Submitted:
30 July 2025
Posted:
31 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Thin Layer Chromatography Analysis (TLC)
2.3. β-galactosidase Activity Determination
2.4. Biofilm Formation Assays and Flocs Visualization
2.5. Statistical analysis
3. Results
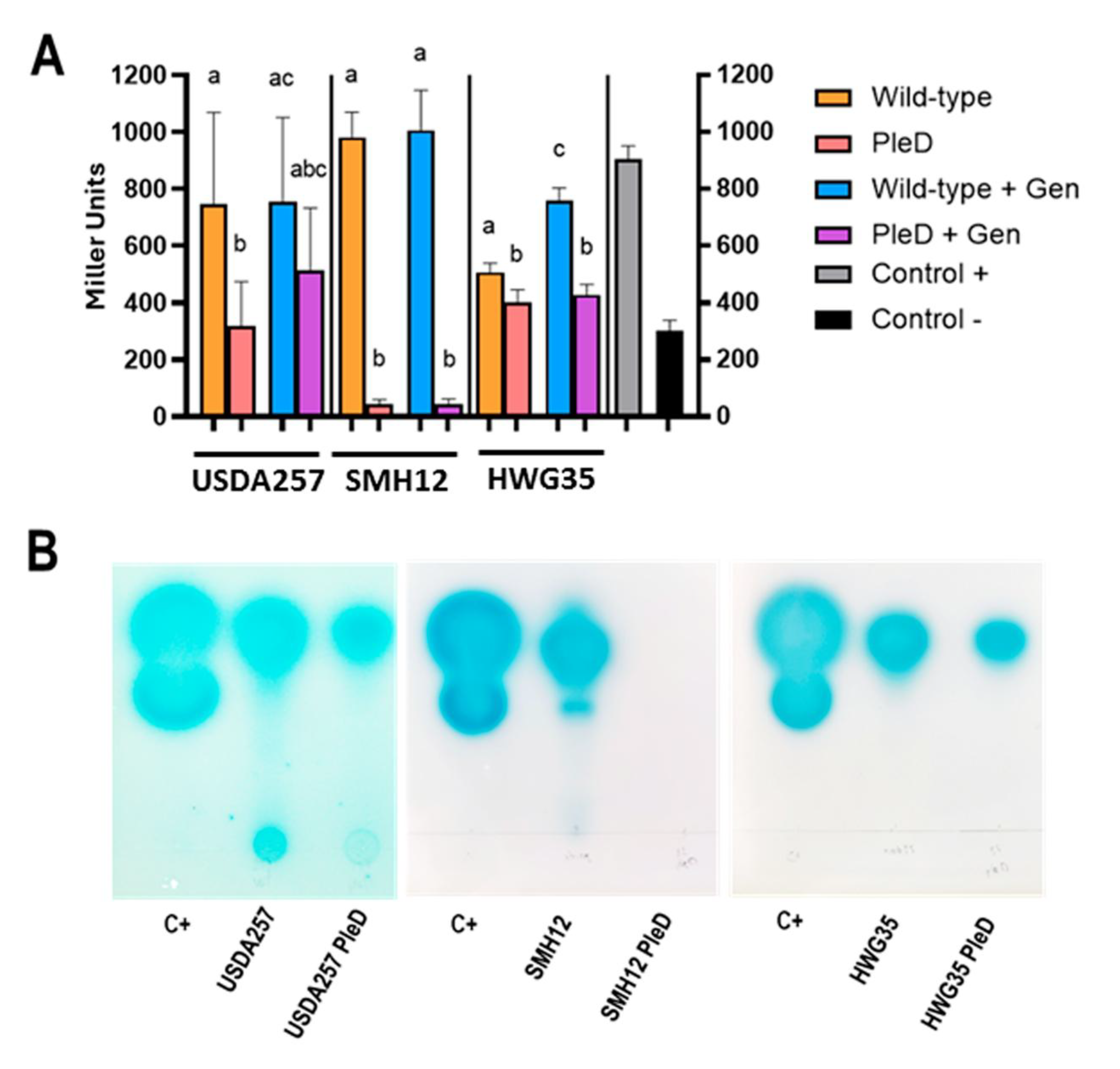
3.1. High Levels of Cyclic di-GMP Reduce AHL Production in S. fredii
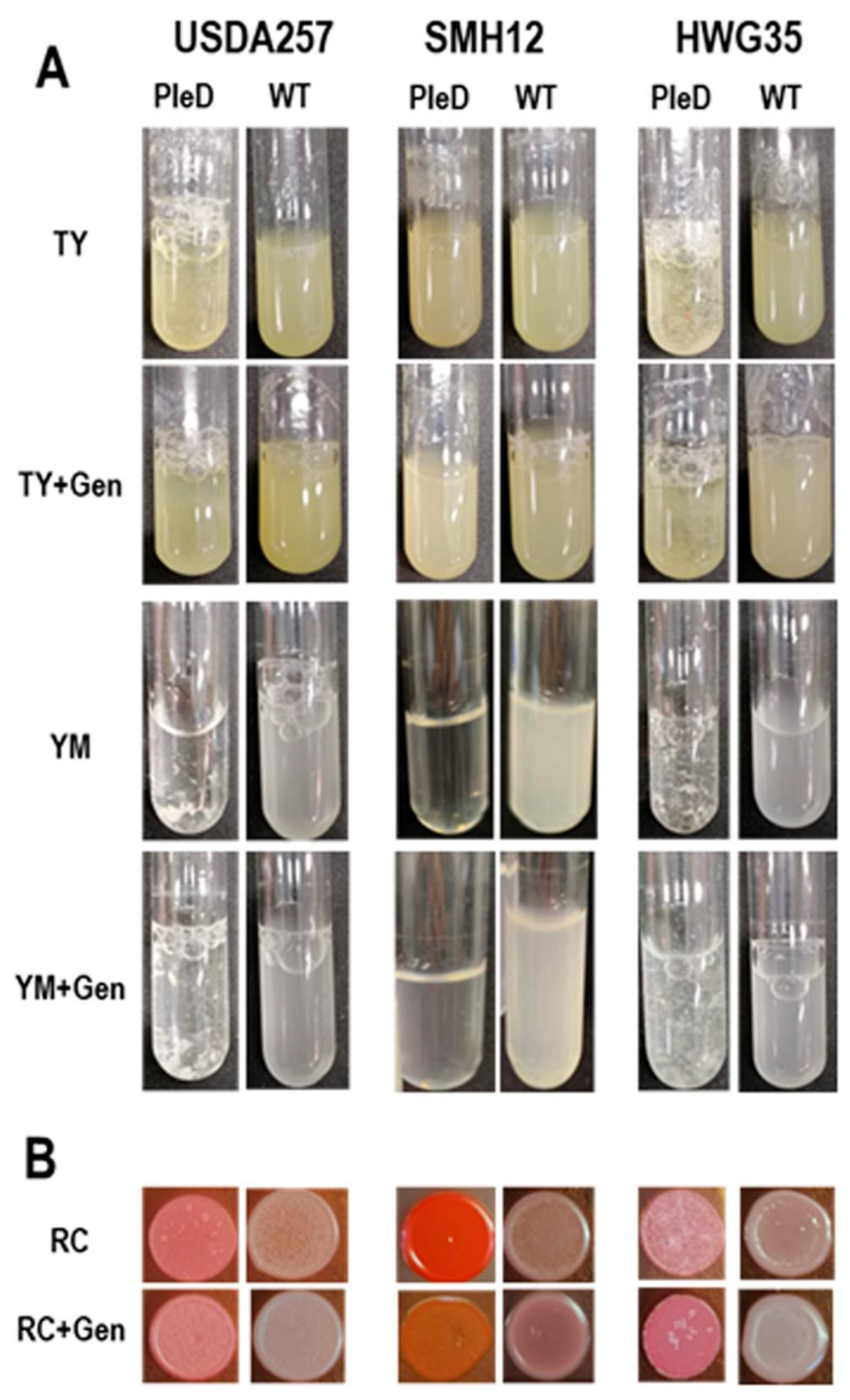
3.2. High Levels of Cyclic di-GMP Diferentially Affect Autoagregation and Adhesion to Solid Surfaces in S. fredii
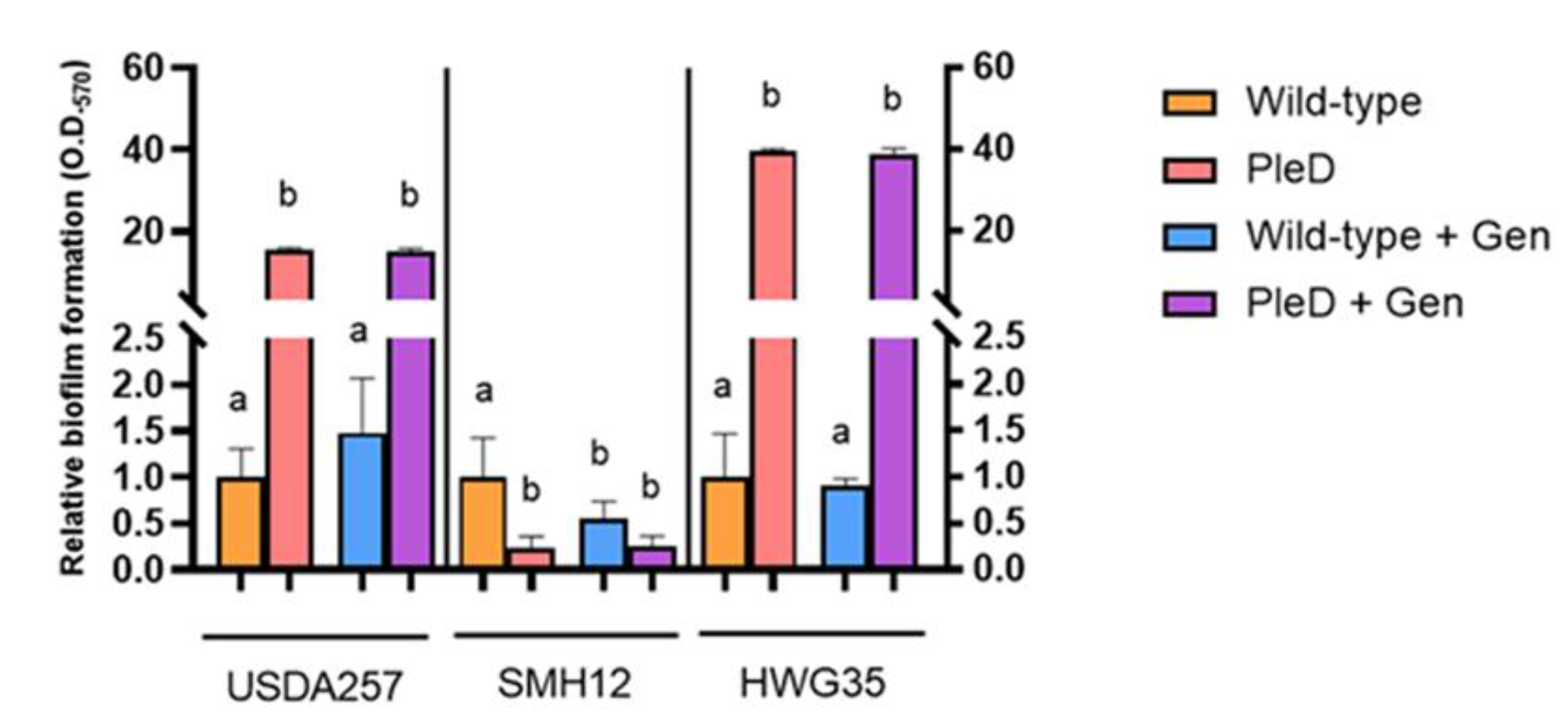
3.3. Biofilm Formation is Differentially Regulated by Elevated Cyclic di-GMP Levels Across S. Fredii Strains
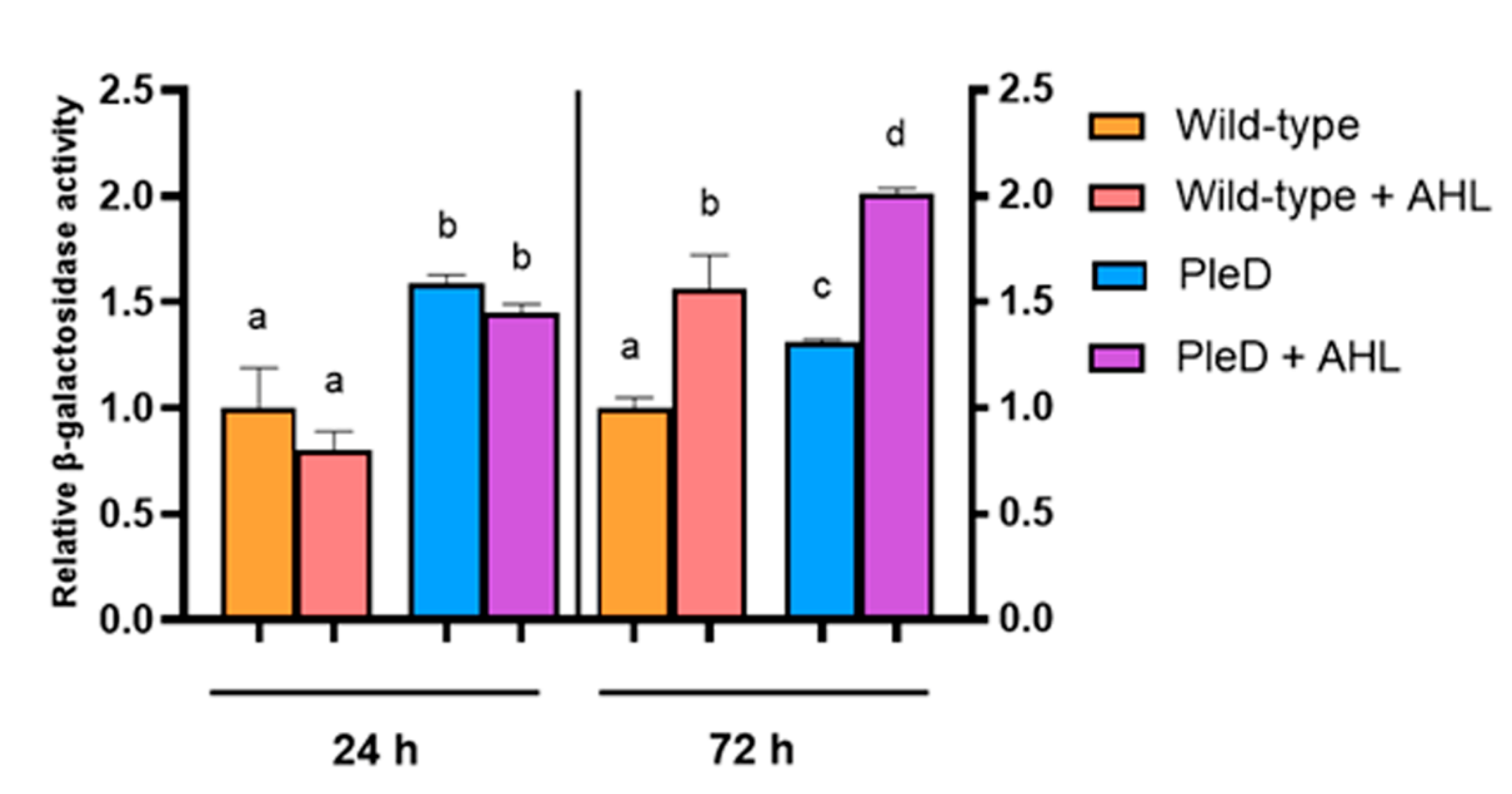
3.4. High Levels of AHLs and Cyclic di-GMP Activate the Expression of the S. fredii USDA257 T6SS Genes
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Oldroyd, G.E.D.; Downie, J.A. Coordinating Nodule Morphogenesis with Rhizobial Infection in Legumes. Ann. Rev. Gen. 2008, 42, 381–409. [Google Scholar] [CrossRef]
- Shamsul, M.; Song, Y.; Liu, Q. Symbiotic Nitrogen Fixation for Sustainable Agriculture: A Review of Recent Advances. Cleaner and Sustainable Energy 2023, 100404. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Muñoz, S.; Sanjuán, J.; Ollero, F.J.; Igual, J.M.; Muñoz-Beltrán, J.M.; Pérez-Montaño, F. Surface Polysaccharides Influence the Legume-Rhizobium Symbiosis. Int. J. Mol. Sci. 2021, 22, 6233. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Medina, C.; Vinardell, J.M.; Ollero, F.J.; López-Baena, F.J. The Rhizobial Type 3 Secretion System: The Dr. Jekyll and Mr. Hyde in the Rhizobium-Legume Symbiosis. Int. J. Mol. Sci 2023, 23, 11089. [Google Scholar] [CrossRef]
- Boyer, F.; Fichant, G.; Berthod, J.; Vandenbrouck, Y.; Attree, I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- De Campos, S.; Riedel, K.; Heckel, B.C.; Wilhelm, S.; Riedel, C.U.; Eberl, L. The Type VI Secretion System: A Versatile Bacterial Weapon. Front. Microbiol. 2017, 8, 2473. [Google Scholar] [CrossRef]
- Lin, L.; Ling, Z.; Zhang, B.; Xu, N.; Lin, Y.; Huang, J.; Xie, Y.; Guan, S.; Guo, K.; Peng, B. Type VI Secretion System of Bradyrhizobium Contributes to Symbiotic Performance and Interbacterial Competition. Mol. Plant-Microb. Interact. 2018, 31, 1249–1261. [Google Scholar] [CrossRef]
- Salinero-Lanzarote, J.; Rezzonico, F.; Otten, L.; Pedraza, J.M. Cross-Talk between Cyclic di-GMP and Quorum Sensing Regulates Motility and Biofilm Formation in Pseudomonas putida. FEMS Microbiol. Ecol. 2019, 95, fiz054. [Google Scholar] [CrossRef]
- Tighilt, M.; Mochel, S.; Mokhtari, M.; Lazzari, A.L.; Benjouad, A.; Hani, F. Involvement of the Type VI Secretion System in Bradyrhizobium Symbiosis and Bacterial Competition. Microb. Ecol. 2022, 84, 999–1012. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-Blocking Genes of Rhizobium NGR234 Identified by Signature-Tagged Mutagenesis. Mol. Plant-Microb. Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef]
- Reyes-Pérez, P.J.; Jiménez-Guerrero, I.; Sánchez-Reina, A.; Civantos, C.; Moreno-de Castro, N.; Ollero, F.J.; Gandullo, J.; Bernal, P.; Pérez-Montaño, F. The Type VI Secretion System of Sinorhizobium fredii USDA257 Is Required for Successful Nodulation with Glycine max cv Pekin. Microb. Biotech. 2025, 18, e70112. [Google Scholar] [CrossRef]
- Jenal, U.; Malone, J. Mechanisms of Cyclic-di-GMP Signaling in Bacteria. Ann. Rev. Gen. 2006, 40, 385–407. [Google Scholar] [CrossRef]
- Hall, C.L.; Lee, V.T. Cyclic di-GMP Signaling in Bacteria: Recent Advances and Perspectives. Wiley Interdisciplinary Reviews: RNA 2019, 10, e1454. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D.; Muñoz, S.; Santamaría, R.I.; Olivares, J.; Soto, M.J.; Sanjuán, J. The Symbiotic Plasmid of Sinorhizobium fredii CCBAU45436 Is Influenced by Quorum Sensing and Regulates Exopolysaccharide Production. PLoS ONE 2014, 9, e91645. [Google Scholar] [CrossRef]
- Krol, E.; Becker, A. The Regulatory Network Mediated by Cdi-GMP in Sinorhizobium meliloti and Other Rhizobia. Hist. Cell Biol. 2020, 153, 219–235. [Google Scholar] [CrossRef]
- Escobar, M.R.; Lepek, V.C.; Basile, L.A. Influence of cyclic di-GMP metabolism to T3SS expression, biofilm formation and symbiosis efficiency in Mesorhizobium japonicum MAFF303099. FEMS Microbiol. Lett. 17:370. [CrossRef]
- Romero-Jiménez, L.; Rodríguez-Carbonell, D.; Gallegos, M.T.; Sanjuán, J.; Pérez-Mendoza, D. Mini-Tn7 vectors for stable expression of diguanylate cyclase PleD* in Gram-negative bacteria. BMC Microbiol. 2015, 15, 190. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D. Rodríguez-Carvajal, M.A., Romero-Jiménez, L., de Araujo-Farias, G., Lloret, J., Gallegos, M.T., Sanjuán, J. Novel mixed-linkage β-glucan activated by c-di-GMP in Sinorhizobium meliloti. Proc. Nat. Acad. Sci. 2015, 112, E757-65. [Google Scholar] [CrossRef]
- Azimi, S.; Klementiev, A.D.; Whiteley, M.; Diggle, S.P. Bacterial Quorum Sensing During Infection. Ann. Rev. Microbiol. 2020, 74, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Calatrava-Morales, N.; McIntosh, M.; Soto, M.J. Regulation Mediated by N-Acyl Homoserine Lactone Quorum Sensing Signals in the Rhizobium-Legume Symbiosis. Genes 2018, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Waters, C.M. A Tangled Web: Regulatory Connections between Quorum Sensing and Cyclic di-GMP. J. Bacteriol. 2012, 194, 4485–4493. [Google Scholar] [CrossRef]
- Schäper, S.; Krol, E.; Skotnicka, D.; Kaever, V.; Hilker, R.; Søgaard-Andersen, L.; Becker, A. Cyclic Di-GMP Regulates Multiple Cellular Functions in the Symbiotic Alphaproteobacterium Sinorhizobium meliloti. J. Bacteriol. 2015, 198, 521–535. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Guasch-Vidal, B.; González-Barroso, S.; López-Baena, F.J.; Cubo, T.; Ollero, F.J.; Gil-Serrano, A.M.; Rodríguez-Carvajal, M.Á.; Bellogín, R.A.; Espuny, M.R. Nodulation-Gene-Inducing Flavonoids Increase Overall Production of Autoinducers and Expression of N-Acyl Homoserine Lactone Synthesis Genes in Rhizobia. Res. Microbiol. 2011, 162, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Contreras Sánchez-Matamoros, R.; López-Baena, F.J.; Ollero, F.J.; Rodríguez-Carvajal, M.A.; Bellogín, R.A.; Espuny, M.R. Rice and Bean AHL-Mimic Quorum-Sensing Signals Specifically Interfere with the Capacity to Form Biofilms by Plant-Associated Bacteria. Res. Microbiol. 2013, 164, 749–760. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Del Cerro, P.; Baena-Ropero, I.; López-Baena, F.J.; Ollero, F.J.; Bellogín, R.; Lloret, J.; Espuny, R. The Symbiotic Biofilm of Sinorhizobium fredii SMH12, Necessary for Successful Colonization and Symbiosis of Glycine max cv Osumi, Is Regulated by Quorum Sensing Systems and Inducing Flavonoids via NodD1. PLoS ONE 2014, 9, e105901. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alías-Villegas, C.; Almozara, A.; Espuny, M.R.; Vinardell, J.M.; Pérez-Montaño, F. Deciphering the Symbiotic Significance of Quorum Sensing Systems of Sinorhizobium fredii HH103. Microorganisms 2020, 8, 68. [Google Scholar] [CrossRef]
- Yang, S.S.; Bellogín, R.A.; Buendía, A.; Camacho, M.; Chen, M.; Cubo, T.; Daza, A.; Díaz, C.L.; Espuny, M.R.; Gutiérrez, R.; et al. Effect of pH and Soybean Cultivars on the Quantitative Analyses of Soybean Rhizobia Populations. J. Biotech. 2001, 91, 243–255. [Google Scholar] [CrossRef]
- Rodríguez-Carvajal, M.A.; Rodrigues, J.A.; Soria-Díaz, M.E.; Tejero-Mateo, P.; Buendía-Clavería, A.; Gutiérrez, R.; Ruiz-Sainz, J.E.; Thomas-Oates, J.; Gil-Serrano, A.M. Structural Analysis of the Capsular Polysaccharide from Sinorhizobium fredii HWG35. Biomacromolecules 2005, 6, 1448–1456. [Google Scholar] [CrossRef]
- Beringer, J.E. R Factor Transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.M. The modified Fåhraeus slide technique. In A Manual for the Practical Study of Root Nodule Bacteria; Blackwell Scientific Publications: Oxford, UK, 1970; pp. 144–145. [Google Scholar]
- Lamrabet, Y.; Bellogín, R.A.; Cubo, T.; Espuny, M.R.; Gil-Serrano, A.; Krishnan, H.B.; Megias, M.; Ollero, F.J.; Pueppke, S.G.; Ruiz-Sainz, J.E.; et al. Mutation in GDP fucose synthesis genes of Sinorhizobium fredii alters Nod factors and significantly decreases competitiveness to nodulate soybeans. Mol. Plant Microb. Interact. 1999, 12, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Keyser, H.H.; Bohlool, B.B.; Hu, T.S.; Weber, D.F. Fast-Growing Rhizobia Isolated from Root Nodules of Soybean. Science 1982, 215, 1631–1632. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, D.N.; Bellogín, R.; Camacho, M.; Daza, A.; Medina, C.; Ollero, F.J.; Santamaría, C.; Ruíz-Saínz, J.E.; Vinardell, J.M.; Temprano, F.J. Field Assessment and Genetic Stability of Sinorhizobium fredii Strain SMH12 for Commercial Soybean Inoculants. Syst. Appl. Microbiol. 2002, 25, 466–474. [Google Scholar] [CrossRef]
- Cha, C.; Gao, P.; Chen, Y.-C.; Shaw, P.D.; Farrand, S.K. Production of Acyl-Homoserine Lactone Quorum-Sensing Signals by Gram-Negative Plant-Associated Bacteria. Mol. Plant-Microb. Interact. 1998, 11, 1119. [Google Scholar] [CrossRef]
- Demarre, G.; Guérout, A.-M.; Matsumoto-Mashimo, C.; Rowe-Magnus, D.A.; Marlière, P.; Mazel, D. A New Family of Mobilizable Suicide Plasmids Based on Broad Host Range R388 Plasmid (IncW) and RP4 Plasmid (IncPα) Conjugative Machineries and Their Cognate Escherichia coli Host Strains. Res. Microbiol. 2005, 156, 245–255. [Google Scholar] [CrossRef]
- Bao, Y.; Lies, D.P.; Fu, H.; Roberts, G.P. An Improved Tn7-Based System for the Single-Copy Insertion of Cloned Genes into Chromosomes of Gram-Negative Bacteria. Gene 1991, 109, 167–168. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York (USA). 1972.
- O'Toole, G.A.; Kolter, R. Initiation of Biofilm Formation in Pseudomonas fluorescens WCS365 Proceeds via Multiple, Convergent Signalling Pathways: A Genetic Analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Navarro-Gómez, P.; Del Socorro Murdoch, P.; Crespo-Rivas, J.-C.; Jie, S.; Cuesta-Berrio, L.; Ruiz-Sainz, J.-E.; Rodríguez-Carvajal, M.-Á.; Vinardell, J.M. Exopolysaccharide Production by Sinorhizobium fredii HH103 Is Repressed by Genistein in a NodD1-Dependent Manner. PLoS ONE 2016, 11, e0160499. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Kahn, S.G.; Bohannon, J.; Travisano, M.; Rainey, P.B. Adaptive Divergence in Experimental Populations of Pseudomonas fluorescens. I. Genetic and Phenotypic Bases of Wrinkly Spreader Fitness. Genetics 2002, 161, 33–46. [Google Scholar] [CrossRef]
- Thomas-Oates, J.; Bereszczak, J.; Edwards, E.; Gill, A.; Noreen, S.; Zhou, J.C.; Chen, M.Z.; Miao, L.H.; Xie, F.L.; Yang, J.K.; et al. A Catalogue of Molecular, Physiological and Symbiotic Properties of Soybean-Nodulating Rhizobial Strains from Different Soybean Cropping Areas of China. Syst. App. Microbiol. 2003, 26, 453–465. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, MB-0011-2014. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Coulthurst, S.J.; Sanjuán, J.; Salmond, G.P.C. N-Acetylglucosamine-dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c-di-GMP levels. Microbiol. 2011, 157(Pt 12) Pt 12, 3340–3348. [Google Scholar] [CrossRef]
- Allsopp, L.P.; Bernal, P. Killing in the name of: T6SS structure and effector diversity. Microbiol. 2023, 169, 001367. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, H.; Ma, J.; Li, H.; Ma, X.; Tang, Y.; Li, J.; Chi, X.; Deng, Y.; Zeng, S.; Liu, Z. Role of Type VI secretion system in pathogenic remodeling of host gut microbiota during Aeromonas veronii infection. ISME J. 2024, 18, wrae053. [Google Scholar] [CrossRef]
- Salinero-Lanzarote, A.; Pacheco-Moreno, A.; Domingo-Serrano, L.; Durán, D.; Ormeño-Orrillo, E.; Martínez-Romero, E.; Albareda, M.; Palacios, J.M.; Rey, L. The Type VI secretion system of Rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol. Ecol. 2019, 95, fiz054. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.; Schwarz, S.; Mougous, J.D. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol. Microbiol. 2009, 72, 1111–1125. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Sauer, K. Controlling biofilm development through cyclic di-GMP signaling. Adv. Exp. Med. Biol. 2022, 1386, 69–94. [Google Scholar] [CrossRef]
- Huang, W.; Wang, D.; Zhang, X.-X.; Zhao, M.; Sun, L.; Zhou, Y.; Guan, X.; Xie, Z. Regulatory roles of the second messenger c-di-GMP in beneficial plant-bacteria interactions. Microbiol. Res. 2024, 285, 127748. [Google Scholar] [CrossRef] [PubMed]
- Römling, U. Cyclic di-GMP signaling—Where did you come from and where will you go? Mol. Microbiol. 2023, 120, 564–574. [Google Scholar] [CrossRef]
- Hespanhol, J. T.; Nóbrega-Silva, L.; Bayer-Santos, E. Regulation of Type VI Secretion Systems at the Transcriptional, Posttranscriptional and Posttranslational Level. Microbiology 2023, 169, 001376. [Google Scholar] [CrossRef]
- López-Baena, F.J.; Vinardell, J.M.; Medina, C. Regulation of protein secretion systems mediated by cyclic diguanylate in plant-interacting bacteria. Front. Microbiol. 2019, 10, 1289. [Google Scholar] [CrossRef]
- Valentini, M.; Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Lu, W.; Rabinowitz, J.D.; Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008, 190, 2527–2536. [Google Scholar] [CrossRef] [PubMed]
- Prentice, J.A.; Bridges, A.A.; Bassler, B.L. Synergy between c-di-GMP and quorum-sensing signaling in Vibrio cholerae biofilm morphogenesis. J. Bacteriol. 2022, 204, e0024922. [Google Scholar] [CrossRef] [PubMed]
| Strain or Plasmid | Relevant Characteristics | Reference |
| S. fredii | ||
| USDA257 | Wild-type strain, RifR | [33] |
| USDA257 miniTn7-Km | USDA257 miniTn7, KmR | This work |
| USDA257-PleD | USDA257 miniTn7-pleD*, KmR | This work |
| USDA257- miniTn7-Km pMUS1483 | USDA257 miniTn7, KmR, containing a plasmid with a PppkA::lacZ fusion. See below | This work |
| USDA257-PleD pMUS1483 | USDA257 miniTn7-pleD*, KmR containing a plasmid with a PppkA::lacZ fusion. See below | This work |
| SMH12 | Wild-type strain, RifR | [34] |
| SMH12 miniTn7-Tc | SMH12 miniTn7-TcR | This work |
| SMH12-PleD | SMH12 miniTn7-pleD*, TcR | This work |
| HWG35 | Wild-type strain, RifR | [27] |
| HWG35 miniTn7-Km | HWG35 miniTn7-KmR | This work |
| HWG35-PleD | HWG35 miniTn7-pleD*, KmR | This work |
| A. tumefaciens | ||
| NT1 (pZRL4) | A. tumefaciens devoid of pTiC58 and harboring pZRL4, which carries the fusion traG::lacZ and the traR gene | [35] |
| E. coli | ||
| β2163 | MG1655::ΔdapA::(erm-pir)RP4-2, Tc::Mu, KmR, EmR | [36] |
| β2155 | RP4-2-Tc::Mu ΔdapA::(erm-pir) thrB1004, pro, thi, strA, hsdS, lacZ ΔM15, (F´ lacZ ΔM15,lacIq, traD36, proA+, proB+) KmR, SmR, EmR | [36] |
| Plasmids | ||
| mini-Tn7-pleD*-Km | mini-Tn7pleD* containing Km marker ApR, KmR | [17] |
| mini-Tn7-Km | mini-Tn7pleD*Km with an internal deletion of pleD* ApR, KmR | [17] |
| mini-Tn7pleD*Tc | mini-Tn7pleD* containing Tc marker ApR, TcR | [17] |
| mini-Tn7-Tc | mini-Tn7pleD*Tc with an internal deletion, ApR, TcR | [17] |
| pUX-BF13 | Helper plasmid providing the Tn7 transposition functions in trans, ApR, mob+, ori-R6K | [37] |
| pMUS1483 | S. fredii USDA257 ppkA promoter region cloned into pMP220 as a transcriptional fusion, PppkA::lacZ, RK2 ori, TcR. [pMP220::PppkA] | [11] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





