Submitted:
18 June 2025
Posted:
24 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Evolution and Fundamentals of CRISPR/Cas Systems
3. Mechanisms of Multiplex Genome Editing
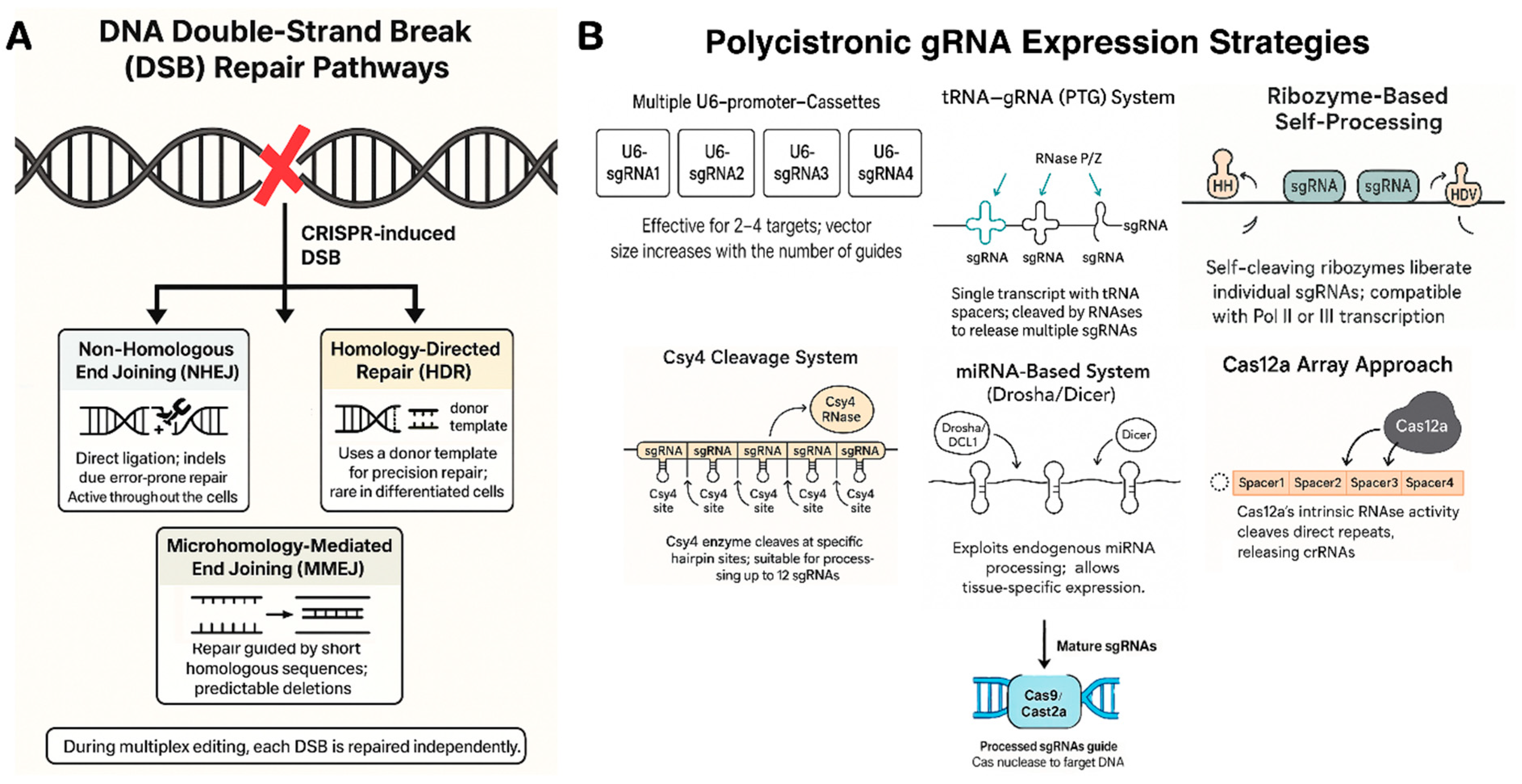
3.1. DNA Double-Strand Break Repair Pathways
3.2. Polycistronic Guide RNA Strategies
- Tandem CRISPR array with direct repeats: In their native systems, Cas nucleases process long precursor transcripts containing multiple spacers separated by repeats [51]. This natural mechanism is exploited in Cas12a-based multiplexing, where a single Pol III promoter (e.g., U3 or U6) drives a compact crRNA array formatted as repeat–spacer1–repeat–spacer2––repeat–spacerN. Cas12a’s built-in RNase activity autonomously cleaves the repeats, releasing mature crRNAs for each target [52]. This enables efficient, simultaneous editing at 6–8 loci using a minimal construct. In contrast, Cas9 lacks self-processing ability, requiring a separate tracrRNA for each spacer [53]. Therefore, multiplexing with Cas9 demands more complex synthetic arrays or alternative strategies involving external processing systems.
- tRNA–gRNA system (PTG): Xie et al. (2015) developed the PTG (polycistronic tRNA–gRNA) system, where tRNAs flank sgRNAs in a single transcript under a U6 promoter [54]. Plant RNases process the tRNAs, releasing functional sgRNAs [55]. This strategy enabled efficient multiplex editing in rice and Arabidopsis, with only a slight drop in efficiency as unit number increased [56]. PTG remains popular for its simplicity and native processing.
- Ribozyme-based self-processing: This method uses self-cleaving ribozymes (hammerhead and HDV) flanking each sgRNA, enabling a single Pol II transcript to yield multiple sgRNAs [57]. Tang et al. (2016) showed a maize ubiquitin promoter-driven cassette expressing Cas9 + 6 ribozyme-flanked sgRNAs, achieving high editing efficiency [58]. Ribozymes cleave in cis, work under Pol II or III, and allow compact, all-in-one constructs ideal for viral or single-vector delivery.
- Csy4 cleavage system: Csy4, an RNA endoribonuclease from Pseudomonas, recognizes a 20-nt hairpin and cleaves 3’ of it [59]. When Csy4 hairpins are appended to sgRNAs in a Pol II transcript, co-expressed Csy4 precisely releases individual sgRNAs [60,61]. Tsai et al. and Cermák et al. applied this for plant multiplexing; Cermák found Csy4 outperformed tRNA and ribozyme systems, efficiently processing up to 12 sgRNAs in tomato [62]. The trade-off is the need to co-express the ~600 bp csy4 gene, but its clean cleavage and minimal scar make it well-suited for complex editing [63].
- MicroRNA-based systems (Drosha/Dicer): miRNA-based sgRNA expression leverages the plant’s own miRNA processing machinery. sgRNAs are embedded in native miRNA backbones (e.g., OsMIR528, AtMIR390), allowing Drosha/DCL1 and Dicer to process the pri-miRNA into functional sgRNAs [64]. Multiple sgRNAs can be multiplexed in a single Pol II transcript. Though sometimes less efficient than tRNA or ribozyme systems, this method enables tissue-specific or inducible expression and has been successfully used in Arabidopsis and rice for multi-gene knockouts.
3.3. Ultra-Multiplexing Capacity
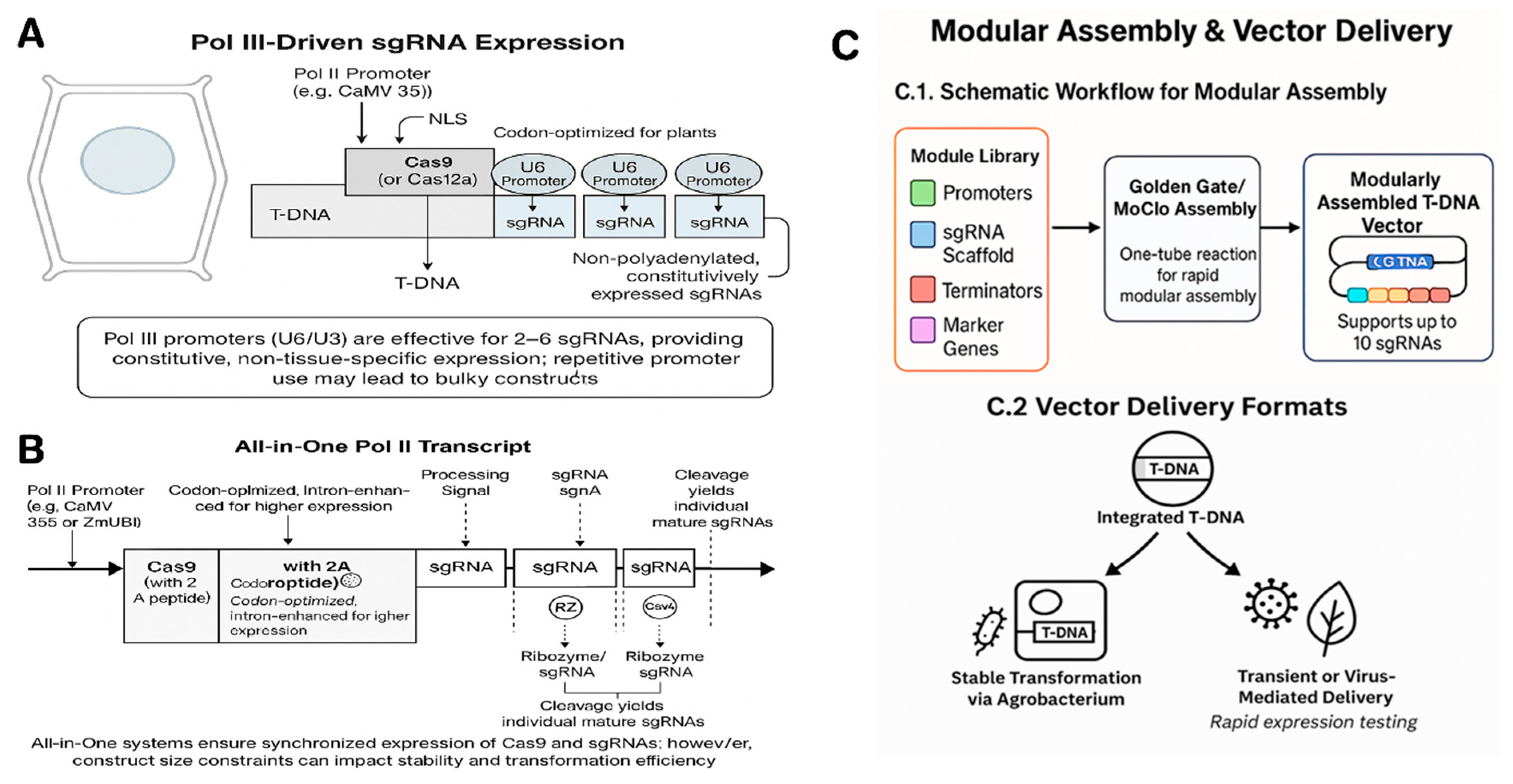
4. Expression Systems for Multiple gRNAs
4.1. Pol III vs. Pol II Promoters
4.2. Single Versus Multiple Transcripts
4.3. Codon Optimization, Introns, and NLS
4.4. Promoter choices for Cas9
4.5. Golden Gate and Modular Assembly Systems
4.6. Vector Delivery Formats
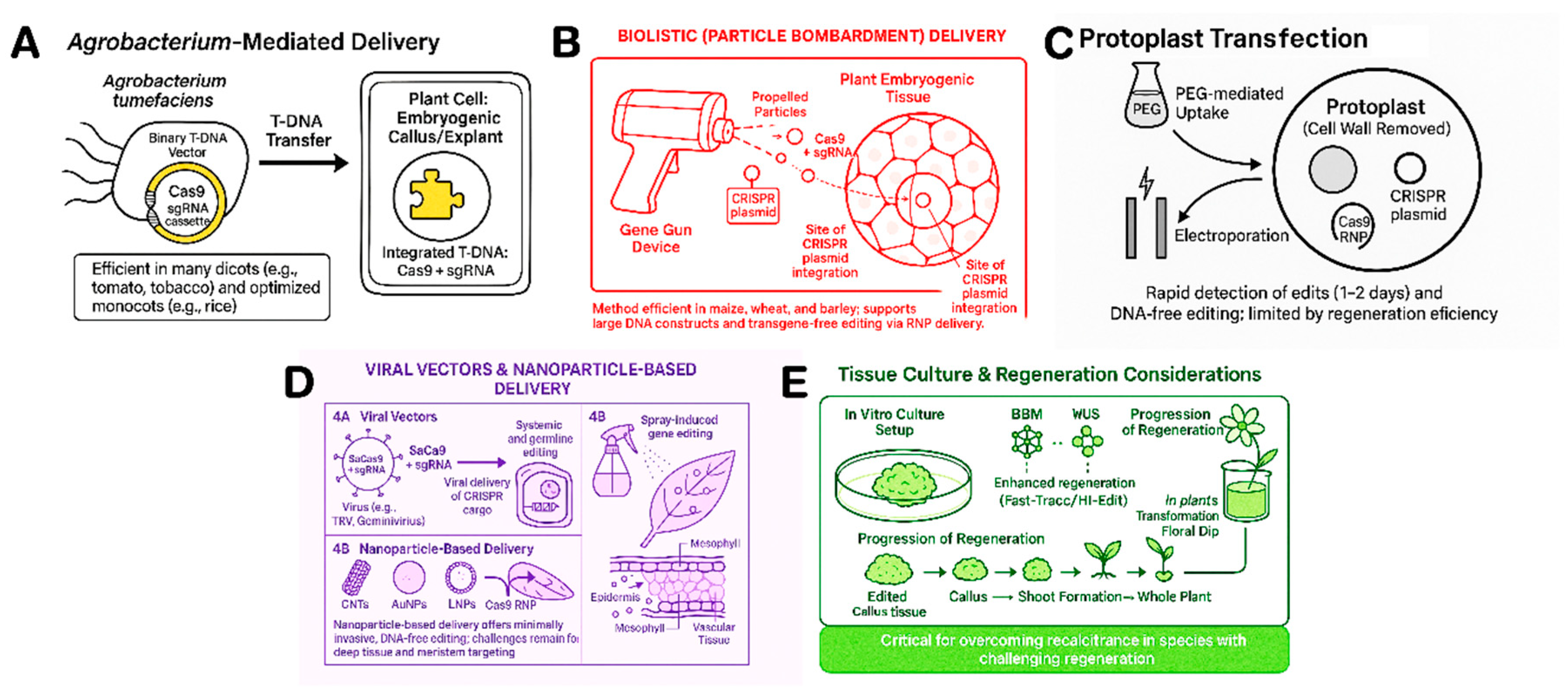
5. Delivery Systems for Mulplex-CRISPR Components in Plants
6. CRISPR Multiplexing in Crop Improvement and Trait Engineering
6.1. Yield and Biomass
6.2. Disease Resistance
6.3. Abiotic Stress Tolerance
6.4. Nutritional Quality
6.5. De Novo Domestication of Wild Plants
7. Challenges and Limitations
8. Computational and AI Integration in Multiplex-CRISPR Design
9. Conclusions
Funding
Conflicts of Interest
References
- Hamdan, M.F.; Tan, B.C. Genetic modification techniques in plant breeding: A comparative review of CRISPR/Cas and GM technologies. Hortic. Plant J. 2024. [CrossRef]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [CrossRef]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014 Apr;32(4):347–55.
- Xiong, X.; Chen, M.; Lim, W.A.; Zhao, D.; Qi, L.S. CRISPR/Cas9 for Human Genome Engineering and Disease Research. Annu. Rev. Genom. Hum. Genet. 2016, 17, 131–154. [CrossRef]
- Abdul Aziz M, Masmoudi K. Molecular breakthroughs in modern plant breeding techniques. Hortic Plant J. 2025 Jan;11(1):15–41.
- Abdelrahman, M.; Wei, Z.; Rohila, J.S.; Zhao, K. Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Front. Plant Sci. 2021, 12. [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2014 31, 397–405. [CrossRef]
- Le Rhun, A.; Escalera-Maurer, A.; Bratovič, M.; Charpentier, E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019, 16, 380–389. [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 1–12. [CrossRef]
- Abe, F.; Haque, E.; Hisano, H.; Tanaka, T.; Kamiya, Y.; Mikami, M.; Kawaura, K.; Endo, M.; Onishi, K.; Hayashi, T.; et al. Genome-Edited Triple-Recessive Mutation Alters Seed Dormancy in Wheat. Cell Rep. 2019, 28, 1362–1369.e4. [CrossRef]
- Singer, S.D.; Laurie, J.D.; Bilichak, A.; Kumar, S.; Singh, J. Genetic Variation and Unintended Risk in the Context of Old and New Breeding Techniques. Crit. Rev. Plant Sci. 2021, 40, 68–108. [CrossRef]
- Hua K, Zhang J, Botella JR, Ma C, Kong F, Liu B, et al. Perspectives on the Application of Genome-Editing Technologies in Crop Breeding. Mol Plant. 2019 Aug;12(8):1047–59.
- Iohannes, S.D.; Jackson, D. Tackling redundancy: genetic mechanisms underlying paralog compensation in plants. New Phytol. 2023, 240, 1381–1389. [CrossRef]
- Chavhan, R.L.; Jaybhaye, S.G.; Hinge, V.R.; Deshmukh, A.S.; Shaikh, U.S.; Jadhav, P.K.; Kadam, U.S.; Hong, J.C. Emerging applications of gene editing technologies for the development of climate-resilient crops. Front. Genome Ed. 2025, 7, 1524767. [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [CrossRef]
- Karginov, F.V.; Hannon, G.J. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol. Cell 2010, 37, 7–19. [CrossRef]
- Zhu, Y.; Klompe, S.E.; Vlot, M.; van der Oost, J.; Staals, R.H. Shooting the messenger: RNA-targetting CRISPR-Cas systems. Biosci. Rep. 2018, 38. [CrossRef]
- Liu, T.Y.; Doudna, J.A. Chemistry of Class 1 CRISPR-Cas effectors: Binding, editing, and regulation. J. Biol. Chem. 2020, 295, 14473–14487. [CrossRef]
- Kim, D.Y.; Lee, S.Y.; Ha, H.J.; Park, H.H. Structural basis of Cas3 activation in type I-C CRISPR-Cas system. Nucleic Acids Res. 2024, 52, 10563–10574. [CrossRef]
- Ebrahimi, S.; Khosravi, M.A.; Raz, A.; Karimipoor, M.; Parvizi, P. CRISPR-Cas Technology as a Revolutionary Genome Editing tool: Mechanisms and Biomedical Applications. Iran. Biomed. J. 2023, 27, 219–246. [CrossRef]
- Das, U.; Chandramouli, L.; Uttarkar, A.; Kumar, J.; Niranjan, V. Discovery of natural compounds as novel FMS-like tyrosine kinase-3 (FLT3) therapeutic inhibitors for the treatment of acute myeloid leukemia: An in-silico approach. Asp. Mol. Med. 2024, 5. [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [CrossRef]
- Tang, Y.; Fu, Y. Class 2 CRISPR/Cas: an expanding biotechnology toolbox for and beyond genome editing. Cell Biosci. 2018, 8, 1–13. [CrossRef]
- Agudelo, D.; Carter, S.; Velimirovic, M.; Duringer, A.; Rivest, J.-F.; Levesque, S.; Loehr, J.; Mouchiroud, M.; Cyr, D.; Waters, P.J.; et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020, 30, 107–117. [CrossRef]
- Asmamaw M, Zawdie B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biol Targets Ther. 2021;15:353–61.
- Karvelis, T.; Gasiunas, G.; Young, J.; Bigelyte, G.; Silanskas, A.; Cigan, M.; Siksnys, V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015, 16, 1–13. [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [CrossRef]
- Li, J.; Xu, R.; Qin, R.; Liu, X.; Kong, F.; Wei, P. Genome editing mediated by SpCas9 variants with broad non-canonical PAM compatibility in plants. Mol. Plant 2021, 14, 352–360. [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [CrossRef]
- Yang, C.; Zhou, Z.; Sun, X.; Ju, H.; Yue, X.; Rao, S.; Xue, C. PAMless SpRY exhibits a preference for the seed region for efficient targeting. Cell Rep. 2024, 43, 114225. [CrossRef]
- Kaya, H.; Ishibashi, K.; Toki, S. A Split Staphylococcus aureus Cas9 as a Compact Genome-Editing Tool in Plants. Plant Cell Physiol. 2017, 58, 643–649. [CrossRef]
- Bandyopadhyay, A.; Kancharla, N.; Javalkote, V.S.; Dasgupta, S.; Brutnell, T.P. CRISPR-Cas12a (Cpf1): A Versatile Tool in the Plant Genome Editing Tool Box for Agricultural Advancement. Front. Plant Sci. 2020, 11. [CrossRef]
- Ming, M.; Ren, Q.; Pan, C.; He, Y.; Zhang, Y.; Liu, S.; Zhong, Z.; Wang, J.; Malzahn, A.A.; Wu, J.; et al. CRISPR–Cas12b enables efficient plant genome engineering. Nat. Plants 2020, 6, 202–208. [CrossRef]
- Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019 Oct;14(10):2986–3012.
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 2022, 11, 2665. [CrossRef]
- Kato, K.; Zhou, W.; Okazaki, S.; Isayama, Y.; Nishizawa, T.; Gootenberg, J.S.; Abudayyeh, O.O.; Nishimasu, H. Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex. Cell 2022, 185, 2324–2337.e16. [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [CrossRef]
- Xin, C.; Yin, J.; Yuan, S.; Ou, L.; Liu, M.; Zhang, W.; Hu, J. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption. Nat. Commun. 2022, 13, 1–10. [CrossRef]
- Urbaitis T, Gasiunas G, Young JK, Hou Z, Paulraj S, Godliauskaite E, et al. A new family of CRISPR -type V nucleases with C-rich PAM recognition. EMBO Rep. 2022 Dec 6;23(12):e55481.
- Al-Shayeb, B.; Skopintsev, P.; Soczek, K.M.; Stahl, E.C.; Li, Z.; Groover, E.; Smock, D.; Eggers, A.R.; Pausch, P.; Cress, B.F.; et al. Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell 2022, 185, 4574–4586.e16. [CrossRef]
- Das, U.; Banerjee, S.; Sarkar, M.; L, F.M.; Soni, T.K.; Saha, M.; Pradhan, G.; Chatterjee, B. Circular RNA vaccines: Pioneering the next-gen cancer immunotherapy. Cancer Pathog. Ther. 2024. [CrossRef]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018, 36, 265–271. [CrossRef]
- Matsumoto D, Kishi K, Matsugi E, Inoue Y, Nigorikawa K, Nomura W. CAS9-GEMININ and Cdt1-fused ANTI-CRISPR protein synergistically increase editing accuracy. FEBS Lett. 2023 Apr;597(7):985–94.
- Stinson, B.M.; Loparo, J.J. Repair of DNA Double-Strand Breaks by the Nonhomologous End Joining Pathway. Annu. Rev. Biochem. 2021, 90, 137–164. [CrossRef]
- Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015 Nov;40(11):701–14.
- Chen, J.; Li, S.; He, Y.; Li, J.; Xia, L. An update on precision genome editing by homology-directed repair in plants. Plant Physiol. 2022, 188, 1780–1794. [CrossRef]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [CrossRef]
- Hsieh-Feng, V.; Yang, Y. Efficient expression of multiple guide RNAs for CRISPR/Cas genome editing. aBIOTECH 2020, 1, 123–134. [CrossRef]
- Nakanishi, T.; Maekawa, A.; Tabata, H.; Yoshioka, T.; Pei, Z.; Sato, K.; Mori, M.; Kato, M.; Saito, I. Highly multiplex guide RNA expression units of CRISPR/Cas9 were completely stable using cosmid amplification in a novel polygonal structure. J. Gene Med. 2019, 21, e3115. [CrossRef]
- Saito, M.; Ladha, A.; Strecker, J.; Faure, G.; Neumann, E.; Altae-Tran, H.; Macrae, R.K.; Zhang, F. Dual modes of CRISPR-associated transposon homing. Cell 2021, 184, 2441–2453.e18. [CrossRef]
- Zhang, Y.; Ren, Q.; Tang, X.; Liu, S.; Malzahn, A.A.; Zhou, J.; Wang, J.; Yin, D.; Pan, C.; Yuan, M.; et al. Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 2021, 12, 1–11. [CrossRef]
- Ng WA, Reed BH. A method to estimate the frequency of chromosomal rearrangements induced by CRISPR/Cas9 multiplexing in Drosophila [Internet]. 2019 [cited 2025 Apr 17]. Available from: http://biorxiv.org/lookup/doi/10.1101/815431.
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3570–5.
- Safari, F.; Zare, K.; Negahdaripour, M.; Barekati-Mowahed, M.; Ghasemi, Y. CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci. 2019, 9, 1–21. [CrossRef]
- Stuttmann J, Barthel K, Martin P, Ordon J, Erickson JL, Herr R, et al. Highly efficient multiplex editing: one-shot generation of 8× Nicotiana benthamiana and 12× Arabidopsis mutants. Plant J. 2021 Apr;106(1):8–22.
- He, Y.; Zhang, T.; Yang, N.; Xu, M.; Yan, L.; Wang, L.; Wang, R.; Zhao, Y. Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J. Genet. Genom. 2017, 44, 469–472. [CrossRef]
- Tang, X.; Zheng, X.; Qi, Y.; Zhang, D.; Cheng, Y.; Tang, A.; Voytas, D.F.; Zhang, Y. A Single Transcript CRISPR-Cas9 System for Efficient Genome Editing in Plants. Mol. Plant 2016, 9, 1088–1091. [CrossRef]
- Lee, H.Y.; Haurwitz, R.E.; Apffel, A.; Zhou, K.; Smart, B.; Wenger, C.D.; Laderman, S.; Bruhn, L.; Doudna, J.A. RNA–protein analysis using a conditional CRISPR nuclease. Proc. Natl. Acad. Sci. 2013, 110, 5416–5421. [CrossRef]
- Huang, S.; Zhang, Z.; Tao, W.; Liu, Y.; Li, X.; Wang, X.; Harati, J.; Wang, P.-Y.; Huang, X.; Lin, C.-P. Broadening prime editing toolkits using RNA-Pol-II-driven engineered pegRNA. Mol. Ther. 2022, 30, 2923–2932. [CrossRef]
- Das U, Banerjee S, Sarkar M. Bibliometric analysis of circular RNA cancer vaccines and their emerging impact. Vacunas. 2025 Mar;500391.
- ermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 2017, 29, 1196–1217. [CrossRef]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608. [CrossRef]
- Xie, C.; Chen, Y.-L.; Wang, D.-F.; Wang, Y.-L.; Zhang, T.-P.; Li, H.; Liang, F.; Zhao, Y.; Zhang, G.-Y. SgRNA Expression of CRIPSR-Cas9 System Based on MiRNA Polycistrons as a Versatile Tool to Manipulate Multiple and Tissue-Specific Genome Editing. Sci. Rep. 2017, 7, 1–12. [CrossRef]
- Wu, S.; Kyaw, H.; Tong, Z.; Yang, Y.; Wang, Z.; Zhang, L.; Deng, L.; Zhang, Z.; Xiao, B.; Quick, W.P.; et al. A simple and efficient CRISPR/Cas9 system permits ultra-multiplex genome editing in plants. Crop. J. 2024, 12, 569–582. [CrossRef]
- Kannan, B.; Jung, J.H.; Moxley, G.W.; Lee, S.; Altpeter, F. TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 2017, 16, 856–866. [CrossRef]
- Yang, T.; Ali, M.; Lin, L.; Li, P.; He, H.; Zhu, Q.; Sun, C.; Wu, N.; Zhang, X.; Huang, T.; et al. Recoloring tomato fruit by CRISPR/Cas9-mediated multiplex gene editing. Hortic. Res. 2022, 10, uhac214. [CrossRef]
- Oh, Y.; Kim, S.-G. RPS5A Promoter-Driven Cas9 Produces Heritable Virus-Induced Genome Editing in Nicotiana attenuata. Mol. Cells 2021, 44, 911–919. [CrossRef]
- Kim, H.K.; Lee, S.; Kim, Y.; Park, J.; Min, S.; Choi, J.W.; Huang, T.P.; Yoon, S.; Liu, D.R.; Kim, H.H. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat. Biomed. Eng. 2020, 4, 111–124. [CrossRef]
- Kor, S.D.; Chowdhury, N.; Keot, A.K.; Yogendra, K.; Chikkaputtaiah, C.; Reddy, P.S. RNA Pol III promoters—key players in precisely targeted plant genome editing. Front. Genet. 2023, 13, 989199. [CrossRef]
- Kuzmin, A.A.; Tomilin, A.N. Building Blocks of Artificial CRISPR-Based Systems beyond Nucleases. Int. J. Mol. Sci. 2022, 24, 397. [CrossRef]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019 Mar;37(3):276–82.
- McCarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1–13. [CrossRef]
- Geilenkeuser, J.; Armbrust, N.; Steinmaßl, E.; Du, S.W.; Schmidt, S.; Binder, E.M.H.; Li, Y.; Warsing, N.W.; Wendel, S.V.; von der Linde, F.; et al. Engineered nucleocytosolic vehicles for loading of programmable editors. Cell 2025. [CrossRef]
- Wang, L.; Han, H. Strategies for improving the genome-editing efficiency of class 2 CRISPR/Cas system. Heliyon 2024, 10, e38588. [CrossRef]
- Shui, S.; Wang, S.; Liu, J. Systematic Investigation of the Effects of Multiple SV40 Nuclear Localization Signal Fusion on the Genome Editing Activity of Purified SpCas9. Bioengineering 2022, 9, 83. [CrossRef]
- Tanaka, T.; Maeda, Y.; Suhaimi, N.; Tsuneoka, C.; Nonoyama, T.; Yoshino, T.; Kato, N.; Lauersen, K.J. Intron-mediated enhancement of transgene expression in the oleaginous diatom Fistulifera solaris towards bisabolene production. Algal Res. 2021, 57. [CrossRef]
- Grützner, R.; Martin, P.; Horn, C.; Mortensen, S.; Cram, E.J.; Lee-Parsons, C.W.; Stuttmann, J.; Marillonnet, S. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2021, 2, 100135. [CrossRef]
- Khakhar, A.; Voytas, D.F. RNA Viral Vectors for Accelerating Plant Synthetic Biology. Front. Plant Sci. 2021, 12. [CrossRef]
- Amack, S.C.; Antunes, M.S. CaMV35S promoter – A plant biology and biotechnology workhorse in the era of synthetic biology. Curr. Plant Biol. 2020, 24. [CrossRef]
- Wang, X.; Xu, L.; Liu, X.; Xin, L.; Wu, S.; Chen, X. Development of potent promoters that drive the efficient expression of genes in apple protoplasts. Hortic. Res. 2021, 8, 211. [CrossRef]
- Ren, Q.; Zhong, Z.; Wang, Y.; You, Q.; Li, Q.; Yuan, M.; He, Y.; Qi, C.; Tang, X.; Zheng, X.; et al. Bidirectional Promoter-Based CRISPR-Cas9 Systems for Plant Genome Editing. Front. Plant Sci. 2019, 10, 1173. [CrossRef]
- Collonnier C, Epert A, Mara K, Maclot F, Guyon-Debast A, Charlot F, et al. CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the moss Physcomitrella patens. Plant Biotechnol J. 2017 Jan;15(1):122–31.
- Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, et al. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2016 Feb;14(2):519–32.
- Das, U.; Chanda, T.; Kumar, J.; Peter, A. Discovery of natural MCL1 inhibitors using pharmacophore modelling, QSAR, docking, ADMET, molecular dynamics, and DFT analysis. Comput. Biol. Chem. 2025, 117, 108427. [CrossRef]
- Zheng, N.; Li, T.; Dittman, J.D.; Su, J.; Li, R.; Gassmann, W.; Peng, D.; Whitham, S.A.; Liu, S.; Yang, B. CRISPR/Cas9-Based Gene Editing Using Egg Cell-Specific Promoters in Arabidopsis and Soybean. Front. Plant Sci. 2020, 11, 800. [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [CrossRef]
- Bird, J.E.; Marles-Wright, J.; Giachino, A. A User’s Guide to Golden Gate Cloning Methods and Standards. ACS Synth. Biol. 2022, 11, 3551–3563. [CrossRef]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S.; Peccoud, J. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLOS ONE 2011, 6, e16765. [CrossRef]
- Sarrion-Perdigones, A.; Falconi, E.E.; Zandalinas, S.I.; Juárez, P.; Fernández-Del-Carmen, A.; Granell, A.; Orzaez, D.; Peccoud, J. GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. PLOS ONE 2011, 6, e21622. [CrossRef]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985. [CrossRef]
- Vasudevan, R.; Gale, G.A.; Schiavon, A.A.; Puzorjov, A.; Malin, J.; Gillespie, M.D.; Vavitsas, K.; Zulkower, V.; Wang, B.; Howe, C.J.; et al. CyanoGate: A Modular Cloning Suite for Engineering Cyanobacteria Based on the Plant MoClo Syntax. Plant Physiol. 2019, 180, 39–55. [CrossRef]
- Fulco, C.P.; Nasser, J.; Jones, T.R.; Munson, G.; Bergman, D.T.; Subramanian, V.; Grossman, S.R.; Anyoha, R.; Doughty, B.R.; Patwardhan, T.A.; et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 2019, 51, 1664–1669. [CrossRef]
- Costa, L.D.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020, 10, 1–14. [CrossRef]
- Knopp, Y.; Geis, F.K.; Heckl, D.; Horn, S.; Neumann, T.; Kuehle, J.; Meyer, J.; Fehse, B.; Baum, C.; Morgan, M.; et al. Transient Retrovirus-Based CRISPR/Cas9 All-in-One Particles for Efficient, Targeted Gene Knockout. Mol. Ther. - Nucleic Acids 2018, 13, 256–274. [CrossRef]
- Thompson, M.G.; Moore, W.M.; Hummel, N.F.; Pearson, A.N.; Barnum, C.R.; Scheller, H.V.; Shih, P.M. Agrobacterium tumefaciens: A Bacterium Primed for Synthetic Biology. 2020, 2020. [CrossRef]
- Tang, Y.; Zhang, Z.; Yang, Z.; Wu, J.; Manavella, P. CRISPR/Cas9 and Agrobacterium tumefaciens virulence proteins synergistically increase efficiency of precise genome editing via homology directed repair in plants. J. Exp. Bot. 2023, 74, 3518–3530. [CrossRef]
- He, Y.; Mudgett, M.; Zhao, Y. Advances in gene editing without residual transgenes in plants. Plant Physiol. 2021, 188, 1757–1768. [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K.; A Wilson, R. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLOS ONE 2016, 11, e0154027–e0154027. [CrossRef]
- Zhong, H.; Li, C.; Yu, W.; Zhou, H.-P.; Lieber, T.; Su, X.; Wang, W.; Bumann, E.; Castro, R.M.L.; Jiang, Y.; et al. A fast and genotype-independent in planta Agrobacterium-mediated transformation method for soybean. Plant Commun. 2024, 5, 101063. [CrossRef]
- Lacroix, B.; Citovsky, V. Biolistic Approach for Transient Gene Expression Studies in Plants. Methods Mol Biol Clifton NJ. 2020;2124:125–39.
- Das, U. Emerging trends and research landscape of the tumor microenvironment in head-and-neck cancer: A comprehensive bibliometric analysis. Cancer Plus 2025, 7, 64. [CrossRef]
- Vollen, K.; Alonso, J.M.; Stepanova, A.N. Beyond a few bases: methods for large DNA insertion and gene targeting in plants. Plant J. 2025, 121, e70099. [CrossRef]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Cigan, A.M. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [CrossRef]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.-S.; et al. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527. [CrossRef]
- Reed, K.M.; Bargmann, B.O.R. Protoplast Regeneration and Its Use in New Plant Breeding Technologies. Front. Genome Ed. 2021, 3. [CrossRef]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018, 164, 378–384. [CrossRef]
- Wu X, Zhang Y, Jiang X, Ma T, Guo Y, Wu X, et al. Considerations in engineering viral vectors for genome editing in plants. Virology. 2024 Jan;589:109922.
- Oh, Y.; Nagalakshmi, U.; Dahlbeck, D.; Koehler, N.; Cho, M.; Dinesh-Kumar, S.P.; Staskawicz, B.J. Heritable virus-induced germline editing in tomato. Plant J. 2025, 122, e70115. [CrossRef]
- Zhang, C.; Liu, S.; Li, X.; Zhang, R.; Li, J. Virus-Induced Gene Editing and Its Applications in Plants. Int. J. Mol. Sci. 2022, 23, 10202. [CrossRef]
- Brokowski, C.; Adli, M. CRISPR Ethics: Moral Considerations for Applications of a Powerful Tool. J. Mol. Biol. 2019, 431, 88–101. [CrossRef]
- Bertier LD, Ron M, Huo H, Bradford KJ, Britt AB, Michelmore RW. High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce ( Lactuca sativa ). G3 GenesGenomesGenetics. 2018 May 1;8(5):1513–21.
- Vats, S.; Kumawat, S.; Brar, J.; Kaur, S.; Yadav, K.; Magar, S.G.; Jadhav, P.V.; Salvi, P.; Sonah, H.; Sharma, S.; et al. Opportunity and challenges for nanotechnology application for genome editing in plants. Plant Nano Biol. 2022, 1, 100001. [CrossRef]
- Liu, S.; Zheng, Y.; Pan, L.; Wang, W.; Li, Y.; Liu, Z.; Zhang, X. Nanodelivery of nucleic acids for plant genetic engineering. Nanoscale Res. Lett. 2025, 20, 1–15. [CrossRef]
- Zhi, H.; Zhou, S.; Pan, W.; Shang, Y.; Zeng, Z.; Zhang, H. The Promising Nanovectors for Gene Delivery in Plant Genome Engineering. Int. J. Mol. Sci. 2022, 23, 8501. [CrossRef]
- Zhao, X.; Meng, Z.; Wang, Y.; Chen, W.; Sun, C.; Cui, B.; Cui, J.; Yu, M.; Zeng, Z.; Guo, S.; et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 2017, 3, 956–964. [CrossRef]
- Babaeianjelodar, N.; Trivedi, J.; Uhde-Stone, C. Eliminating Tissue Culture from Plant Gene Editing in the Near Future: A Wish or Reality?. Curr. Plant Biol. 2025, 41. [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [CrossRef]
- Bent, A. Arabidopsis thaliana Floral Dip Transformation Method. In: Agrobacterium Protocols [Internet]. New Jersey: Humana Press; 2006 [cited 2025 Apr 17]. p. 87–104. Available from: http://link.springer.com/10.1385/1-59745-130-4:87.
- Saini, H.; Thakur, R.; Gill, R.; Tyagi, K.; Goswami, M. CRISPR/Cas9-gene editing approaches in plant breeding. GM Crop. Food 2023, 14, 1–17. [CrossRef]
- Quiroz, L.F.; Khan, M.; Gondalia, N.; Lai, L.; McKeown, P.C.; Brychkova, G.; Spillane, C. Tissue culture-independent approaches to revolutionizing plant transformation and gene editing. Hortic. Res. 2024, 12, uhae292. [CrossRef]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.-K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. 2018, 115, 6058–6063. [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR–Cas9 System. Front. Plant Sci. 2020, 10, 1663. [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [CrossRef]
- Brant EJ, Eid A, Kannan B, Baloglu MC, Altpeter F. The extent of multiallelic, co-editing of LIGULELESS1 in highly polyploid sugarcane tunes leaf inclination angle and enables selection of the ideotype for biomass yield. Plant Biotechnol J. 2024 Oct;22(10):2660–71.
- Zaman, Q.U.; Chu, W.; Hao, M.; Shi, Y.; Sun, M.; Sang, S.-F.; Mei, D.; Cheng, H.; Liu, J.; Li, C.; et al. CRISPR/Cas9-Mediated Multiplex Genome Editing of JAGGED Gene in Brassica napus L.. Biomolecules 2019, 9, 725. [CrossRef]
- Shi, J.; Ni, X.; Huang, J.; Fu, Y.; Wang, T.; Yu, H.; Zhang, Y. CRISPR/Cas9-Mediated Gene Editing of BnFAD2 and BnFAE1 Modifies Fatty Acid Profiles in Brassica napus. Genes 2022, 13, 1681. [CrossRef]
- Zhu, X.; Gong, H.; He, Q.; Zeng, Z.; Busse, J.S.; Jin, W.; Bethke, P.C.; Jiang, J. Silencing of vacuolar invertase and asparagine synthetase genes and its impact on acrylamide formation of fried potato products. Plant Biotechnol. J. 2015, 14, 709–718. [CrossRef]
- Biswas S, Ibarra O, Shaphek M, Molina-Risco M, Faion-Molina M, Bellinatti-Della Gracia M, et al. Increasing the level of resistant starch in ‘Presidio’ rice through multiplex CRISPR–Cas9 gene editing of starch branching enzyme genes. Plant Genome. 2023 Jun;16(2):e20225.
- Peng A, Chen S, Lei T, Xu L, He Y, Wu L, et al. Engineering canker-resistant plants through CRISPR /Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol J. 2017 Dec;15(12):1509–19.
- Das, U.; Uttarkar, A.; Kumar, J.; Niranjan, V. In silico exploration natural compounds for the discovery of novel DNMT3A inhibitors as potential therapeutic agents for acute myeloid leukemia. Silico Res. Biomed. 2025, 1. [CrossRef]
- Macioszek, V.K.; Jęcz, T.; Ciereszko, I.; Kononowicz, A.K. Jasmonic Acid as a Mediator in Plant Response to Necrotrophic Fungi. Cells 2023, 12, 1027. [CrossRef]
- Li, M.; Yang, Z.; Chang, C. Susceptibility Is New Resistance: Wheat Susceptibility Genes and Exploitation in Resistance Breeding. Agriculture 2022, 12, 1419. [CrossRef]
- Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L.. Int. J. Mol. Sci. 2018, 19, 2716. [CrossRef]
- Kang, Y.; Zhou, M.; Merry, A.; Barry, K. Mechanisms of powdery mildew resistance of wheat – a review of molecular breeding. Plant Pathol. 2020, 69, 601–617. [CrossRef]
- Shawky, A.; Hatawsh, A.; Al-Saadi, N.; Farzan, R.; Eltawy, N.; Francis, M.; Abousamra, S.; Ismail, Y.Y.; Attia, K.; Fakhouri, A.S.; et al. Revolutionizing Tomato Cultivation: CRISPR/Cas9 Mediated Biotic Stress Resistance. Plants 2024, 13, 2269. [CrossRef]
- Xie, Z.; Jin, L.; Sun, Y.; Zhan, C.; Tang, S.; Qin, T.; Liu, N.; Huang, J. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice. Plant Commun. 2023, 5, 100782. [CrossRef]
- Ding, F.; Wang, C.; Xu, N.; Wang, M. The ethylene response factor SlERF.B8 triggers jasmonate biosynthesis to promote cold tolerance in tomato. Environ. Exp. Bot. 2022, 203. [CrossRef]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021, 21, 1–15. [CrossRef]
- Abdallah, N.A.; Elsharawy, H.; Abulela, H.A.; Thilmony, R.; Abdelhadi, A.A.; Elarabi, N.I. Multiplex CRISPR/Cas9-mediated genome editing to address drought tolerance in wheat. GM Crop. Food 2022, 16, 1–17. [CrossRef]
- Patel-Tupper, D.; Kelikian, A.; Leipertz, A.; Maryn, N.; Tjahjadi, M.; Karavolias, N.G.; Cho, M.-J.; Niyogi, K.K. Multiplexed CRISPR-Cas9 mutagenesis of rice PSBS1 noncoding sequences for transgene-free overexpression. Sci. Adv. 2024, 10, eadm7452. [CrossRef]
- Rahim, A.A.; Uzair, M.; Rehman, N.; Fiaz, S.; Attia, K.A.; Abushady, A.M.; Yang, S.H.; Khan, M.R. CRISPR/Cas9 mediated TaRPK1 root architecture gene mutagenesis confers enhanced wheat yield. J. King Saud Univ. - Sci. 2024, 36. [CrossRef]
- Khan MS, Ahmad D, Khan MA. Utilization of genes encoding osmoprotectants in transgenic plants for enhanced abiotic stress tolerance. Electron J Biotechnol. 2015 Jul;18(4):257–66.
- Zhang, J.; Lyu, H.; Chen, J.; Cao, X.; Du, R.; Ma, L.; Wang, N.; Zhu, Z.; Rao, J.; Wang, J.; et al. Releasing a sugar brake generates sweeter tomato without yield penalty. Nature 2024, 635, 647–656. [CrossRef]
- Li, A.; Jia, S.; Yobi, A.; Ge, Z.; Sato, S.J.; Zhang, C.; Angelovici, R.; Clemente, T.E.; Holding, D.R. Editing of an Alpha-Kafirin Gene Family Increases, Digestibility and Protein Quality in Sorghum. Plant Physiol. 2018, 177, 1425–1438. [CrossRef]
- Wang L, O’Conner S, Tanvir R, Zheng W, Cothron S, Towery K, et al. CRISPR /Cas9-based editing of NF-YC4 promoters yields high-protein rice and soybean. New Phytol. 2025 Mar;245(5):2103–16.
- Liu, H.; Lin, B.; Ren, Y.; Hao, P.; Huang, L.; Xue, B.; Jiang, L.; Zhu, Y.; Hua, S. CRISPR/Cas9-mediated editing of double loci of BnFAD2 increased the seed oleic acid content of rapeseed (Brassica napus L.). Front. Plant Sci. 2022, 13, 1034215. [CrossRef]
- Sánchez-León, S.; Marín-Sanz, M.; Guzmán-López, M.H.; Gavilán-Camacho, M.; Simón, E.; Barro, F.; Trafford, K. CRISPR/Cas9-mediated multiplex gene editing of gamma and omega gliadins: paving the way for gliadin-free wheat. J. Exp. Bot. 2024, 75, 7079–7095. [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [CrossRef]
- Feng, X.; Lin, K.; Zhang, W.; Nan, J.; Zhang, X.; Wang, C.; Wang, R.; Jiang, G.; Yuan, Q.; Lin, S. Improving the blast resistance of the elite rice variety Kongyu-131 by updating the pi21 locus. BMC Plant Biol. 2019, 19, 1–12. [CrossRef]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, et al. The Tomato Aux / IAA Transcription Factor IAA9 Is Involved in Fruit Development and Leaf Morphogenesis. Plant Cell. 2005 Oct 3;17(10):2676–92.
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [CrossRef]
- Lei, Y.; Lu, L.; Liu, H.-Y.; Li, S.; Xing, F.; Chen, L.-L. CRISPR-P: A Web Tool for Synthetic Single-Guide RNA Design of CRISPR-System in Plants. Mol. Plant 2014, 7, 1494–1496. [CrossRef]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [CrossRef]
- Concordet, J.-P.; Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [CrossRef]
- Son, S.; Park, S.R. Challenges Facing CRISPR/Cas9-Based Genome Editing in Plants. Front. Plant Sci. 2022, 13, 902413. [CrossRef]
- Zhou, J.; Luan, X.; Liu, Y.; Wang, L.; Wang, J.; Yang, S.; Liu, S.; Zhang, J.; Liu, H.; Yao, D. Strategies and Methods for Improving the Efficiency of CRISPR/Cas9 Gene Editing in Plant Molecular Breeding. Plants 2023, 12, 1478. [CrossRef]
- Wen, W.; Zhang, X.-B. CRISPR–Cas9 gene editing induced complex on-target outcomes in human cells. Exp. Hematol. 2022, 110, 13–19. [CrossRef]
- Yin, J.; Lu, R.; Xin, C.; Wang, Y.; Ling, X.; Li, D.; Zhang, W.; Liu, M.; Xie, W.; Kong, L.; et al. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing. Nat. Commun. 2022, 13, 1–14. [CrossRef]
- Iyer, S.V.; Goodwin, S.; McCombie, W.R. Leveraging the power of long reads for targeted sequencing. Genome Res. 2024, 34, 1701–1718. [CrossRef]
- Santelices, B. Mosaicism and chimerism as components of intraorganismal genetic heterogeneity. J. Evol. Biol. 2004, 17, 1187–1188. [CrossRef]
- Wang, Z.-P.; Xing, H.-L.; Dong, L.; Zhang, H.-Y.; Han, C.-Y.; Wang, X.-C.; Chen, Q.-J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 1–12. [CrossRef]
- Tenorio Berrío R, Nelissen H, Inzé D, Dubois M. Increasing yield on dry fields: molecular pathways with growing potential. Plant J Cell Mol Biol. 2022 Jan;109(2):323–41.
- Chuai, G.; Ma, H.; Yan, J.; Chen, M.; Hong, N.; Xue, D.; Zhou, C.; Zhu, C.; Chen, K.; Duan, B.; et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018, 19, 1–18. [CrossRef]
- Kim HK, Kim Y, Lee S, Min S, Bae JY, Choi JW, et al. SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci Adv. 2019 Nov;5(11):eaax9249.
- Xu, H.; Xiao, T.; Chen, C.-H.; Li, W.; Meyer, C.A.; Wu, Q.; Wu, D.; Cong, L.; Zhang, F.; Liu, J.S.; et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015, 25, 1147–1157. [CrossRef]
- Anthon C, Corsi GI, Gorodkin J. CRISPRon/off: CRISPR/Cas9 on- and off-target gRNA design. Valencia A, editor. Bioinformatics. 2022 Dec 13;38(24):5437–9.
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [CrossRef]
- Hsu, J.Y.; Grünewald, J.; Szalay, R.; Shih, J.; Anzalone, A.V.; Lam, K.C.; Shen, M.W.; Petri, K.; Liu, D.R.; Joung, J.K.; et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 2021, 12, 1–6. [CrossRef]
- Hwang, G.-H.; Jeong, Y.K.; Habib, O.; Hong, S.-A.; Lim, K.; Kim, J.-S.; Bae, S. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 2021, 49, W499–W504. [CrossRef]
- Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. Maas S, editor. PLOS ONE. 2015 Apr 24;10(4):e0124633.
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; A Wolfe, S. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [CrossRef]
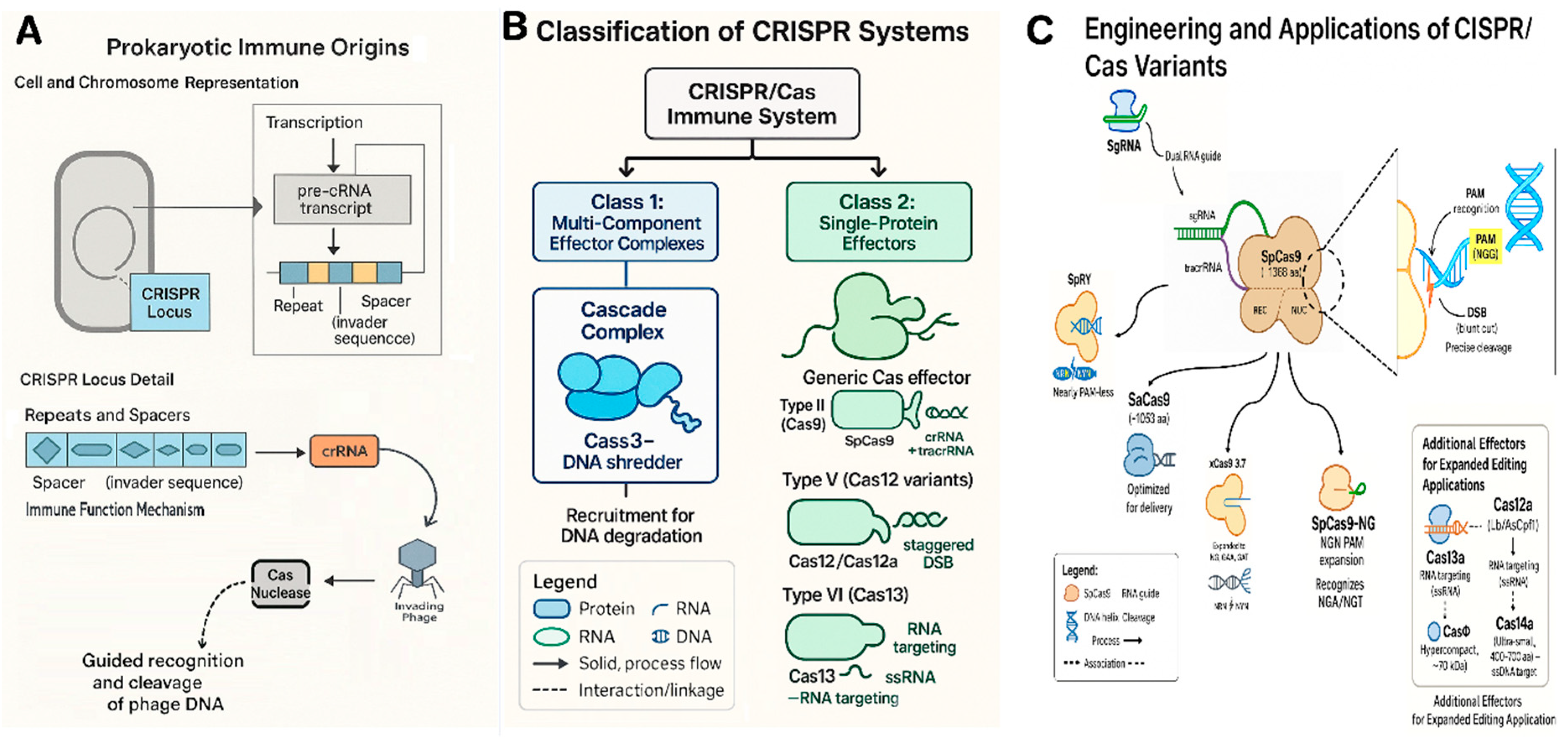
| Cas Protein | Type (Class) | Size (aa) / Complex | Target & Cut | PAM / Target Motif | Notable Features |
| SpCas9 (wild-type) | II-A (Class 2) | ~1368 aa, sgRNA:crRNA+tracrRNA | DNA, blunt DSB (3 bp from PAM) | NGG (also NAG at lower efficiency) | Most widely used; numerous variants (e.g. SpCas9-NG, SpRY) expand PAM to NRN/NYN. Requires tracrRNA. |
| SaCas9 | II-A (Class 2) | ~1053 aa, sgRNA | DNA, blunt DSB | NNGRRT | ~25% smaller than SpCas9, useful for delivery; different PAM. Efficient in some plants after codon optimization. |
| Cas12a (Lb/AsCpf1) | V-A (Class 2) | ~1300 aa, crRNA (42 nt) | DNA, staggered DSB (5’ overhang) | TTTV (e.g. TTTA, TTTG) | No tracrRNA needed; processes its own array of crRNAs. Cuts distal to PAM (~, allowing large fragment deletions or repeat cutting. High AT-targeting ability and specificity. |
| Cas12b (BhCas12b) | V-B (Class 2) | ~1100 aa, crRNA | DNA, staggered DSB | TBD (e.g. TTN) | Smaller type V; engineered variants (e.g. BhCas12b v4) active at 37°C for plants. Potential for compact multiplex tools. |
| CasΦ | V? (Class 2, phage) | ~70 kDa (~600 aa), crRNA | DNA, DSB (overhang unclear) | Minimal PAM (e.g. TBN) | Hypercompact single-effector from phages; single active site for crRNA processing & cleavage. Active in plant cells; easier delivery due to small size. |
| Cas14a (Cas12f) | V-F (Class 2) | ~400–700 aa, crRNA | ssDNA, multiple cuts (collateral activity) | None (no strict PAM) | Extremely small; cleaves single-stranded DNA with high single-base mismatch sensitivity. Useful for diagnostics; potential to target ssDNA viruses. |
| CasX | V-E (Class 2) | ~980 aa, crRNA | DNA, DSB (5’ overhang) | TTCN (for DxCasX variant) | Discovered in Duggariela bacteria; independently evolved from Cas9/Cas12. Compact and non-immunogenic (from non-human microbes). Efficient editing in E. coli and human cells; being adapted for plants. |
| Cas13a (LwaCas13a) | VI (Class 2) | ~1250 aa, crRNA | ssRNA, multiple cleavage (collateral RNase) | Requires protospacer-flanking A/U/C (no DNA PAM) | RNA-specific editing (transient knockdowns). Collateral RNase used for viral RNA detection. Cas13 variants (a–d) can be repurposed for virus resistance in plants without genomic changes. |
| dCas9 or dCas12a | (engineered) | As above but catalytically dead | DNA (no cleavage) | NGG (SpCas9) or TTTV (Cas12a) | DNA-binding platform for base editors (e.g. CBE, ABE) and epigenome modifiers. dCas9 fused to deaminase yields precise single-base changes without DSB. Fusions to activators (CRISPRa) or repressors (CRISPRi) enable gene regulation without mutation. |
| Strategy | Mechanism & Components | Example Usage and Performance |
| Multiple U6 cassettes(Pol III promoters) | Each sgRNA is expressed from a separate U6/U3 small RNA promoter. Simple but increases vector size linearly with guide number. | Common in early multiplex experiments (2–4 targets). E.g., 4 U6-driven sgRNAs in tomato to mutate four loci [67]. Efficient for few targets, but >6 cassettes becomes cumbersome. |
| tRNA–sgRNA array(Pol III, PTG) | Tandem tRNA–sgRNA units transcribed as one RNA. Endogenous RNase P/Z cleave at tRNA ends, releasing sgRNAs. | Xie et al. (2015) edited 4 genes in rice with one PTG transcript [54]. High editing rates, though very large arrays see some efficiency drop-off. Supports >10 guides. |
| Ribozyme flanks(Pol II or Pol III) | Each sgRNA is flanked by self-cleaving ribozymes (HH and HDV). After transcription, ribozymes auto-cleave to yield individual sgRNAs. | Tang et al. (2016) achieved 6 multiplex edits in rice from a single Pol II transcript (UBI promoter) [58]. Very efficient (up to 100% mutant frequency per target). Modular and portable to any promoter. |
| Csy4-mediated(Pol II) | sgRNAs separated by Csy4 recognition sequences. Co-express Csy4 RNase which cleaves and releases sgRNAs. Requires adding Csy4 protein. | Cermák et al. (2017) multiplexed 8 sgRNAs in tomato; Csy4 system gave highest editing efficiency vs tRNA or ribozymes. Also used for large deletions with up to 12 guides. Extra burden of expressing Csy4, but highly effective. |
| miRNA-based(Pol II, Drosha/Dicer) | sgRNAs embedded in endogenous miRNA stem-loops. The pri-miRNA transcript is processed by Drosha/DCL1, releasing sgRNAs (as faux miRNAs). | Used in Arabidopsis: 2–3 sgRNAs designed into AtMIR390 backbone, driven by RPS5a promoter, yielded heritable edits without transgenes (in a haploid inducer system) [68]. Efficiency can be lower, but allows tissue-specific promoters for sgRNA expression. |
| Cas12a array(Pol III, self-process) | A single CRISPR array (repeats and spacers) transcribed; Cas12a cleaves the repeats to release crRNAs. No tracrRNA needed. | FnCas12a edited 4 targets in soybean from one array (driven by LjU6) with 20–50% efficiency per locus [69]. Very compact – one promoter for many guides – but limited to Cas12a or Cas12j systems. |
| Plant Species | Target Genes | Trait Improved | Specific Improvement | Key Outcomes | Reference |
| Sugarcane (Saccharum) | LG1 | Yield | Adjusted leaf angle | Mutated line showed a 56% decrease in leaf inclination angle and an 18% increase in yield. | [125] |
| Brassica napus(Rapeseed) | BnFAD2, BnFAE1 | Oil Quality | Increased oleic acid, reduced erucic acid | Significant increase in oleic acid content (70-80%) and dramatic reduction in erucic acid levels in seed oil. | [127] |
| Potato (Solanum tuberosum) | Vacuolar Invertase (VInv), Asparagine Synthetase 1 (AS1) | Processing Quality & Safety | Reduced browning and acrylamide formation | Tubers from edited events showed reduced fructose and glucose concentrations after cold storage. Crisps made from these tubers were lighter in color and contained significantly reduced levels of acrylamide. Multiplex CRISPR-Cas9 technology demonstrated its ability to generate improved potato cultivars for healthier processed products. | [128] |
| Rice (Oryza sativa) | Starch Branching Enzyme (SBE) genes (four subunits) | Nutritional Value | Increased resistant starch content | Knockout mutations were identified at all four SBE genes across multiple transgenic rice plants. This multiplex editing led to an increase in the level of resistant starch in the rice grains, enhancing their nutritional value for human consumption. | [129] |
| Plant Species | Target Genes | Pathogen/Pest | Edited Trait (Resistance/Susceptibility) | Key Outcomes | Reference |
| Brassica napus(Rapeseed) | BnWRKY11, BnWRKY70 | Sclerotinia sclerotiorum | Enhanced Resistance (BnWRKY70 mutants) | BnWRKY70 mutants showed enhanced resistance; BnWRKY11 mutants showed no significant difference. Overexpression of BnWRKY70 increased sensitivity. | [134] |
| Wheat (Triticum) | MLO gene family, dormant gene | Powdery Mildew | Resistance | Full resistance to powdery mildew achieved by simultaneously knocking out multiple MLO susceptibility genes and activating a dormant resistance gene in elite wheat varieties. | [135] |
| Tomato (Solanum lycopersicum) | Various | Various | Resistance | Potential for developing lines resistant to common bacterial, fungal, and viral diseases by inducing resistance genes or knocking out susceptibility genes. Multiplexing can further enhance this by targeting multiple factors. | [136] |
| Plant Species | Target Genes | Edited Trait | Key Outcomes | Reference |
| Wheat (Triticum) | TaSal1 gene family | Drought Tolerance | Improved germination and growth under simulated drought conditions. | [140] |
| Rice (Oryza sativa) | PsbS regulatory DNA | Water-Use Efficiency | Increased expression of PsbS, leading to enhanced water-use efficiency without foreign DNA integration. | [141] |
| Wheat (Triticum) | TaRPK1 | Water Absorption | Potential for enhanced water absorption through deeper root system development. | [142] |
| Various Plant Species | Sal1 | Osmo-protectant Production | Improved production of osmoprotectants like proline, enhancing drought resistance through precise modulation of gene expression. | [143] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





