Submitted:
29 July 2025
Posted:
30 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Retrieval of Gene Sequences and Analysis
2.2. Phylogenetic and Selection Pressure Analyses of HA, NP, and NA Genes
2.3. Protein Sequence Retrieval and Phylogenetic Analysis of HA, NP, and NA Proteins
2.4. Structure Prediction and Assessment of HA, NP, and NA Proteins
2.5. MD-Simulation of Predicted Structures of HA, NP, and NA Proteins
2.6. Docking of the H5N1 Clade 2.3.4.4b (HA, NP, and NA) Proteins with Their Ligand-Receptors
3. Results and Discussion
3.1. Sequence Analysis of HA, NP, and NA Genes
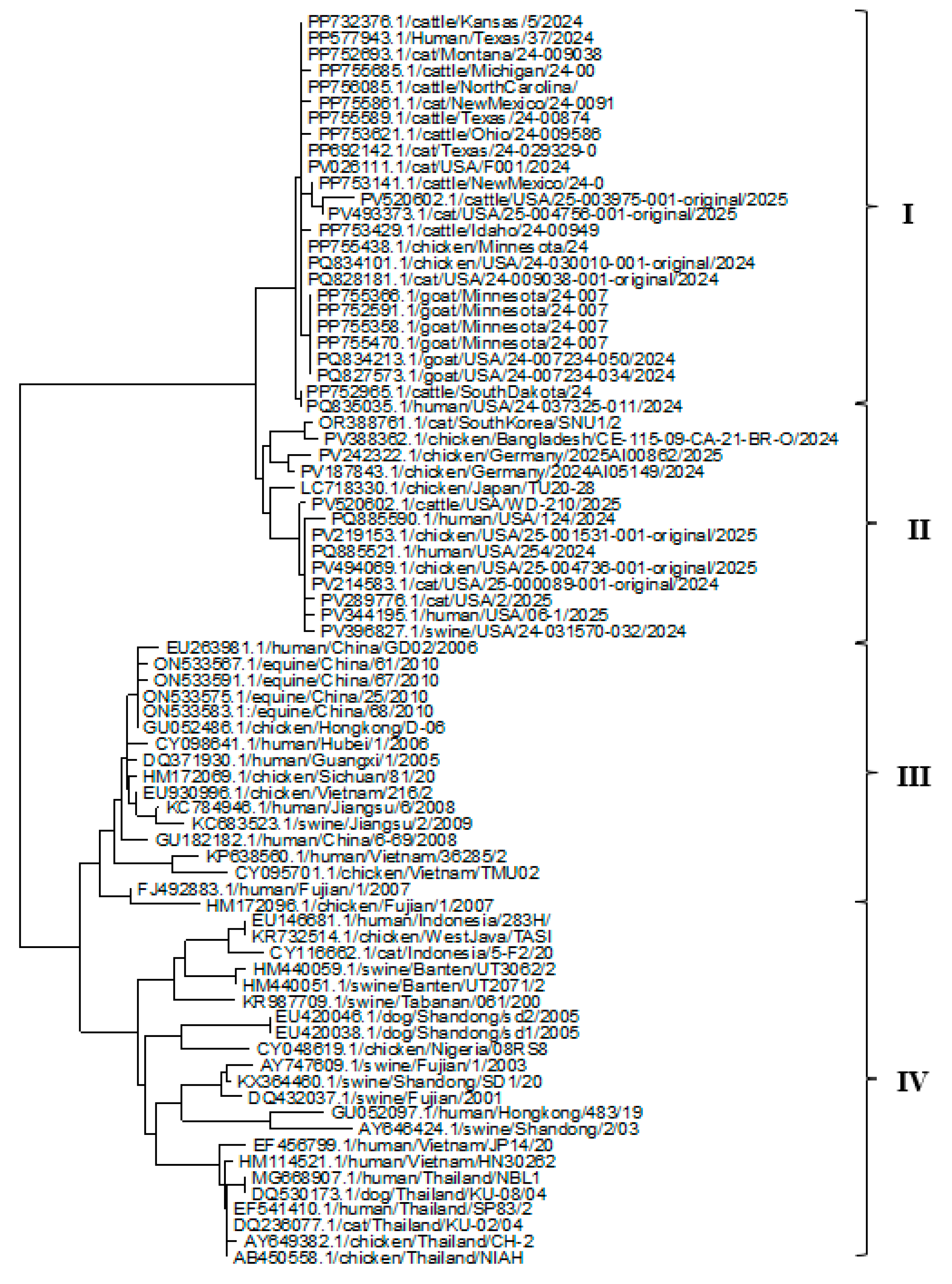
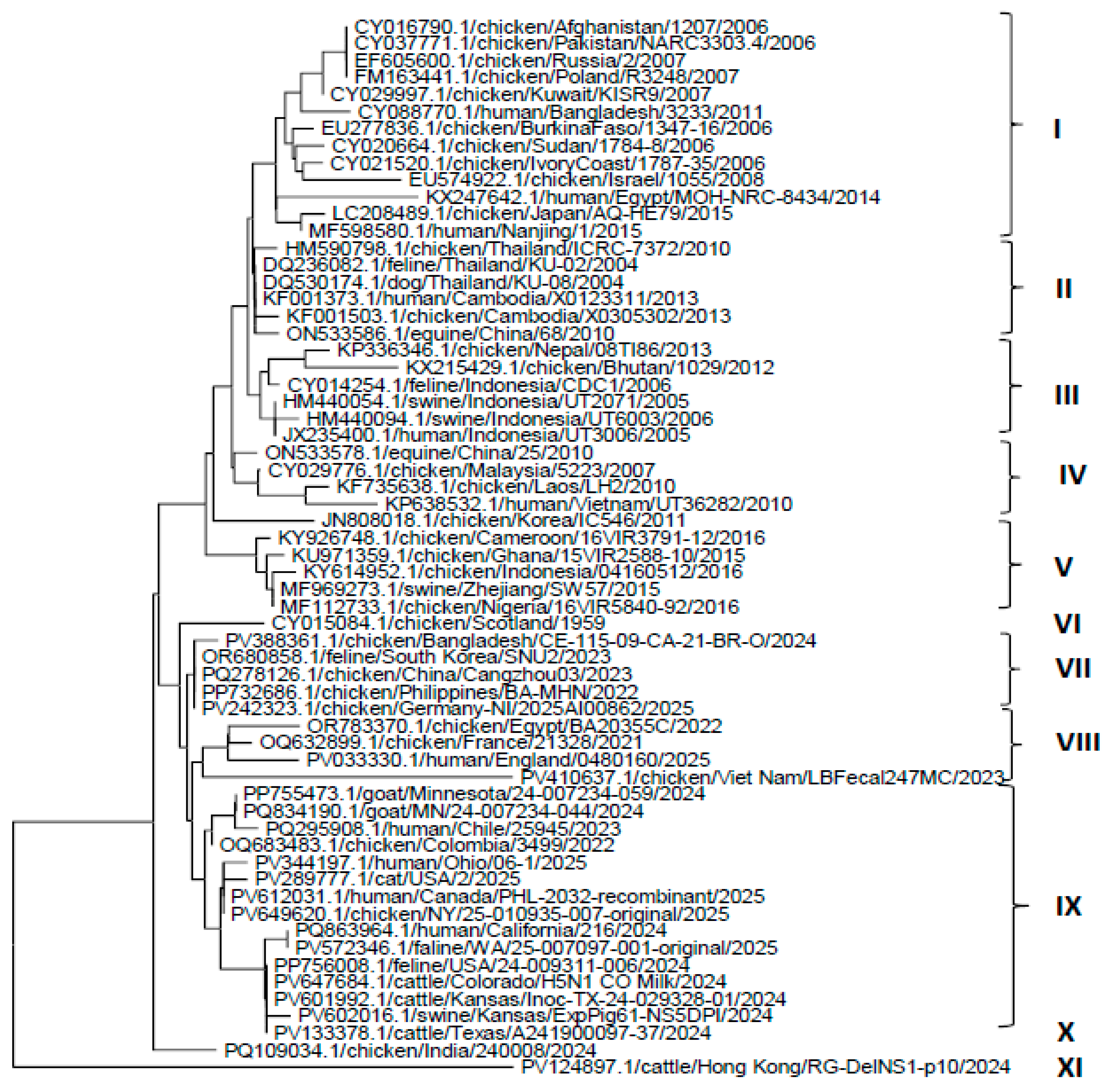
3.2. Phylogenetic Analysis of HA, NP, and NA Genes
3.3. Selection Pressure Analysis of HA, NP, and NA Genes
3.4. Phylogenetic Analysis of the (HA, NP, and NA) Proteins of the H5N1 Clade 2.3.4.4b
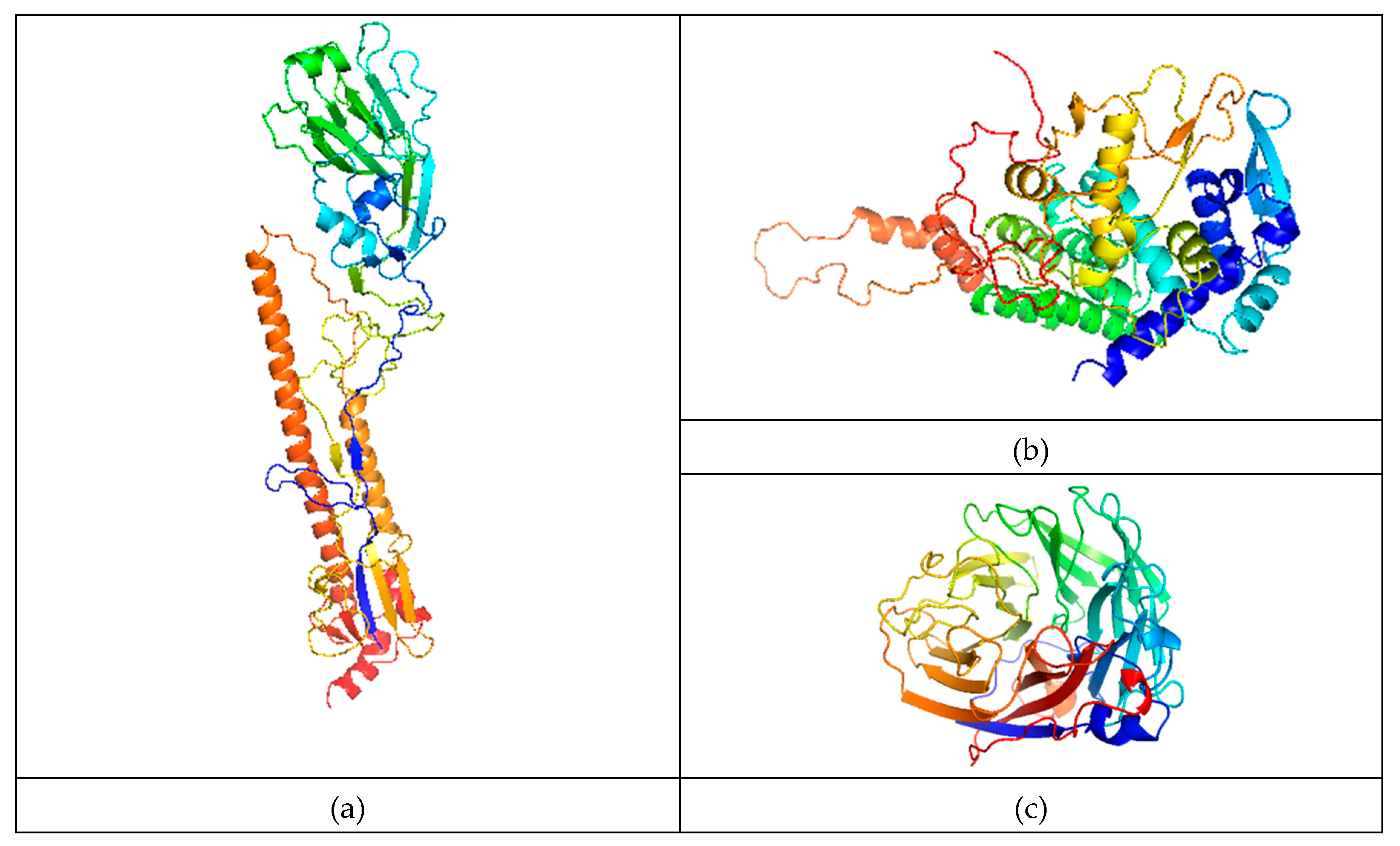
3.5. Protein Structure Prediction, MD-Simulation, and Domain Analysis of the (HA, NP and NA) Proteins
3.6. Molecular Docking of HA, NP, and NA Proteins with Their Ligands
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HA | Hemagglutinin |
| NA | Neuraminidase |
| NP | Nucleoprotein |
| IAV | Influenza A Virus |
| HPAI | Highly Pathogenic Avian Influenza |
| WHO | World Health Organization |
| MD | Molecular Dynamics |
| NCBI | National Center for Biotechnology Information |
| NVT | Constant Number of Particles, Volume, and Temperature |
| NPT | Constant Number of Particles, Pressure, and Temperature |
| OPLS | Optimized Potentials for Liquid Simulations |
| SPC/E | Extended Simple Point Charge (water model) |
| BLASTp | Basic Local Alignment Search Tool for Proteins |
| ProSA | Protein Structure Analysis |
| QMEAN | Qualitative Model Energy Analysis |
| CD-Dock2 | Curvature Detection-based Docking Tool |
| NAG-GAL-SIA | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose |
| SIA | N-acetylneuraminic acid (Sialic acid) |
References
- Alnaeem, A.A.; Al-Shabeb, A.; Hemida, M.G. Evaluation of the immune status of birds and domestic and companion animals for the influenza A virus in Eastern Saudi Arabia. Vet World 2020, 13, 1966–1969. [Google Scholar] [CrossRef]
- Dadonaite, B.; Gilbertson, B.; Knight, M.L.; Trifkovic, S.; Rockman, S.; Laederach, A.; Brown, L.E.; Fodor, E.; Bauer, D.L.V. The structure of the influenza A virus genome. Nat Microbiol 2019, 4, 1781–1789. [Google Scholar] [CrossRef]
- Hemida, M.G.; Chu, D.; Abdelaziz, A.; Alnaeem, A.; Chan, S.M.S.; Peiris, M. Molecular characterisation of an avian influenza (H5N8) outbreak in backyard flocks in Al Ahsa, Eastern Saudi Arabia, 2017-2018. Vet Rec Open 2019, 6, e000362. [Google Scholar] [CrossRef]
- Hemida, M.G.; Perera, R.; Chu, D.K.W.; Alnaeem, A.A.; Peiris, M. Evidence of equine influenza A (H3N8) activity in horses from Eastern and Central Saudi Arabia: 2013-2015. Equine Vet J 2019, 51, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Hermann, E.; Rasmussen, A.L. Highly pathogenic avian influenza H5N1: history, current situation, and outlook. J Virol 2025, 99, e0220924. [Google Scholar] [CrossRef] [PubMed]
- Charostad, J.; Rezaei Zadeh Rukerd, M.; Mahmoudvand, S.; Bashash, D.; Hashemi, S.M.A.; Nakhaie, M.; Zandi, K. A comprehensive review of highly pathogenic avian influenza (HPAI) H5N1: An imminent threat at doorstep. Travel Med Infect Dis 2023, 55, 102638. [Google Scholar] [CrossRef]
- Skeik, N.; Jabr, F.I. Influenza viruses and the evolution of avian influenza virus H5N1. Int J Infect Dis 2008, 12, 233–238. [Google Scholar] [CrossRef]
- Diskin, E.R.; Friedman, K.; Krauss, S.; Nolting, J.M.; Poulson, R.L.; Slemons, R.D.; Stallknecht, D.E.; Webster, R.G.; Bowman, A.S. Subtype Diversity of Influenza A Virus in North American Waterfowl: a Multidecade Study. J Virol 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Shi, J.; Wang, C.; Zhang, Y.; Xing, X.; Kong, H.; Yan, C.; Zeng, X.; Liu, L.; Tian, G.; et al. Global dissemination of H5N1 influenza viruses bearing the clade 2.3.4.4b HA gene and biologic analysis of the ones detected in China. Emerg Microbes Infect 2022, 11, 1693–1704. [Google Scholar] [CrossRef]
- Pardo-Roa, C.; Nelson, M.I.; Ariyama, N.; Aguayo, C.; Almonacid, L.I.; Munoz, G.; Navarro, C.; Avila, C.; Ulloa, M.; Reyes, R.; et al. Cross-species transmission and PB2 mammalian adaptations of highly pathogenic avian influenza A/H5N1 viruses in Chile. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sriwilaijaroen, N.; Suzuki, Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci 2012, 88, 226–249. [Google Scholar] [CrossRef]
- Cohen, M.; Zhang, X.Q.; Senaati, H.P.; Chen, H.W.; Varki, N.M.; Schooley, R.T.; Gagneux, P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 2013, 10, 321. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. Structural Biology of Influenza Hemagglutinin: An Amaranthine Adventure. Viruses 2020, 12. [Google Scholar] [CrossRef]
- Tan, C.C.S.; van Dorp, L.; Balloux, F. The evolutionary drivers and correlates of viral host jumps. Nat Ecol Evol 2024, 8, 960–971. [Google Scholar] [CrossRef]
- Prachanronarong, K.L.; Canale, A.S.; Liu, P.; Somasundaran, M.; Hou, S.; Poh, Y.P.; Han, T.; Zhu, Q.; Renzette, N.; Zeldovich, K.B.; et al. Mutations in Influenza A Virus Neuraminidase and Hemagglutinin Confer Resistance against a Broadly Neutralizing Hemagglutinin Stem Antibody. J Virol 2019, 93. [Google Scholar] [CrossRef]
- Malbranke, C.; Bikard, D.; Cocco, S.; Monasson, R. Improving sequence-based modeling of protein families using secondary-structure quality assessment. Bioinformatics 2021, 37, 4083–4090. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Shi, J.; Cui, P.; Yan, C.; Zhang, Y.; Zhang, Y.; Wang, C.; Chen, Y.; Zeng, X.; Tian, G.; et al. Evolution and biological characterization of H5N1 influenza viruses bearing the clade 2.3.2.1 hemagglutinin gene. Emerg Microbes Infect 2024, 13, 2284294. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Bergner, A.; Schwede, T. Modelling three-dimensional protein structures for applications in drug design. Drug Discov Today 2014, 19, 890–897. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Dushoff, J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc Natl Acad Sci U S A 2003, 100, 7152–7157. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Lipman, D.J.; Pearson, W.R. Rapid and sensitive protein similarity searches. Science 1985, 227, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. Proteins 2007, 66, 778–795. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: the protein sequence classification resource in 2025. Nucleic Acids Res 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Gamblin, S.J.; Vachieri, S.G.; Xiong, X.; Zhang, J.; Martin, S.R.; Skehel, J.J. Hemagglutinin Structure and Activities. Cold Spring Harb Perspect Med 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.K.; Guttman, M.; Ebner, J.L.; Lee, K.K. Dynamic changes during acid-induced activation of influenza hemagglutinin. Structure 2015, 23, 665–676. [Google Scholar] [CrossRef]
- Hosaka, H.; Nakagawa, A.; Tanaka, I.; Harada, N.; Sano, K.; Kimura, M.; Yao, M.; Wakatsuki, S. Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor. Structure 1997, 5, 1199–1208. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates. Virus Genes 2022, 58, 255–269. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res 2022, 50, W159–W164. [Google Scholar] [CrossRef]
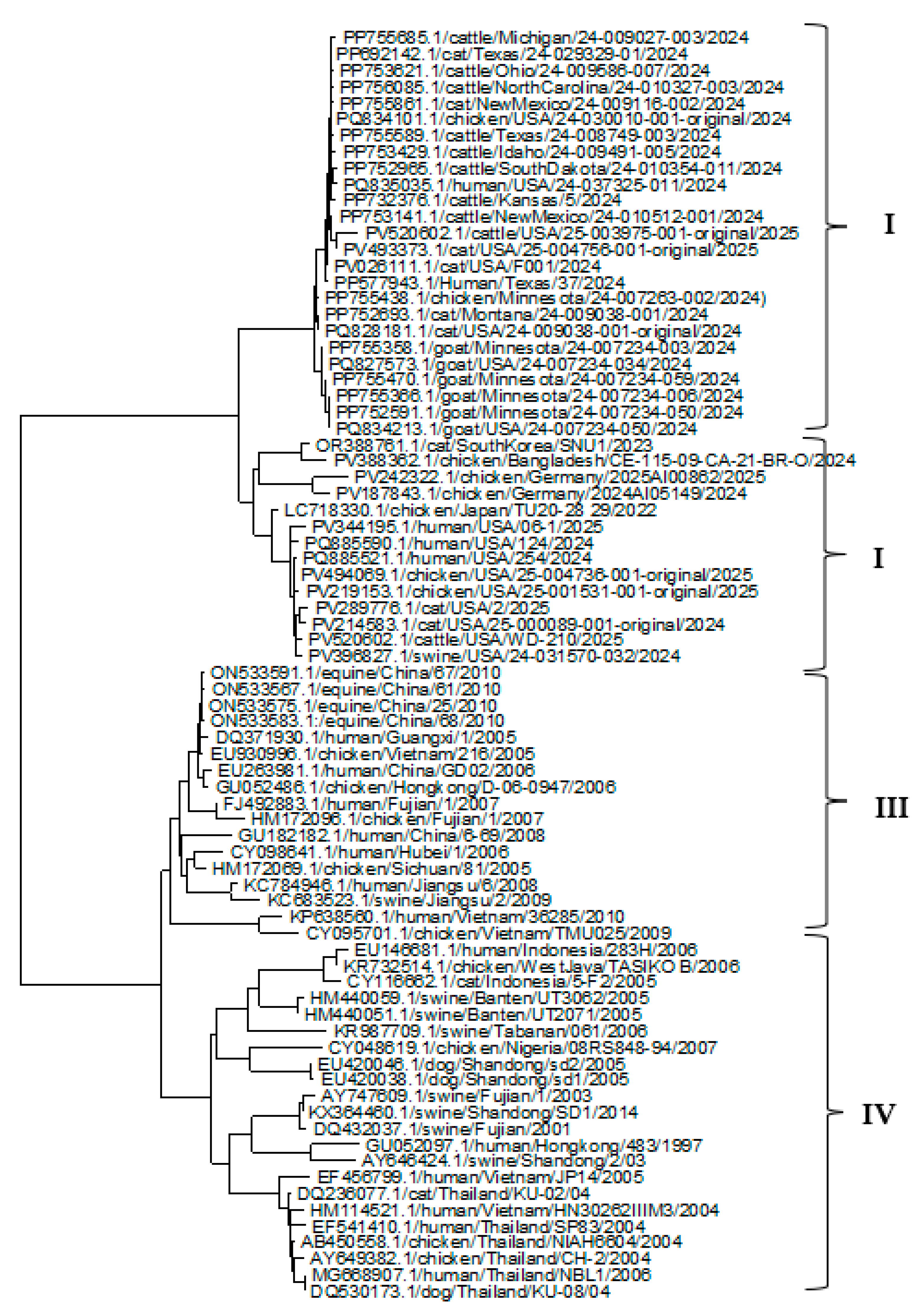
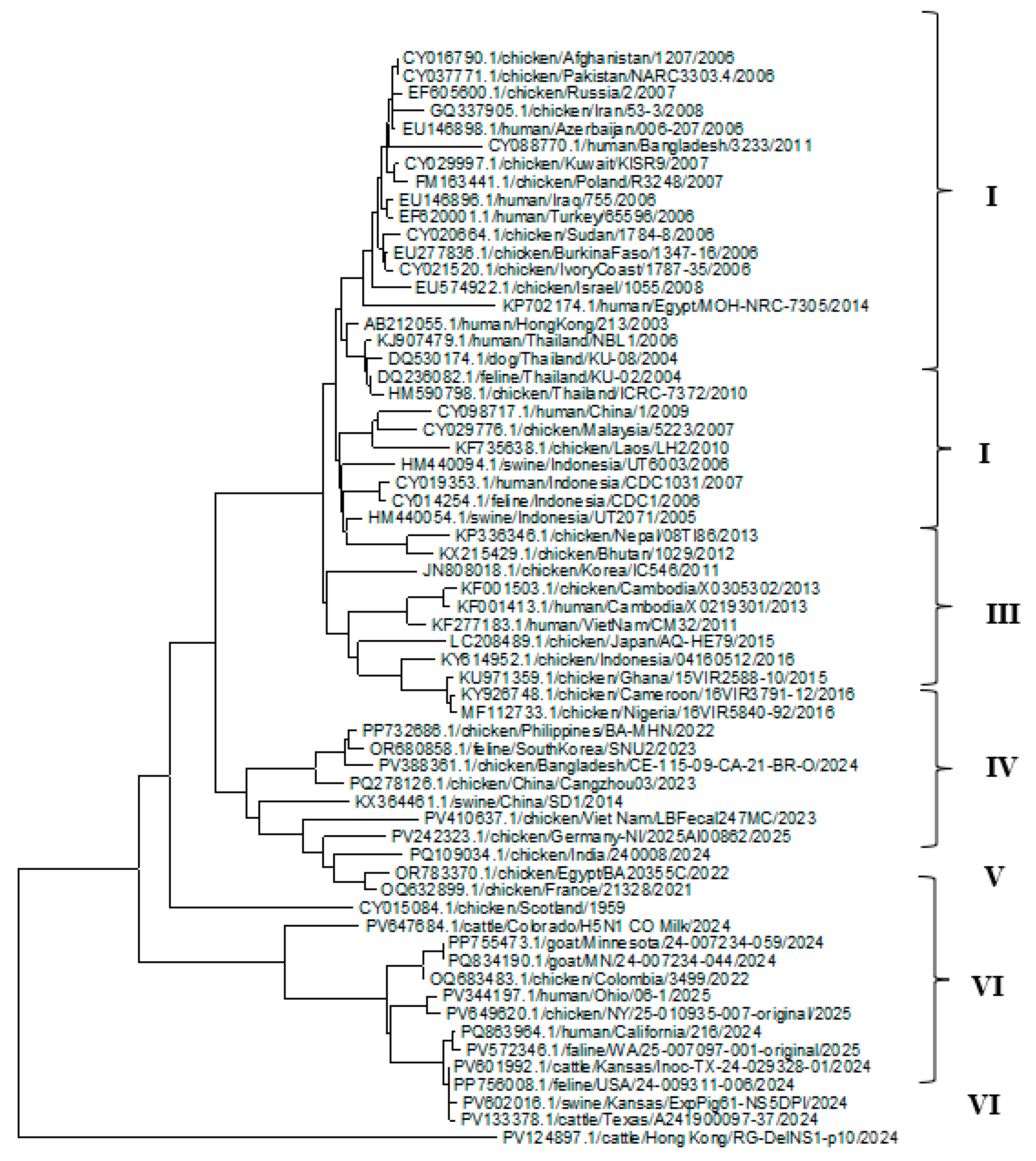


| Gene | No. of sequences | No. of codons | No. of codons under selection | Non-synonymous/ synonymous rate ratio | Selection pressure |
|---|---|---|---|---|---|
| HA gene | 78 | 530 | 3 | 2.7856 | Positive |
| NP gene | 61 | 439 | 2 | 2.4435 | Positive |
| NA gene | 62 | 435 | 13 | 2.4312 | Positive |
| Protein | Codon no. | S | N | dS | dN | Selection pressure |
|---|---|---|---|---|---|---|
| HA | 151 | 0.000 | 6.000 | 0.000 | 3.000 | Positive |
| 191 | 3.000 | 1.000 | 3.752 | 0.454 | Negative | |
| 487 | 2.000 | 0.000 | 2.499 | 0.000 | Negative | |
| NP | 10 | 0 | 4 | 0 | 2.507 | Positive |
| 138 | 1 | 9 | 0 | 9 | Negative | |
| NA | 17 | 1 | 5 | 0.678 | 3.538 | Positive |
| 45 | 6 | 5 | 7.237 | 2.313 | Negative | |
| 91 | 0 | 6 | 0 | 2.979 | Positive | |
| 105 | 0 | 6 | 0 | 3.02 | Positive | |
| 112 | 0 | 7 | 0 | 3.464 | Positive | |
| 175 | 3 | 0 | 3 | 0 | Negative | |
| 181 | 0 | 6 | 0 | 2.986 | Positive | |
| 235 | 0 | 8 | 0 | 4.102 | Positive | |
| 271 | 3 | 1 | 4.117 | 0.44 | Negative | |
| 347 | 1.5 | 5.5 | 0.917 | 4.03 | Positive | |
| 348 | 0 | 9 | 0 | 4.287 | Positive | |
| 352 | 0 | 6 | 0 | 2.997 | Positive | |
| 402 | 0 | 6 | 0 | 3 | Positive |
| Proteins | Assessment scores | Pre MD-simulation | Post MD-simulation |
|---|---|---|---|
| HA | Procheck | 92.06 (Ramachandran score) | 94.79 (Ramachandran score) |
| 79.1% most favoured core | 88.7% most favoured core | ||
| 18.7% additional allowed region | 9.7% additional allowed region | ||
| 1.6% generously allowed region | 0.9% generously allowed region | ||
| 0.7% disallowed region | 0.7% disallowed region | ||
| ERRAT | 91.9913 | 92.5926 | |
| Verify3D | 78.64% | 79.44% | |
| QMEAN | 0.71 | 0.78 | |
| ProSA | -9.8 | -9.5 | |
| NP | Procheck | 89.02 (Ramachandran score) | 91.75 (Ramachandran score) |
| 85.4% most favoured core | 86.1% most favoured core | ||
| 12.9% additional allowed region | 12.4% additional allowed region | ||
| 1.4% generously allowed region | 1.2% generously allowed region | ||
| 0.2% disallowed region | 0.2% disallowed region | ||
| ERRAT | 90.9548 | 96.1276 | |
| Verify3D | 84.21% | 82.32% | |
| QMEAN | 0.69 | 0.79 | |
| ProSA | -9.7 | -9.36 | |
| NA | Procheck | 91.53 (Ramachandran score) | 94.82 (Ramachandran score) |
| 76.9% most favoured core | 86.7% most favoured core | ||
| 20.4% additional allowed region | 13.0% additional allowed region | ||
| 2.8% generously allowed region | 0.3% generously allowed region | ||
| 0.0% disallowed region | 0.0% disallowed region | ||
| ERRAT | 82.9913 | 89.0957 | |
| Verify3D | 78.64% | 84.75% | |
| QMEAN | 0.88 | 0.93 | |
| ProSA | -5.4 | -5.15 |
| Protein | Ligand | Template | Docking score | Binding site residues |
|---|---|---|---|---|
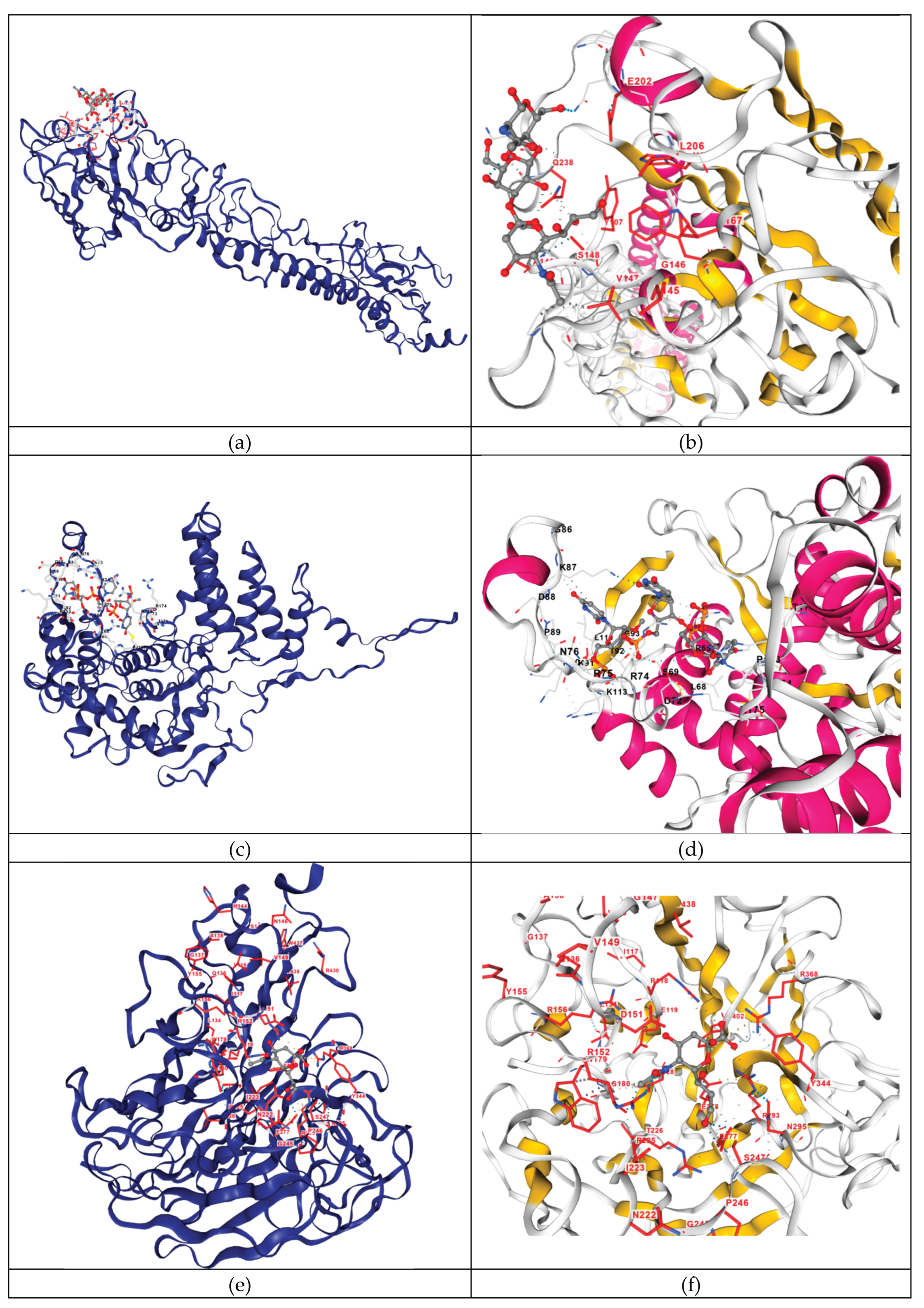
| HA | NAG-GAL-SIA (N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose) | 1hge | -2.8 | V147 S148 A149 A150 P157 W165 N198 N199 E202 L206 Q234 G237 Q238 |
| NP | RNA | 7dxp | -7.5 | Q58 I61 T62 R65 M66 L68 S69 A70 D72 E73 R74 R75 N76 H82 G86 K87 D88 P89 K90 K91 T92 G93 G94 P95 L108 L110 K113 R117 L136 M137 H140 T171 L172 P173 R174 R175 S367 |
| NA | SIA (N-ACETYL-ALPHA-NEURAMINIC ACID) | 4gzq | -5.7 | R118 E119 D151 R152 R156 W179 S180 R225 E228 S247 E277 E278 R293 N295 Y344 G345 R368 Y402 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





