1. Introduction
Viruses are structurally diverse nanocapsules carrying genetic material, that infect host organisms with an array of molecular tricks. The tomato brown rugose fruit virus (ToBRFV) is a positive-sense single-stranded RNA plant virus classified in the genus
Tobamovirus (family
Virgaviridae). It was first recognized as a distinct Tobamovirus species in 2016 [
1]. The spreading of ToBRFV has caused major agricultural damages globally [
1,
2,
3,
4,
5,
6]. ToBRFV does not have any known insect vectors and is transmitted mechanically (
via contact or contaminated tools/seeds), just like other tobamoviruses. ToBRFV virions are non-enveloped, rigid helical rods consisting of many coat protein subunits encapsidating the RNA. A recent transmission electron microscope study measured the dimensions of ToBRFV particles ~274.8 nm by 13.9 nm [
7], which is comparable with the 300×18 nm dimensions reported for related tobamoviruses. The capsid has helical symmetry with each coat protein (CP) subunit bound to the genomic RNA. Similarly to the tobacco mosaic virus (TMV), ~2,130 copies of the CP likely assemble per virion, forming a hollow, helical tube around the RNA. The RNA is tightly intercalated along the inner surface of the protein helix. This “crinkled cylindrical” capsid architecture protects the RNA and gives tobamoviruses their characteristic stability. The CP of ToBRFV comprises 159 amino acids (≈17.5 kDa), very similar to other tobamovirus CPs [
8], and it mediates particle assembly and aids long-distance movement in plants. ToBRFV contains a single-stranded positive-sense RNA genome 6.392 kb in length, with a 5′ methylated cap and a 3′ tRNA-like structure instead of a poly(A) tail. The genome encodes four open reading frames (ORFs). At the 5′ end, ORF1 and ORF2 overlap
via a leaky stop codon and together code for the viral replicase, a 126 kDa protein and a 183 kDa readthrough protein that function as subunits of the RNA-dependent RNA polymerase (RdRP) [
8,
9]. Further downstream, ORF3 encodes the movement protein (MP) (~28–31 kDa), which enables cell-to-cell spreading through plasmodesmata [
8,
9]. The final ORF4 encodes the CP (~17.5 kDa) that forms the capsid [
8,
9]. ORF3 and ORF4 are expressed from subgenomic RNAs produced during replication [
8,
9]. The genomic RNA also has untranslated regions (UTRs) that are important for replication and encapsidation and, notably, a tRNA-like structure at the 3′ end as in other tobamoviruses. ToBRFV isolates are very similar genetically worldwide (over 99% identity) [
10], but if any structural variations exist, they could affect properties such as virulence. It remains unknown whether any mutations in the coat protein or other structural regions of ToBRFV have functional consequences on stability or host interactions. So far, the main resistance-breaking determinant was found in the movement protein [
8], not in the CP, and serologically the CP is cross-reactive with TMV and tomato mosaic virus (ToMV). This suggests that the CP structure is highly conserved. We currently lack detailed knowledge of how ToBRFV disassembles inside the host. In TMV, the process by which the coat protein is stripped off the RNA is only partly understood, and for ToBRFV it is entirely inferred. A recent cryo-electron microscopy study pointed out that changes in the local pH and calcium level may modulate the interaction between key glutamates (called Caspar-carboxylates [
11]) of neighboring CP, serving as a disassembly switch in TMV [
12]. Whether a similar mechanism may operate in ToBRFV and whether structural intermediates are present during disassembly, are not known.
Here we investigated the topographical structure and nanomechanical properties of ToBRFV by using atomic force microscopy and force mapping. We find that ToBRF virions are soft rodlike structures that can be fragmented into unit-length elements which may have important implications on the infection efficiency.
4. Discussion
In the present work we investigated the topographical structure and nanomechanical properties of the tomato brown rugose fruit virus (ToBRFV). ToBRFV, a member of the
Tobamovirus genus, is a recently emerged virus, the spread of which has caused major agricultural damages globally [
1,
2,
3,
4,
5,
6]. Understanding the detailed features of ToBRFV is important in developing preventive and defensive measures. Furthermore, the structural and mechanical information collected may help us better understand the behavior of further pathogenic tobamoviruses.
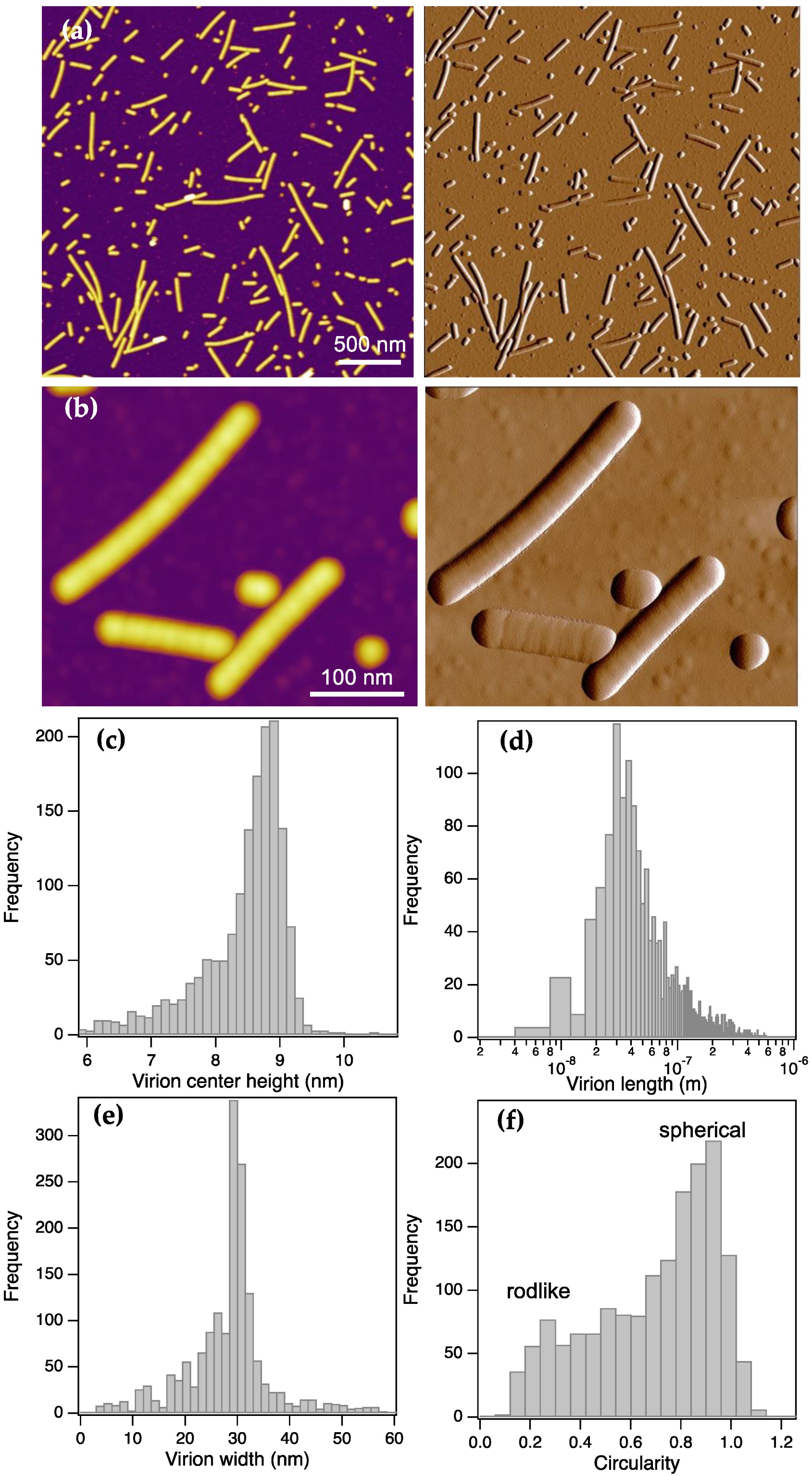
ToBRFV appeared in AFM images (
Figure 1) as rodlike particles with a remarkably wide length distribution (5-1000 nm), and occasionally even longer viruses could be observed (
Figure S1d). The rods displayed rounded ends. However, the rounded end might be the result of convolution between the cylindrical virion and the conical AFM tip. Therefore, at present we are careful with the interpretation, as the ToBRFV virions are likely to be be open-ended cylinders, similarly to TMV [
25]. Indeed, recent electron microscopic analysis of ToBRFV particles indicated that they are blunt ended [
1]. The length histogram displayed a mode at 30 nm, indicating that a ~30-nm-long element may be a structural unit of the virus, from which it assembles or into which it disassembles. We point out, however, that the particle analysis always assigns the long axis of a particle image to its length, but this long axis might not coincide with the long axis of the virus. In other words, if a ToBRFV fragment is shorter than its width, then the width value is assigned to the particle length because this is the longer value. These short fragments will then increase the population of particles with a length equal to the virion width. Altogether, the length of an elementary structural segment of ToBRFV is smaller than or equal to ~30 nm.
The topographical height of the ToBRFV particles was remarkably (more than threefold) smaller than their width, indicating that they spread across the substrate surface. This behavior reflects the capacity for deformation, hence the ToBRFV virions appear to be rather compliant. The virion height
versus length function (
Figure S1e) indicates that the peak height (9 nm) is characteristic of virions with a length between 30-60 nm. Below 30 nm the height drops sharply, which is most likely due to breakage of the cylindrical structure altogether. Above 60 nm the height decreases monotonically by ~0.5 nm in virions several hundred nm long. We speculate that the effect may be related to the length-dependence of bending rigidity. From the width and height data one may compute the cross-sectional perimeter, assuming that it is likely to be a flattened ellipse (
Figure S2). The isotropic (i.e., equivalent distance units along the x- and y-axes) representation of the cross-sectional height profile (
Figure S2) indicates that the width measured at half-maximal topographical height corresponds to the real virion width. Accordingly, the height (
Figure 1c) and width (
Figure 1e) data correspond to the short and long axes of the cross-sectional ellipse. From these data we approximated the perimeter (equation 3) which allowed us to calculate the diameter (equation 4) of the equilibrium cylindrical ToBRFV virion as 22 nm.
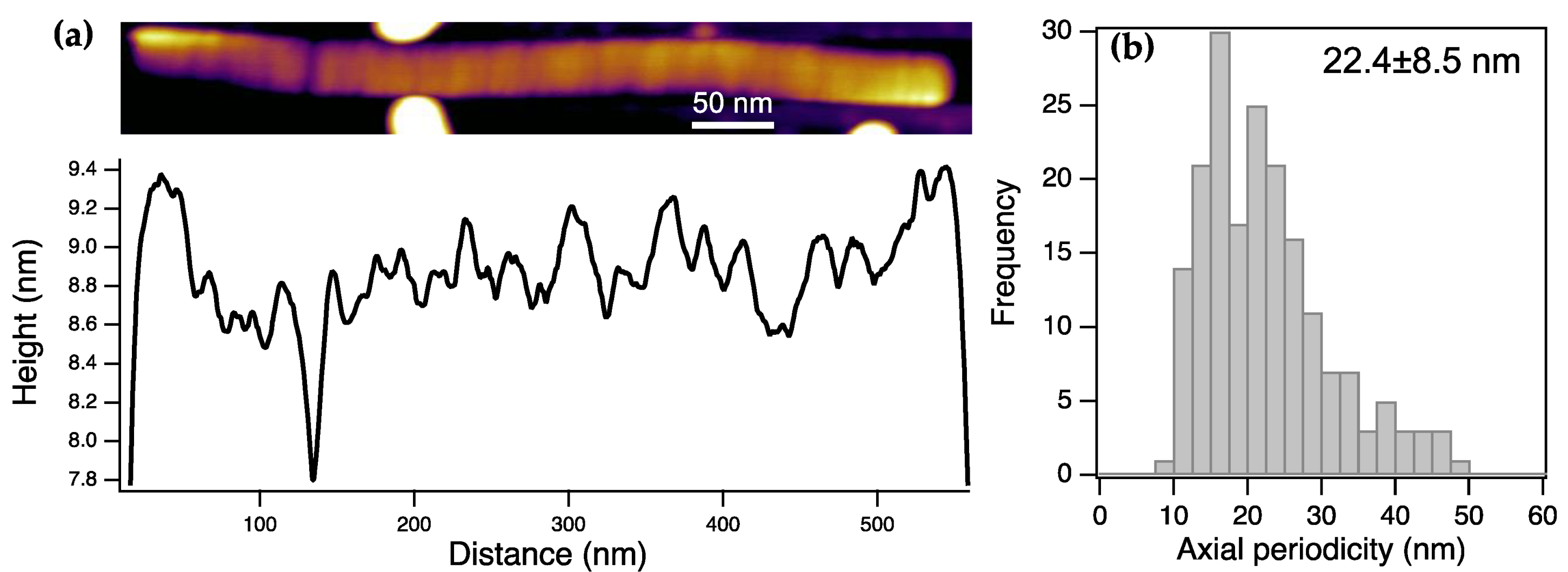
High-resolution AFM images revealed a 22.4 nm (±8.5 SD) periodicity along the long axis of the ToBRFV virions (
Figure 2). We hypothesize that the uncovered periodic transverse ridges reflect boundaries of elementary structural units of ToBRFV either from which it is constructed during viral assembly or to which it fragments during infection. Accordingly, the length of the elementary structural unit is ~ 22 nm. Thus, the unit-length structural element of ToBRFV is a right circular cylinder, the height and diameter of which are identical. Conceivably, this unit-length element represents a highly stable configuration; should its length be significantly higher or lower than its diameter, it would be easily deformed by axial or tangential bending forces, respectively.
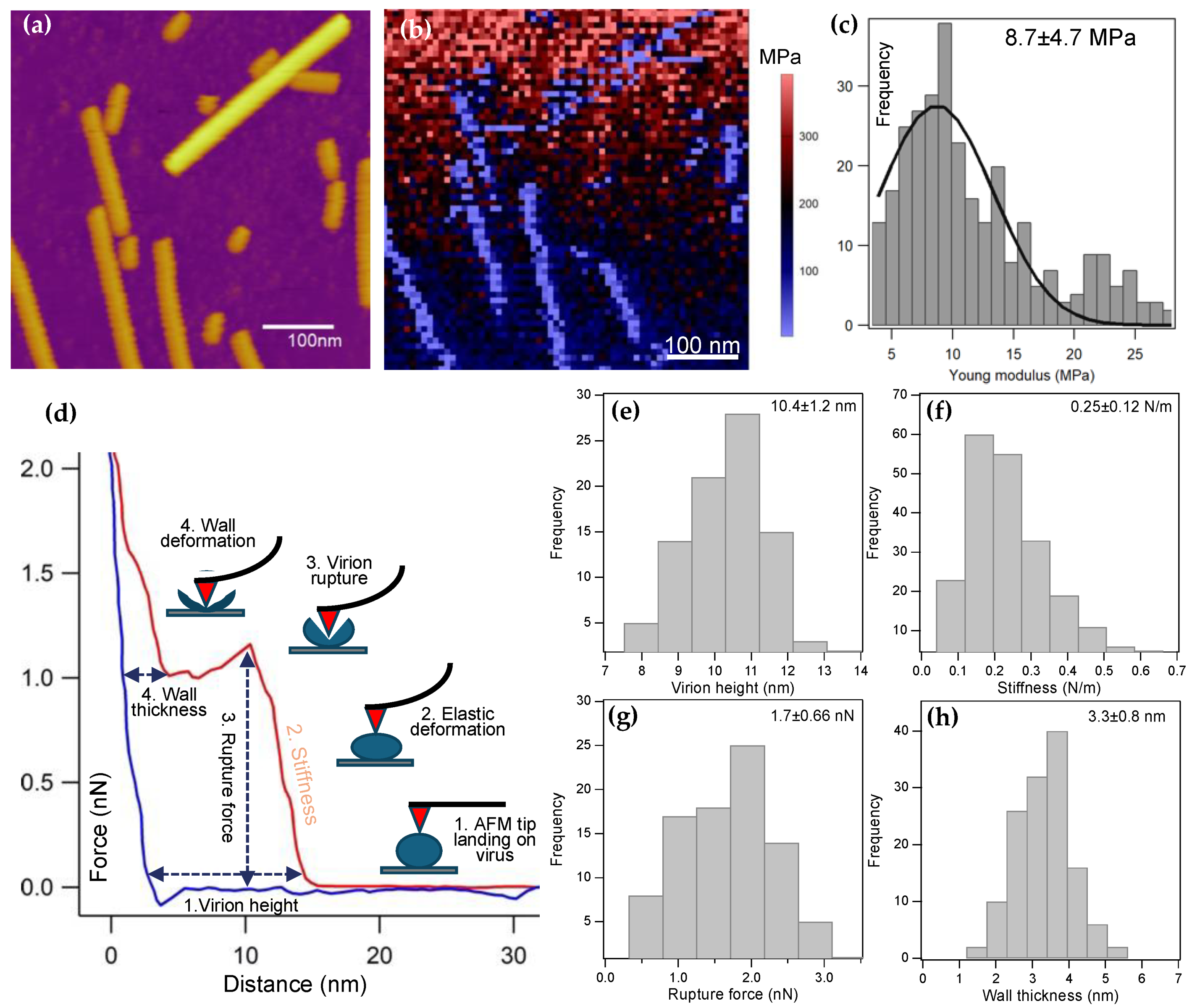
Nanomechanical manipulation of surface-adsorbed ToBRF virions indicated that they are softer and mechanically less stable than numerous icosahedral or helical viruses investigated so far [
20,
22]. Interestingly, the height of the ToBRF virions was nearly 15% greater when measured with nanoindentation (wet conditions) than with particle analysis (dry conditions). We suspect that water,
via hydration of the coat protein and RNA, plays a role in the structure, and probably the mechanical behavior of ToBRFV. The nanoindentation force spectra revealed that following capsid rupture a compressible structure remains, which we assigned to the back wall of the virion. The mean wall thickness was 3.3 nm (±0.8 SD), which is significantly smaller than that of TMV [
25]. Since the measured thickness corresponds to that of the ruptured, broken capsid, it is likely that the structure of the wall has been compromised. Nevertheless, this value provides a lowermost estimate of the capsid wall thickness. Notably, in some of the force spectra, force peaks and non-linear force traces appeared in the retraction phase of the mechanical cycle (
Figure S4). The observation indicates that filamentous structures were pulled out of the broken capsid, and the applied mechanical force induced conformational changes resulting in the release of further filament segments. Considering that the length of these filaments often reached beyond 250 nm, which far exceeds the unfolded length of a single coat protein (~60 nm), the filaments likely correspond to the genomic RNA of ToBRFV. Because the force peaks (sometimes reaching several hundred pN) were much greater than the forces required to unfold RNA hairpins (~15 pN) [
26], they likely reflect the mechanically-driven dissociation of RNA from the coat proteins constituting the capsid. The results suggest that the genomic RNA of ToBRFV is tightly associated with the capsid wall, hence the coat proteins.
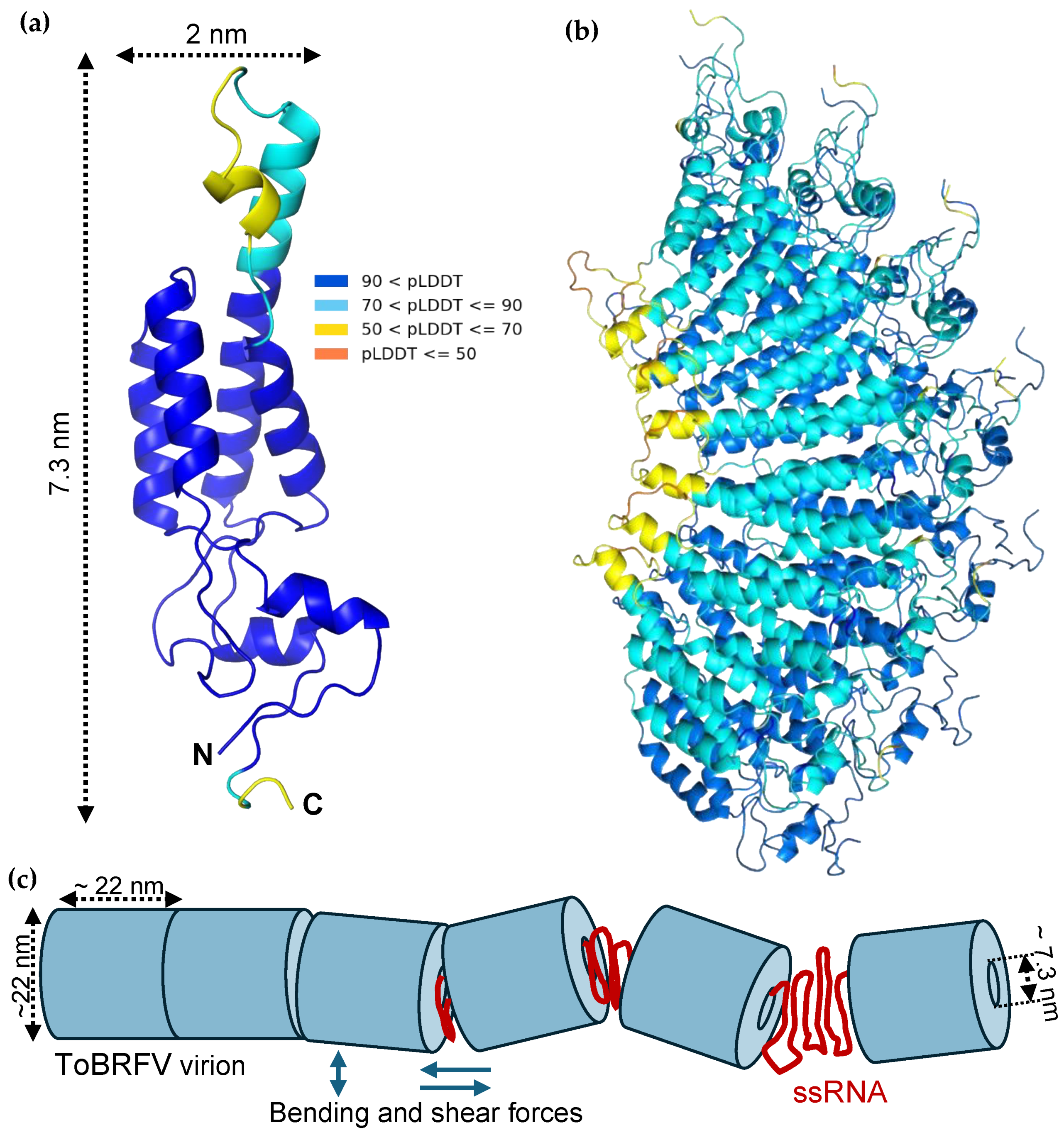
The structural and nanomechanical information gathered in our experiments allow us to propose a model of the ToBRF virion (
Figure 4). Given that the nanoindentation experiments provided only a lowermost estimate of the capsid wall, we carried out a sructural prediction of the 17.5 kDa coat protein (CP) of ToBRFV by using AlphaFold (
Figure 4a-b and Figure S6), so that the wall thickness could be more precisely estimated. Based on the amino-acid sequence of CP (
Figure S6a), AlphaFold predicted a protein dominated by alpha-helices with short disordered regions in the middle and at the C-terminus (
Figure 4a). The length of the protein from the C-terminal SER159 to ALA101 located in a disordered loop at the other end of the molecule, is 7.3 nm. This length provides an uppermost estimate of the capsid wall thickness, as the exact orientation of the CP within the wall is not known. However, predictions involving multiple CP molecules (
Figure 4b and Figure
S6b-d) strongly suggest that this upper estimate represents a plausible
in situ thickness. The lower- and uppermost wall thickness estimates form the basis for two models (a and b) (
Figure S7), the parameters of which are listed in
Table 1. We hypothesize that the genomic ssRNA is helically organized within the core of the cylindrical capsid, similarly to TMV [
25]. Importantly, the capsid's inner diameter far exceeds the persistence length of ssRNA (assumed to be identical to ssDNA, ~0.6 nm [
27]) in either model; therefore, in contrast to bacteriophages [
28], energy need not be invested in the incorporation of the genome. On the contrary, the genomic RNA, aided by its entropic elasticity, likely assists in holding the capsid together. Based on the width of the CP (
Figure 4a), we assume that the pitch of the genomic ssRNA helix is 2 nm. Accordingly, the ToBRFV genome (6392 bases) extends across several unit-length cylindrical elements in both models (four and nine). Considering that a ~200 nm average length has been reported recently for ToBRF virions by using electron microscopy [
1],
model b is likely to more closely reflect the true structural arrangement. We note, however, that the coat proteins must re-arrange considerably to give way to the significant flattening when binging to the substrate surface (
Figure S2). Regardless of the model, the unit-length elements and the fragility of ToBRFV may have important implications for the infection mechanism. Conceivably, during infection the virions become exposed to bending and shear forces, leading to fragmentation (
Figure 4c). Because the genome spans numerous unit-length elements, the fragmentation results in the exposure of the genome several locations at the same time, which is a much more efficient genome release mechanism than diffusion out of a long cylinder. Whether the unit-length cylindrical units play a role during virus assembly, remains to be investigated.
Author Contributions
Conceptualization, P.P. and M.K.; methodology, , L.H., T.H. and M.K.; software, T.H.; validation, P.P., K.S., L.H., T.H. and M.K.; formal analysis, P.P., L.H., T.H. and M.K.; investigation, P.P. and M.K.; resources, S.K.; data curation, P.P., L.J. and M.K.; writing—original draft preparation, P.P. and M.K.; writing—review and editing, P.P., K.S., L.H., T.H. and M.K.; visualization, T.H. and M.K.; supervision, M.K.; project administration, M.K.; funding acquisition, P.P. and M.K. All authors have read and agreed to the published version of the manuscript.