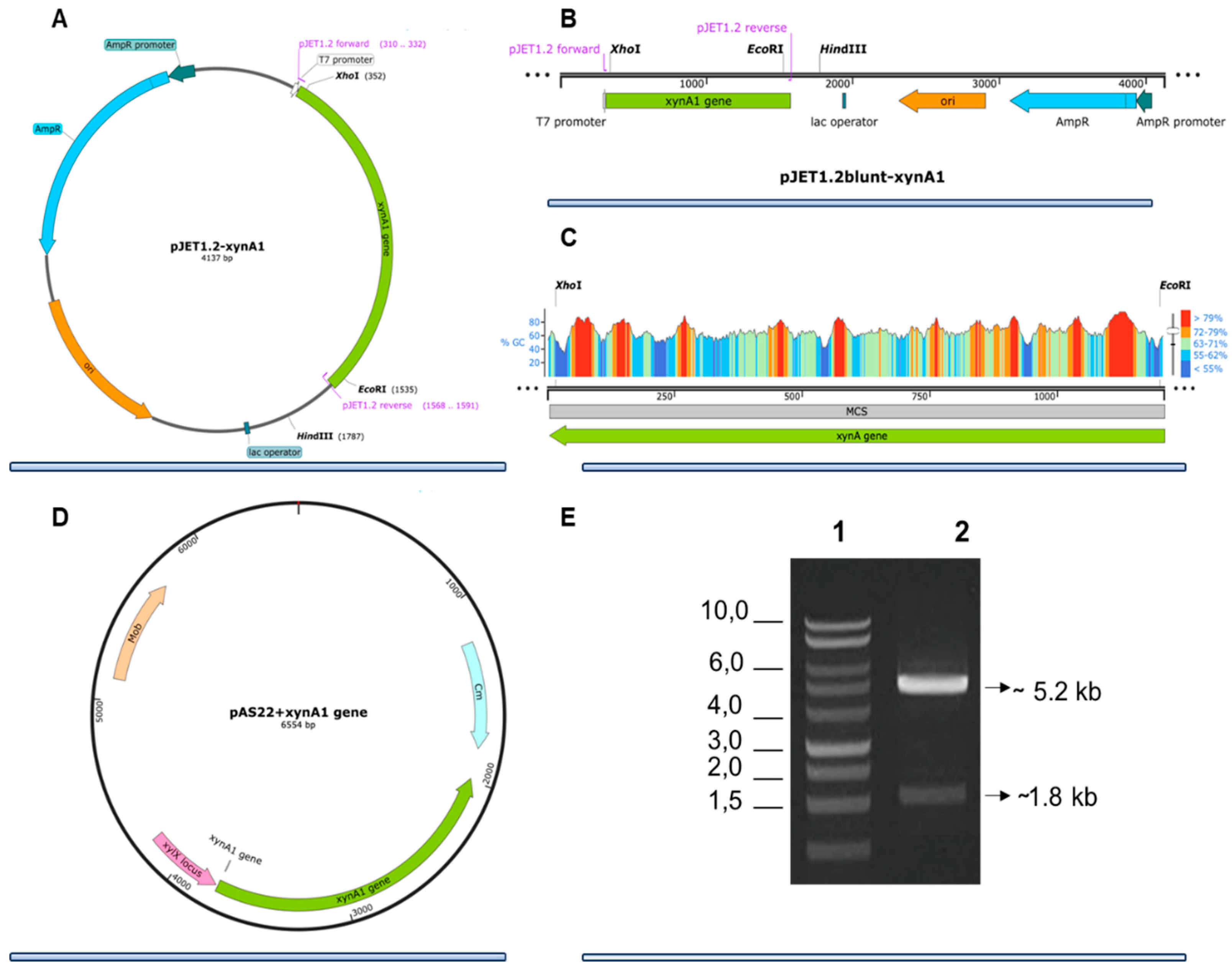
Figure 1.
Strategies for engineering C. crescentus strains that overexpress xylanase I in the strain BS-xynA1. (A) Genetic context of the xynA1 construct in the pJet1.2-Blunt cloning vector. (B) The pJet1.2-Blunt construct containing the xynA1 gene was digested with XhoI and treated to produce blunt-end DNA. Next, pJet1.2-Blunt-xynA1 was digested with EcoRI, and the resulting fragment, containing the complete xynA1 gene (C), was subcloned and inserted into the pAS22 vector (EcoRI/EcoRV) under the control of a promoter induced by xylose (D), generating the synthetic strain BS-xynA1, which expresses Xylanase I in its own bacterium when 0.3% xylose is added. (E) DNA agarose gel electrophoresis on a TAE1X 1% gel. 1: Molecular weight marker 1 kb DNA Ladder (Thermo); 2: Construction of pAS22-xynA1 digested with EcoRI/KpnI enzymes.
Figure 1.
Strategies for engineering C. crescentus strains that overexpress xylanase I in the strain BS-xynA1. (A) Genetic context of the xynA1 construct in the pJet1.2-Blunt cloning vector. (B) The pJet1.2-Blunt construct containing the xynA1 gene was digested with XhoI and treated to produce blunt-end DNA. Next, pJet1.2-Blunt-xynA1 was digested with EcoRI, and the resulting fragment, containing the complete xynA1 gene (C), was subcloned and inserted into the pAS22 vector (EcoRI/EcoRV) under the control of a promoter induced by xylose (D), generating the synthetic strain BS-xynA1, which expresses Xylanase I in its own bacterium when 0.3% xylose is added. (E) DNA agarose gel electrophoresis on a TAE1X 1% gel. 1: Molecular weight marker 1 kb DNA Ladder (Thermo); 2: Construction of pAS22-xynA1 digested with EcoRI/KpnI enzymes.
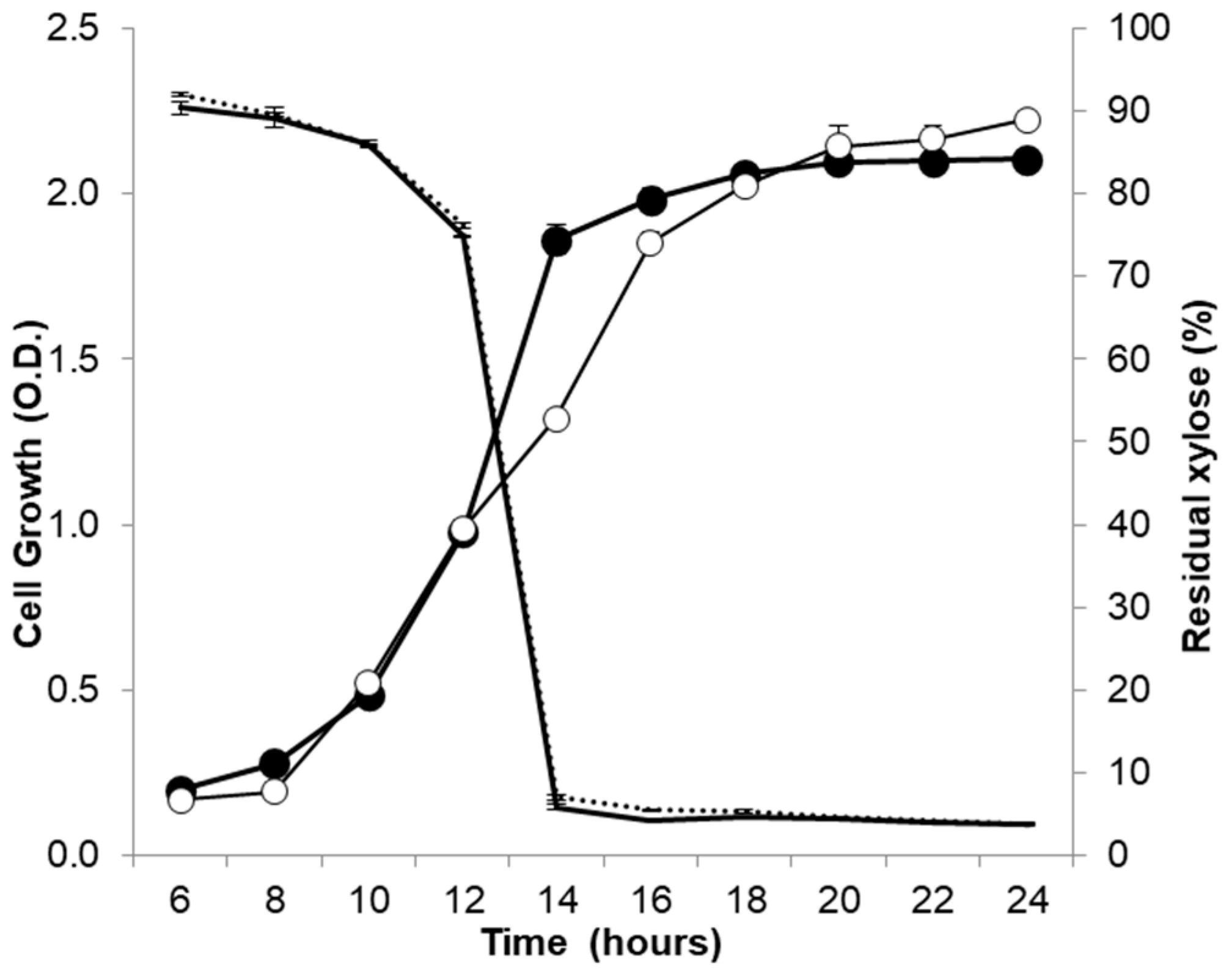
Figure 2.
Cell growth curves and xylose consumption of the control strain Cc-pAS22 and the BS-xynA1 strain in M2 media supplemented with 0.3% xylose at 30 °C and 120 rpm. Cell growth (continuous line) is expressed as the O.D.λ = 600 nm. The residual xylose content (sectioned line) is expressed as a percentage of the total initial sugar content.
Figure 2.
Cell growth curves and xylose consumption of the control strain Cc-pAS22 and the BS-xynA1 strain in M2 media supplemented with 0.3% xylose at 30 °C and 120 rpm. Cell growth (continuous line) is expressed as the O.D.λ = 600 nm. The residual xylose content (sectioned line) is expressed as a percentage of the total initial sugar content.
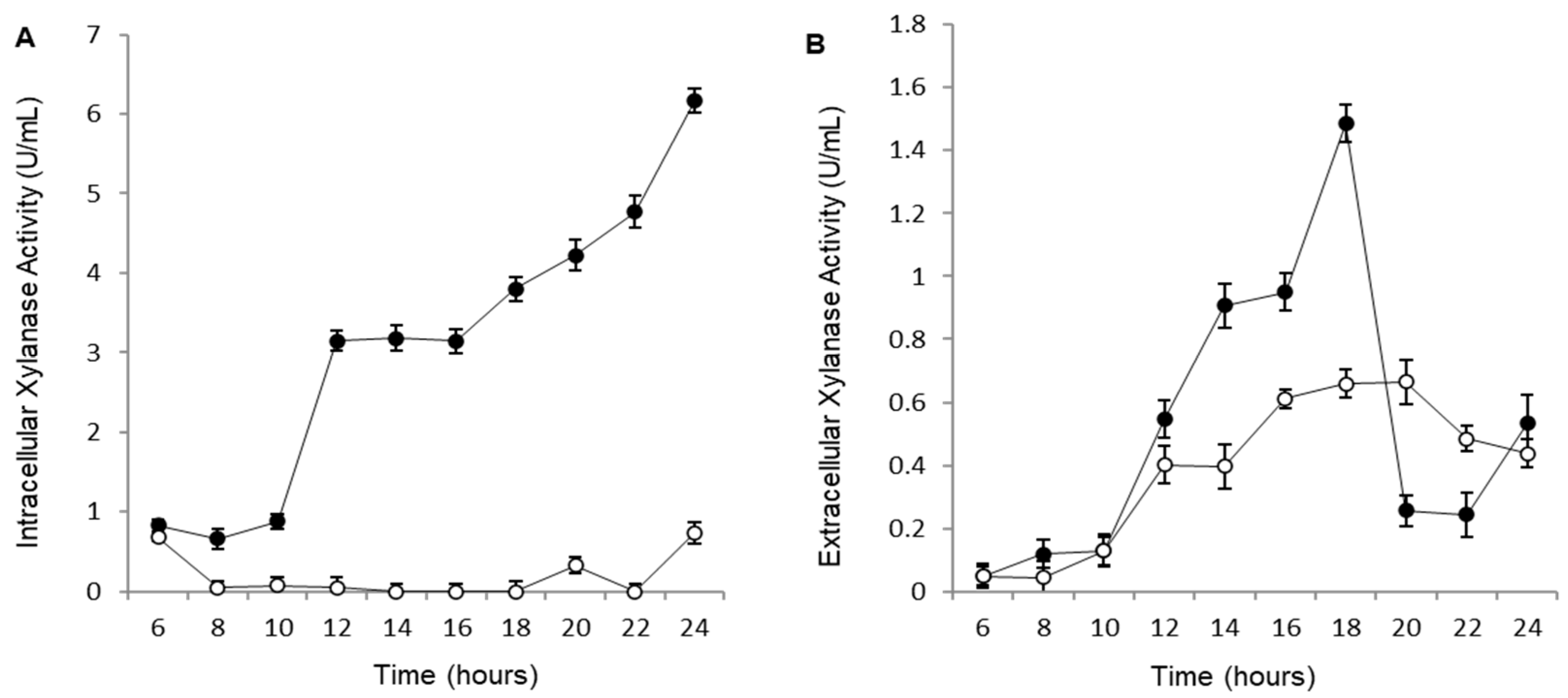
Figure 3.
(A) Intracellular xylanase activity of the control strain Cc-pAS22 (open circle) and BS-xynA1 (closed circle). Bacterial cells were grown in M2 media at 30 °C and 120 rpm. (B) Extracellular xylanase activity of the Cc-pAS22 (open circle) and BS-xynA1 (closed circle) strains of C. crescentus. Bacterial cells were grown in M2 media supplemented with 0.2% (w/v) glucose (Cc-pAS22) and 0.3% (w/v) xylose (BS-xynA1) at 30 °C and 120 rpm for 24 hours. Aliquots of the culture were removed, and xylanolytic activities were measured.
Figure 3.
(A) Intracellular xylanase activity of the control strain Cc-pAS22 (open circle) and BS-xynA1 (closed circle). Bacterial cells were grown in M2 media at 30 °C and 120 rpm. (B) Extracellular xylanase activity of the Cc-pAS22 (open circle) and BS-xynA1 (closed circle) strains of C. crescentus. Bacterial cells were grown in M2 media supplemented with 0.2% (w/v) glucose (Cc-pAS22) and 0.3% (w/v) xylose (BS-xynA1) at 30 °C and 120 rpm for 24 hours. Aliquots of the culture were removed, and xylanolytic activities were measured.
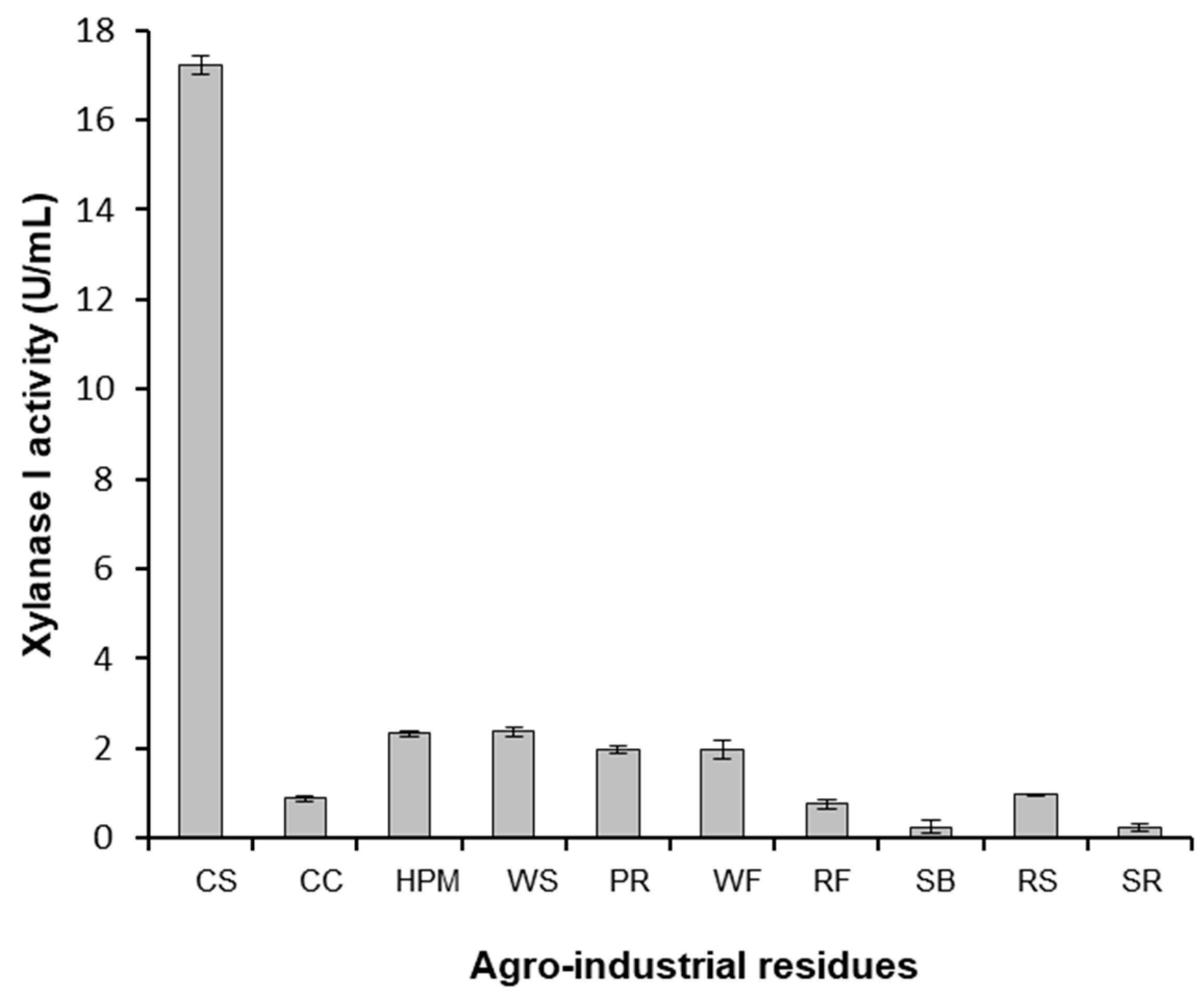
Figure 4.
Xylanase I production in the presence of different agro-industrial residues. C. crescentus BS-xynA1 strain cells were grown in minimal M2 medium supplemented with 0.3% xylose (w/v) and supplemented with 0.2% (w/v) agro-industrial residues: (CS) corn straw; (CC) corn cob; (HPM) hemicellulose from corn straw; (WS) wheat straw; (PR) polisher residue (WF) wheat flour industry vacuum cleaner residue (RF) rice flour (SB) sugarcane bagasse (RS) rice straw; and (SR) soybean residue. The inoculum was generated by diluting the cells at the stationary phase to an O.D.λ of 0.1 at 600 nm in the same culture medium containing different carbon sources. Bacterial growth occurred at 30 °C for 18 h with agitation at 120 rpm.
Figure 4.
Xylanase I production in the presence of different agro-industrial residues. C. crescentus BS-xynA1 strain cells were grown in minimal M2 medium supplemented with 0.3% xylose (w/v) and supplemented with 0.2% (w/v) agro-industrial residues: (CS) corn straw; (CC) corn cob; (HPM) hemicellulose from corn straw; (WS) wheat straw; (PR) polisher residue (WF) wheat flour industry vacuum cleaner residue (RF) rice flour (SB) sugarcane bagasse (RS) rice straw; and (SR) soybean residue. The inoculum was generated by diluting the cells at the stationary phase to an O.D.λ of 0.1 at 600 nm in the same culture medium containing different carbon sources. Bacterial growth occurred at 30 °C for 18 h with agitation at 120 rpm.
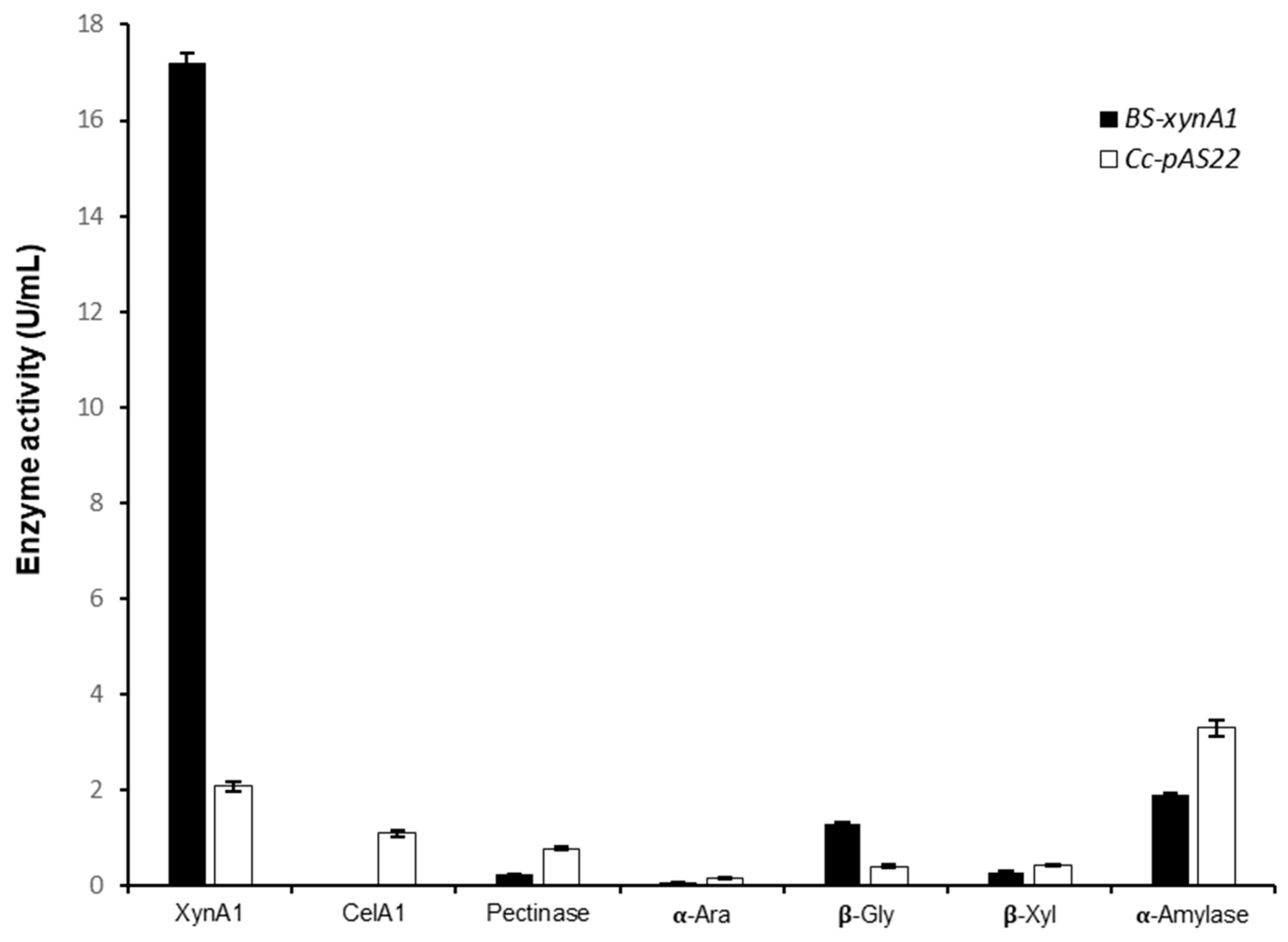
Figure 5.
Enzyme activities produced by different C. crescentus strains (Cc-pAS22 and BS-xynA1) after growth in M2 medium supplemented with 0.2% (w/v) corn straw at 30 °C and 120 rpm for 18 hours. (XynA1) xylanase I; (CelA1) cellulase; (Pect) pectinase; (α-Ara) α-L-arabinosidase; (β-gly) β-glycosidase; (β-xyl) β-xylosidase; (α-amy) α-amylase.
Figure 5.
Enzyme activities produced by different C. crescentus strains (Cc-pAS22 and BS-xynA1) after growth in M2 medium supplemented with 0.2% (w/v) corn straw at 30 °C and 120 rpm for 18 hours. (XynA1) xylanase I; (CelA1) cellulase; (Pect) pectinase; (α-Ara) α-L-arabinosidase; (β-gly) β-glycosidase; (β-xyl) β-xylosidase; (α-amy) α-amylase.
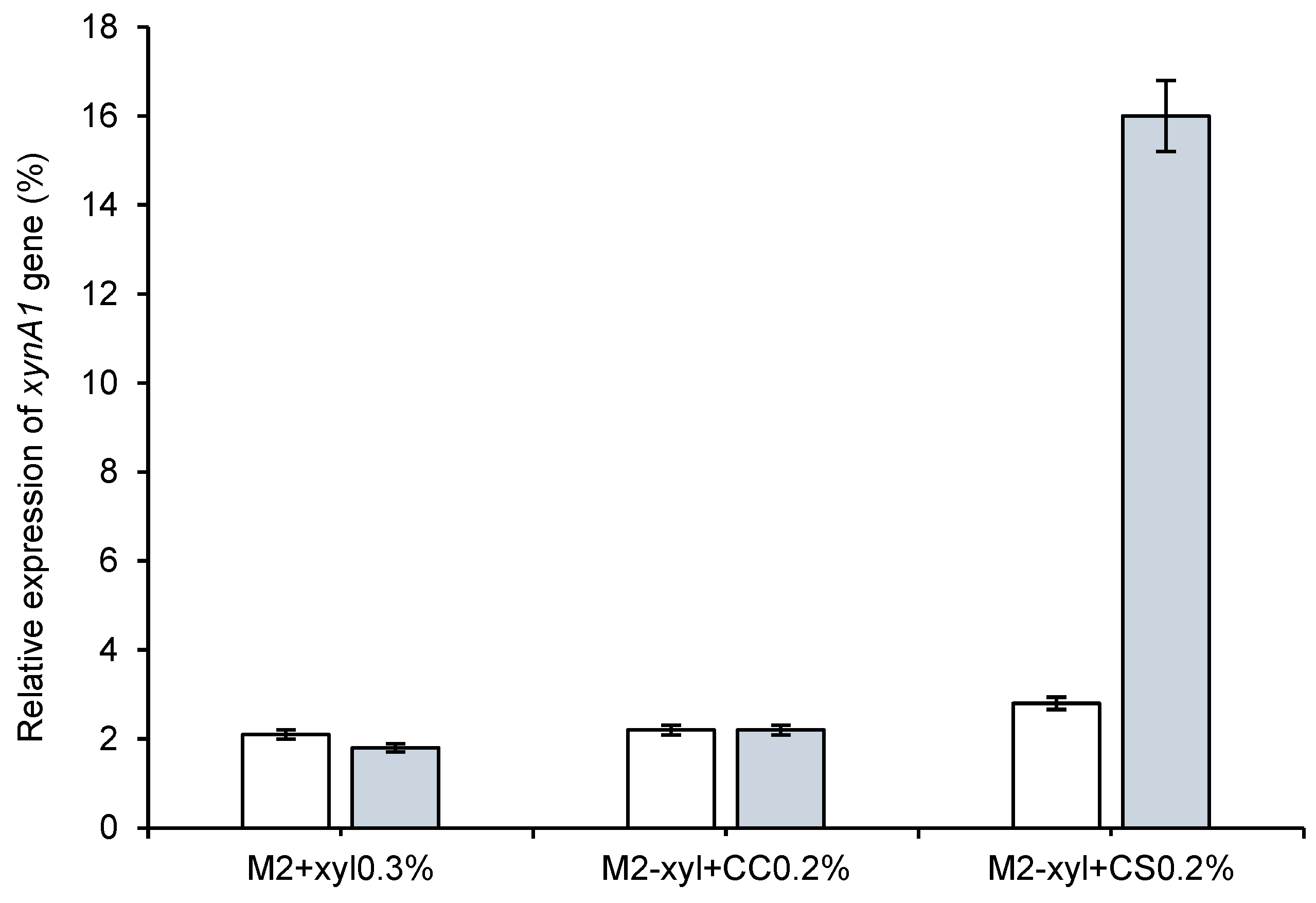
Figure 6.
Relative gene expression of xylanase genes (xynA1-2) in the C. crescentus NA1000 (white bars) and BS-xynA1 (blue bars) strains determined via quantitative real-time PCR (RT‒qPCR). This study aimed to understand how nutrient availability and growth conditions affect the expression of the xynA1 gene in C. crescentus strains. RNA extraction and RT-qPCR protocols followed established procedures. Data normalization was performed using the 16S rRNA gene as an endogenous control, ensuring accuracy and reliability across treatments. Three independent biological experiments with three replicates each were conducted to ensure robustness and consistency. Relative gene expression levels were calculated for treatments under different media conditions, CS: corn straw; CC: corn cob and M2 minimum media.
Figure 6.
Relative gene expression of xylanase genes (xynA1-2) in the C. crescentus NA1000 (white bars) and BS-xynA1 (blue bars) strains determined via quantitative real-time PCR (RT‒qPCR). This study aimed to understand how nutrient availability and growth conditions affect the expression of the xynA1 gene in C. crescentus strains. RNA extraction and RT-qPCR protocols followed established procedures. Data normalization was performed using the 16S rRNA gene as an endogenous control, ensuring accuracy and reliability across treatments. Three independent biological experiments with three replicates each were conducted to ensure robustness and consistency. Relative gene expression levels were calculated for treatments under different media conditions, CS: corn straw; CC: corn cob and M2 minimum media.
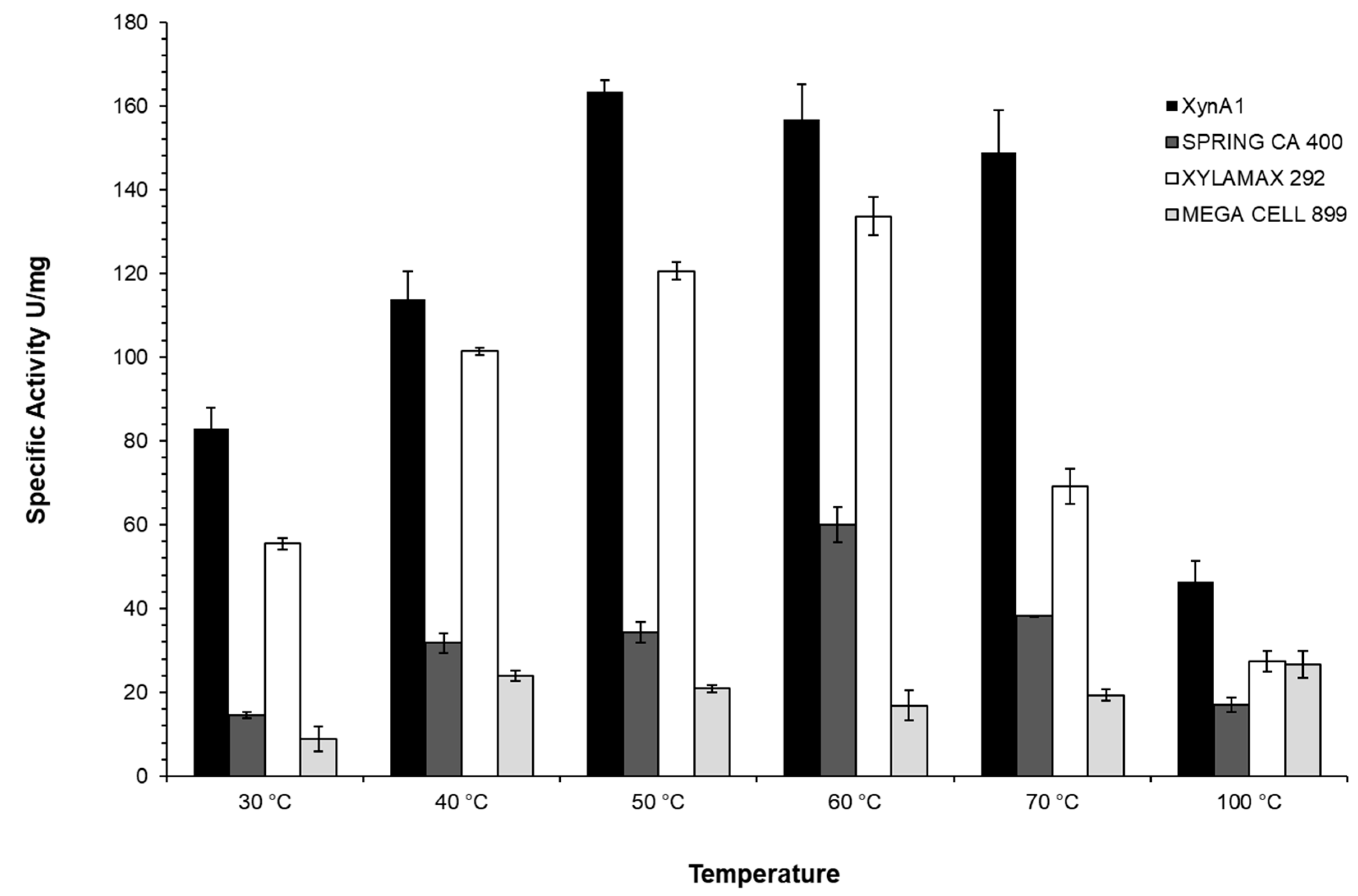
Figure 7.
Comparison between the specific activity (U/mg) of different commercial xylanases and cell-free Xylanase I from C. crescentus BS-xynA1 strain.
Figure 7.
Comparison between the specific activity (U/mg) of different commercial xylanases and cell-free Xylanase I from C. crescentus BS-xynA1 strain.
Figure 8.
Average alveographs provided by the apparatus (alveograph). (A) Control assay. (B) Alveographs obtained after treatment with cell-free Xylanase I (30 mL of extract in 250 g of flour). The axis of the abscissa shows the values of elasticity expressed in millimeters (mm). The ordinate axis expresses the toughness values expressed in millimeters of water (mmH2O). The data represent the arithmetic mean of five independent repetitions.
Figure 8.
Average alveographs provided by the apparatus (alveograph). (A) Control assay. (B) Alveographs obtained after treatment with cell-free Xylanase I (30 mL of extract in 250 g of flour). The axis of the abscissa shows the values of elasticity expressed in millimeters (mm). The ordinate axis expresses the toughness values expressed in millimeters of water (mmH2O). The data represent the arithmetic mean of five independent repetitions.
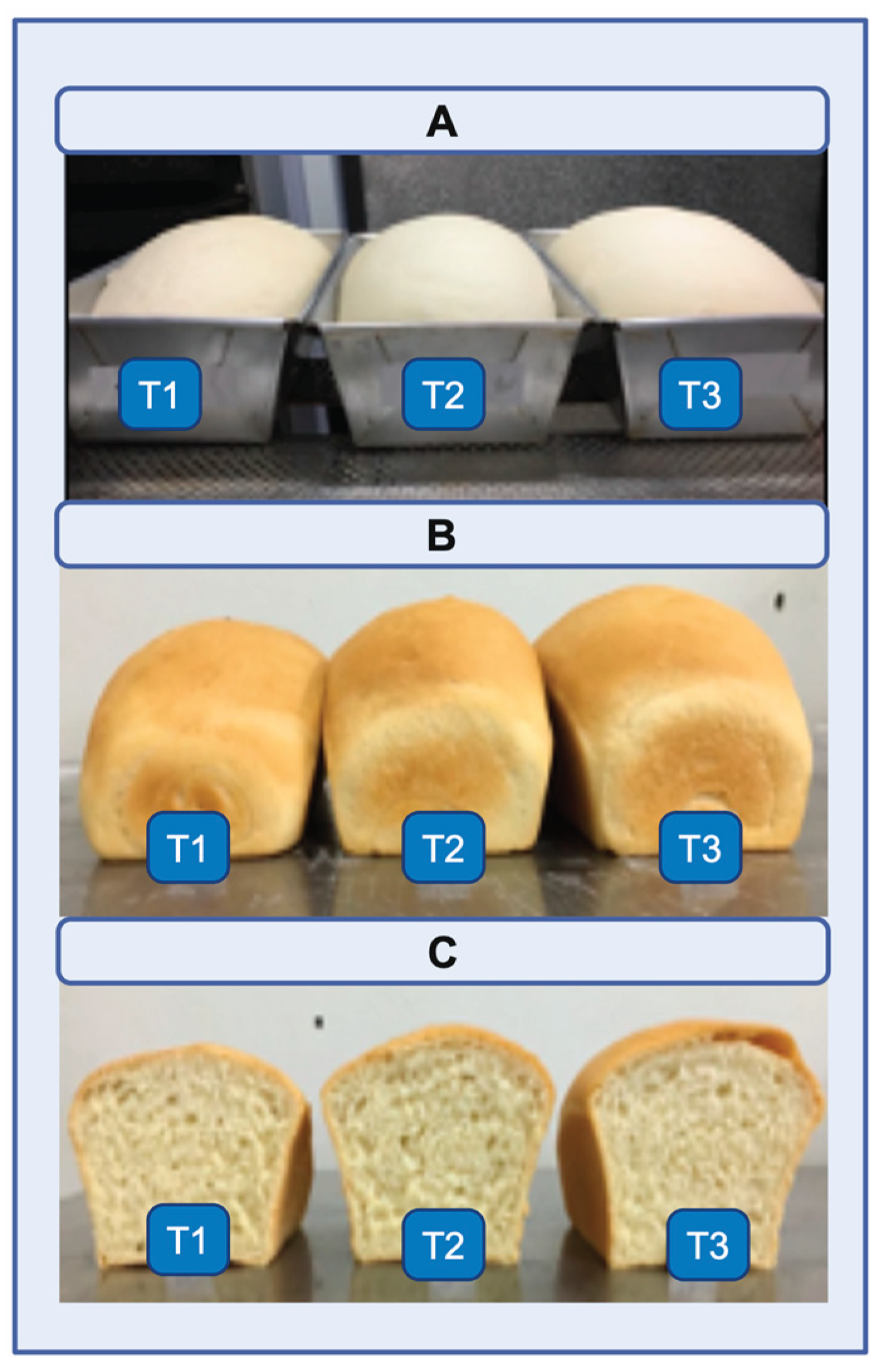
Figure 9.
(A) Representative images of breads after fermentation. (B, C) Images of breads after baking. (B) Whole breads; (C) Breads cut exactly in half. (T1) Control without the addition of the enzyme extract. (T2) Addition of 60 mL of enzyme extract to 1 kg of flour. (T3) A total of 120 mL of enzymatic extract containing cell-free Xylanase I (specific activity = 278.64 U/mg) was added to 1 kg of flour.
Figure 9.
(A) Representative images of breads after fermentation. (B, C) Images of breads after baking. (B) Whole breads; (C) Breads cut exactly in half. (T1) Control without the addition of the enzyme extract. (T2) Addition of 60 mL of enzyme extract to 1 kg of flour. (T3) A total of 120 mL of enzymatic extract containing cell-free Xylanase I (specific activity = 278.64 U/mg) was added to 1 kg of flour.
Table 1.
Strains and plasmids used in the present report.
Table 1.
Strains and plasmids used in the present report.
| Strains/Plasmids |
Genotype/Description |
Source/Reference |
| E. coli |
|
|
| DH5α
|
Δ (lacZYA-argF) U169 deoR recA1 endA1 hsdR17 phoA sup144 thi-1 gyrA96 relA1 (φ80 lacZDM15).
|
Invitrogen®. |
| S17 |
M294::RP4-2 (Tc::Mu) (Km::Tn7)
|
[24] |
| DH5α-pAS22-xynA1
|
E. coli DH5α carrying pAS22-xynA1.
|
This study |
| S17-pAS22-xynA1
|
E. coli S17 carrying pAS22-xynA1
|
This study |
| DH5α-pAS22 |
E. coli DH5α carrying pAS22 |
This study |
| S17-pAS22 |
E. coli S17 carrying pAS22 |
This study |
| C. crescentus |
|
|
| NA1000 |
Holdfast mutant derivative
of wild-type strain CB15 |
[26] |
| BS-xynA1 |
NA1000 carrying pAS22-xynA1
|
This study |
| Cc-pAS22 |
NA1000 carrying pAS22 |
This study |
| Plasmid |
|
|
| pAS22 |
Vector for expression of genes in Caulobacter from the PxylX promoter, ori T, CmR
|
[27] |
| pAS22-xynA1
|
pAS22 containing the xynA1 gene under the control of PxylX promoter |
This study |
Table 2.
Test formulations.
Table 2.
Test formulations.
| Components |
T1 |
T2 |
T3 |
| Flour |
1000 g |
1000 g |
1000 g |
| Salt |
20 g |
20 g |
20 g |
| Sugar |
60 g |
60 g |
60 g |
| Vegetable fat |
40 g |
40 g |
40 g |
| Yeast |
20 g |
20 g |
20 g |
| Water |
500 mL |
440 mL |
380 mL |
| Cell-free XylanaseI * |
- |
60 mL |
120mL |
Table 3.
Composition of alveograph tests.
Table 3.
Composition of alveograph tests.
| XynA |
− |
+* |
| Flour |
250 g |
250 g |
| Water |
127.7 mL |
97.7 mL |
| Sodium Chloride |
3,192.5 g |
3,192.5 g |
| Cell-free XynA1* |
no addition |
30 mL |
Table 4.
Parameters checked on breads.
Table 4.
Parameters checked on breads.
| Parameters |
T1 |
T2 |
T3 |
| Beat time to get the veil point (min) |
11 |
10 |
10 |
| Height of bread after fermentation (cm) |
7.9 ± 0.05 |
8.6 ± 0.04 |
9.3 ± 0.05 |
| Height of bread after furnace (cm) |
8.8 ± 0.07 |
9.8 ± 0.08 |
10.4 ± 0.07 |
Table 5.
Alveography results.
Table 5.
Alveography results.
| Parameters Generated |
XynA1 - |
Cell-free XynA1+ |
| P (tenacity) |
134 mmH2O |
126 mmH2O |
| L (extensibility) |
69 mm |
82 mm |
| W (mass deformation energy) |
326 10-4J |
353 10-4J |
| P/L (tenacity / extensibility) |
1.94 |
1.54 |
| Ie (elasticity index) |
53.6 % |
56.4 % |