Submitted:
28 June 2023
Posted:
03 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plants cultivation
2.2. Agrobacterium-mediated wheat transformation
2.3. Cloning of gRNAs and PCR-confirmation of transgenic plants
2.4. Evaluation of GE at the target site using cleaved amplified polymorphic sequences and qPCR assays
2.5. Estimation of the transgene copy number
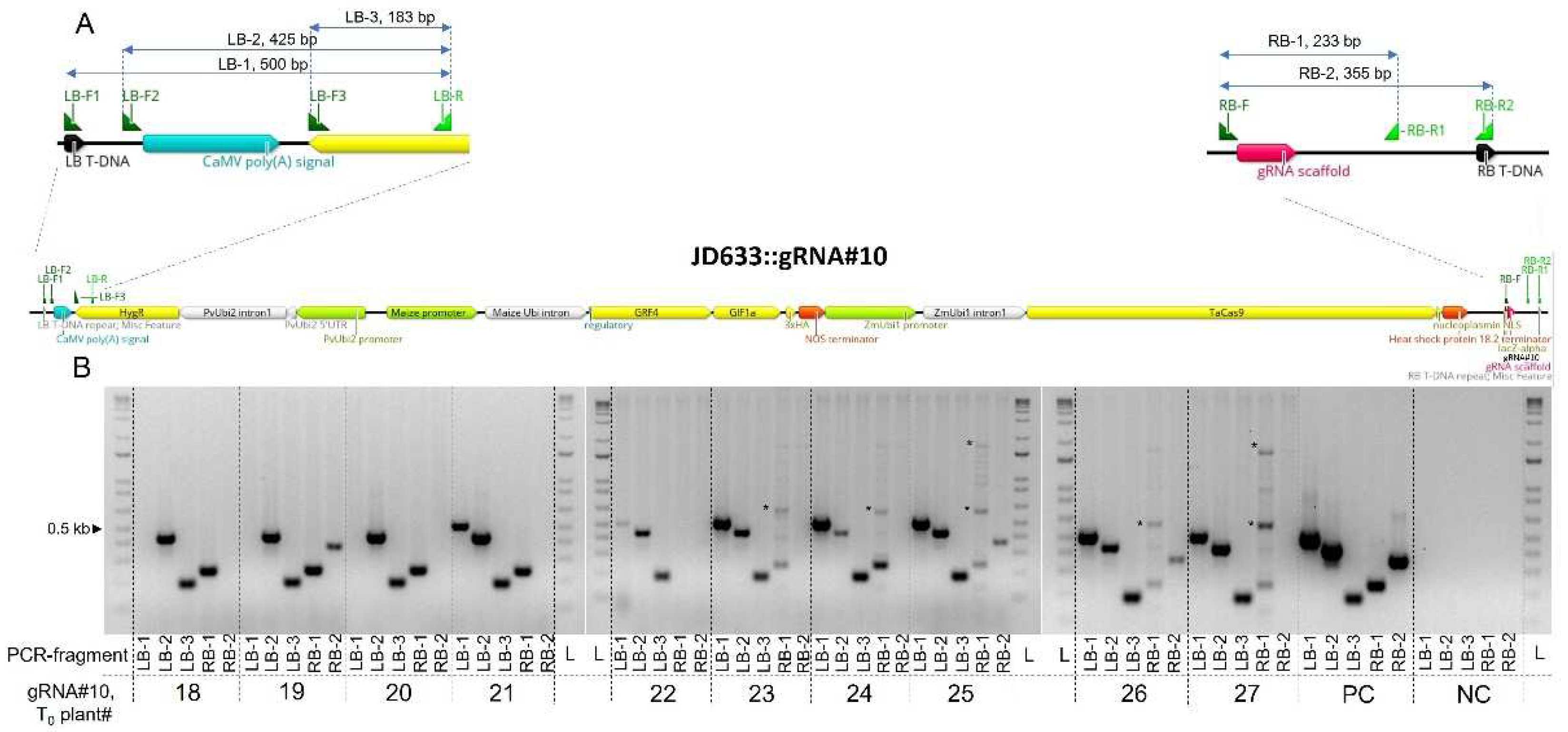
2.6. Evaluation of the left and right T-DNA borders integrity and transgenes co-integration events
2.7. Gene expression analysis using ddPCR
2.8. Characterization of chromatin features
2.9. Statistical treatment of the data
3. Results
3.1. Agrobacterium-mediated co-delivery of the GRF-GIF chimeric gene results in the efficient regeneration of transgenic wheat plants
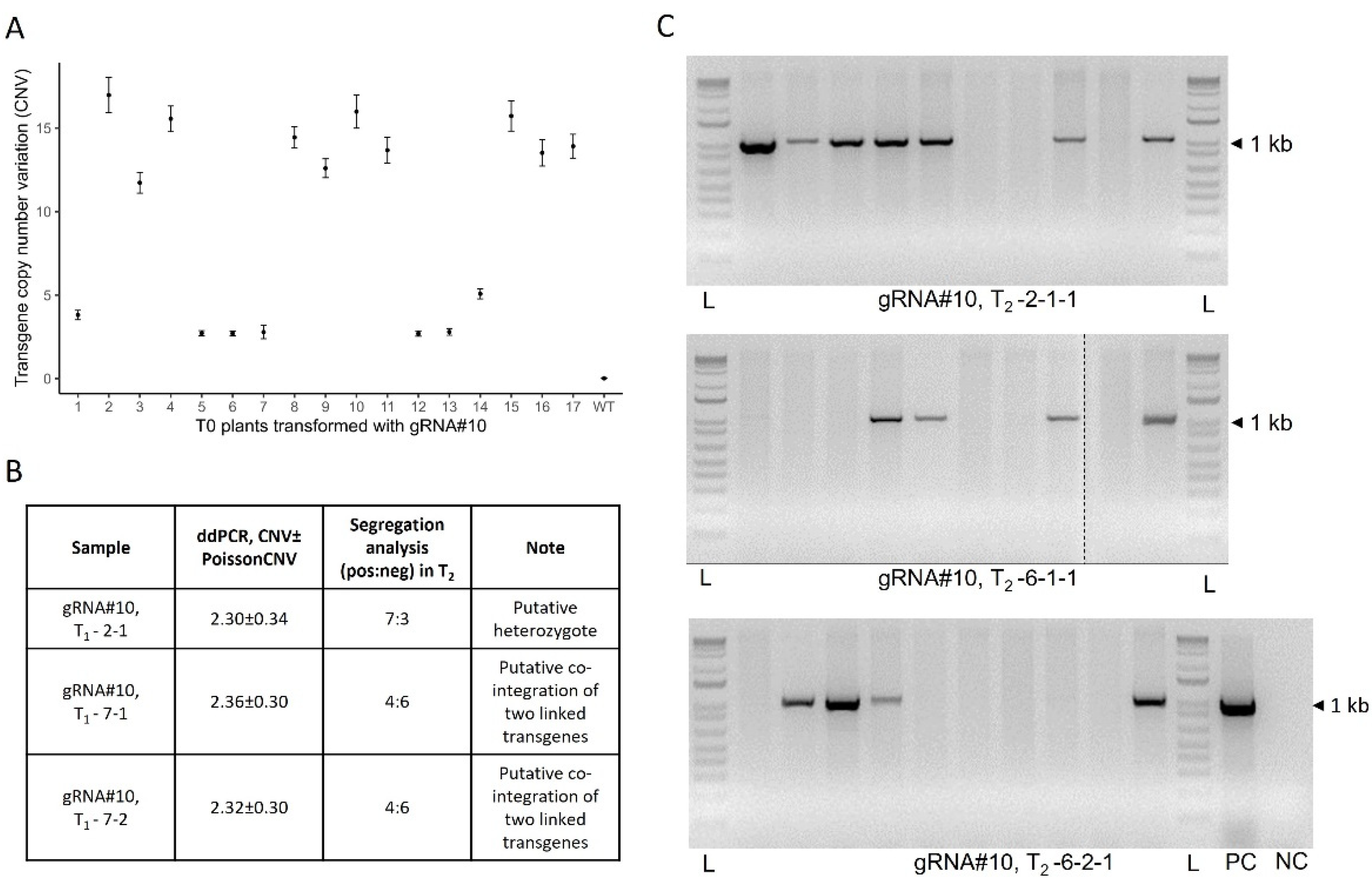
3.2. Low- and high-copy number T-DNA integration events can be recovered in the transgenic population
3.3. Frequency of co-integration events and the integrity of the T-DNA borders
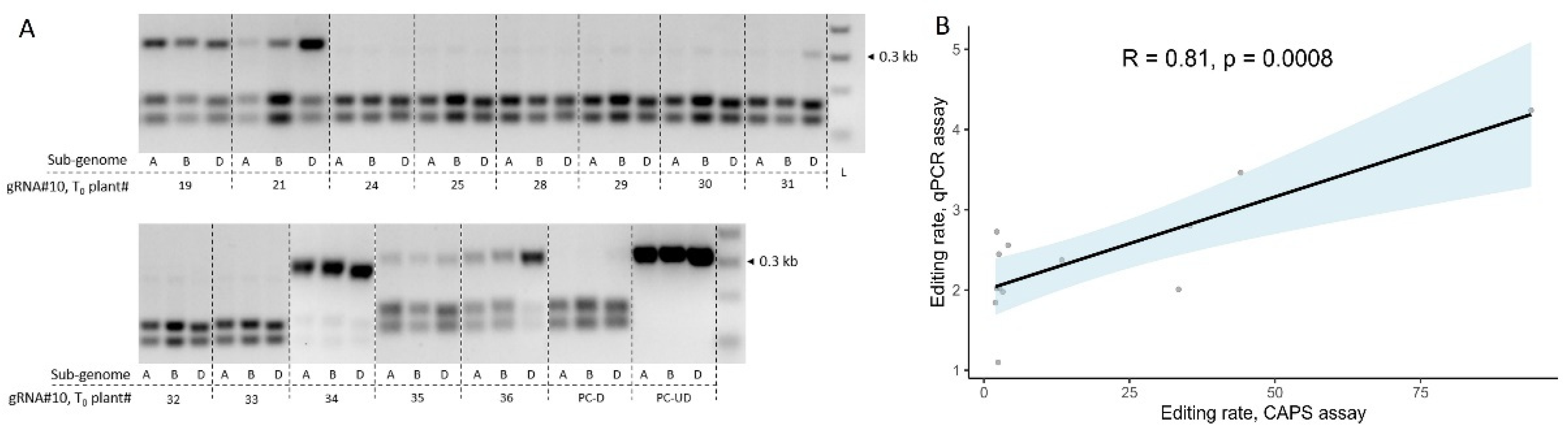
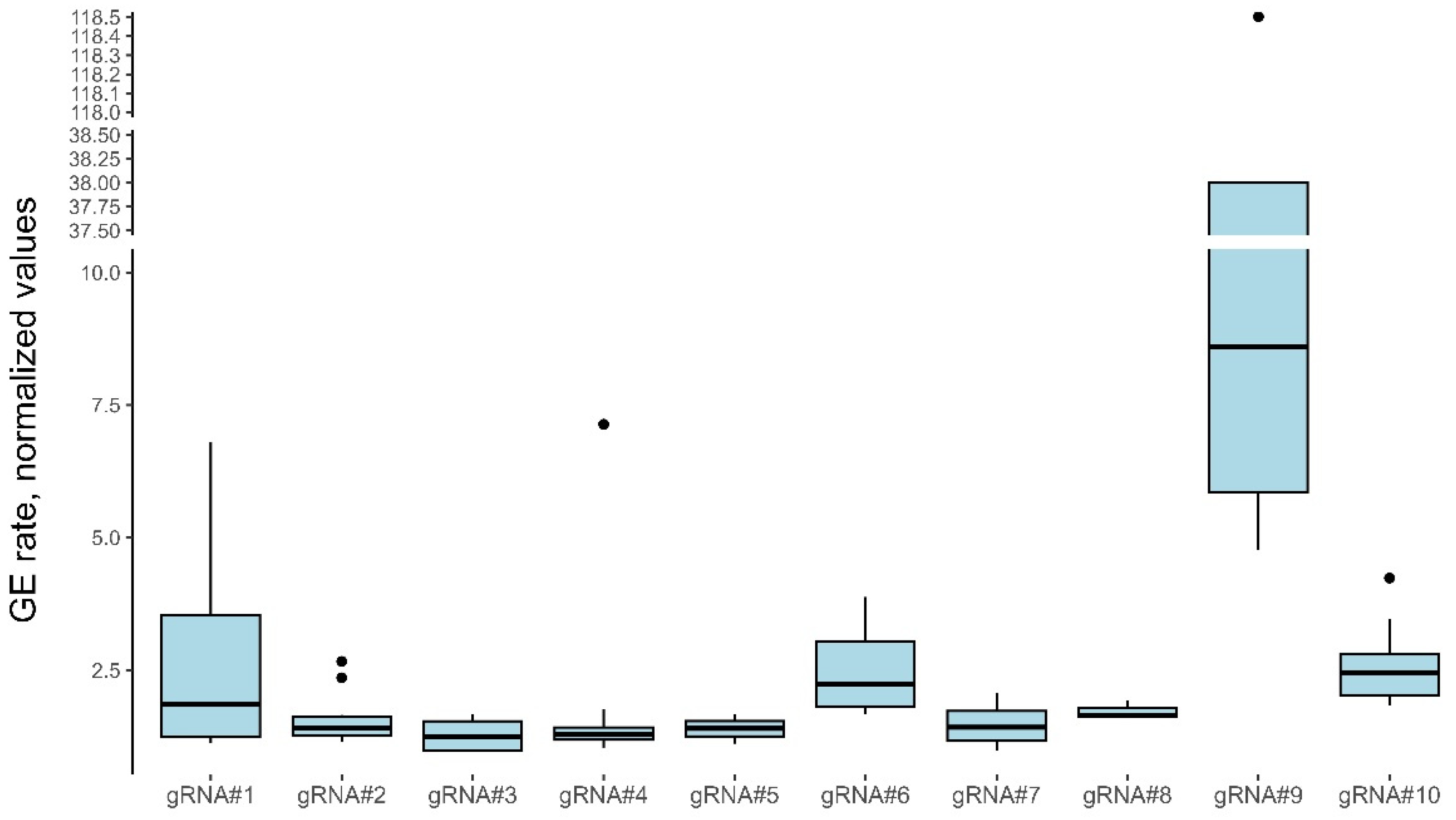
3.4. Agrobacterium-mediated wheat transformation results in the efficient generation of gene-edited plants
| gRNA | Target gene | Homeolog IDs | Activity score, WheatCrispr [32] |
|---|---|---|---|
| gRNA#1 |
TRYPTOPHAN SYNTHASE, ALPHA CHAIN (TaTRP_SYNTHASE_SUA) |
TraesCS1A02G428400, TraesCS1B02G463100, TraesCS1D02G437700 | 0.35 |
| gRNA#2 | INOSITOL TETRA-/PENTAPHOSPHATE 2-KINASE (TaIPK1) | TraesCS2A02G497700, TraesCS2B02G525900 TraesCS2D02G612600LC | 0.45 |
| gRNA#3 | INOSITOL TETRA-/PENTAPHOSPHATE 2-KINASE (TaIPK1) | TraesCS2A02G497700, TraesCS2B02G525900 TraesCS2D02G612600LC | 0.39 |
| gRNA#4 | MANGANESE-SUPEROXIDE DISMUTASE (TaMnSOD) | TraesCS2A02G537100, TraesCS2B02G567600, TraesCS2D02G538300 | 0.42 |
| gRNA#5 | WHEAT PROLAMIN-BOX BINDING FACTOR (TaWPBF) | TraesCS5A02G155900, TraesCS5B02G154100, TraesCS5D02G161000 | 0.44 |
| gRNA#6 | DEMETER (TaDME) | TraesCS5A02G169000, TraesCS5B02G165800, TraesCS5D02G173300 | 0.46 |
| gRNA#7 | DEMETER (TaDME) | TraesCS5A02G169000, TraesCS5B02G165800, TraesCS5D02G173300 | 0.46 |
| gRNA#8 | QUANTITATIVE TRAIT LOCUS ON SEED DORMANCY 1 (TaQSD1) | TraesCS5A02G216200, TraesCS5B02G214700, TraesCS5D02G224200 | 0.47 |
| gRNA#9 | QUANTITATIVE TRAIT LOCUS ON SEED DORMANCY 1 (TaQSD1) | TraesCS5A02G216200, TraesCS5B02G214700, TraesCS5D02G224200 | 0.46 |
| gRNA#10 | GRAIN WIDTH and WEIGHT2 (TaGW2) | TraesCS6A02G189300, TraesCS6B02G215300, TraesCS6D02G176900 | 0.45 |
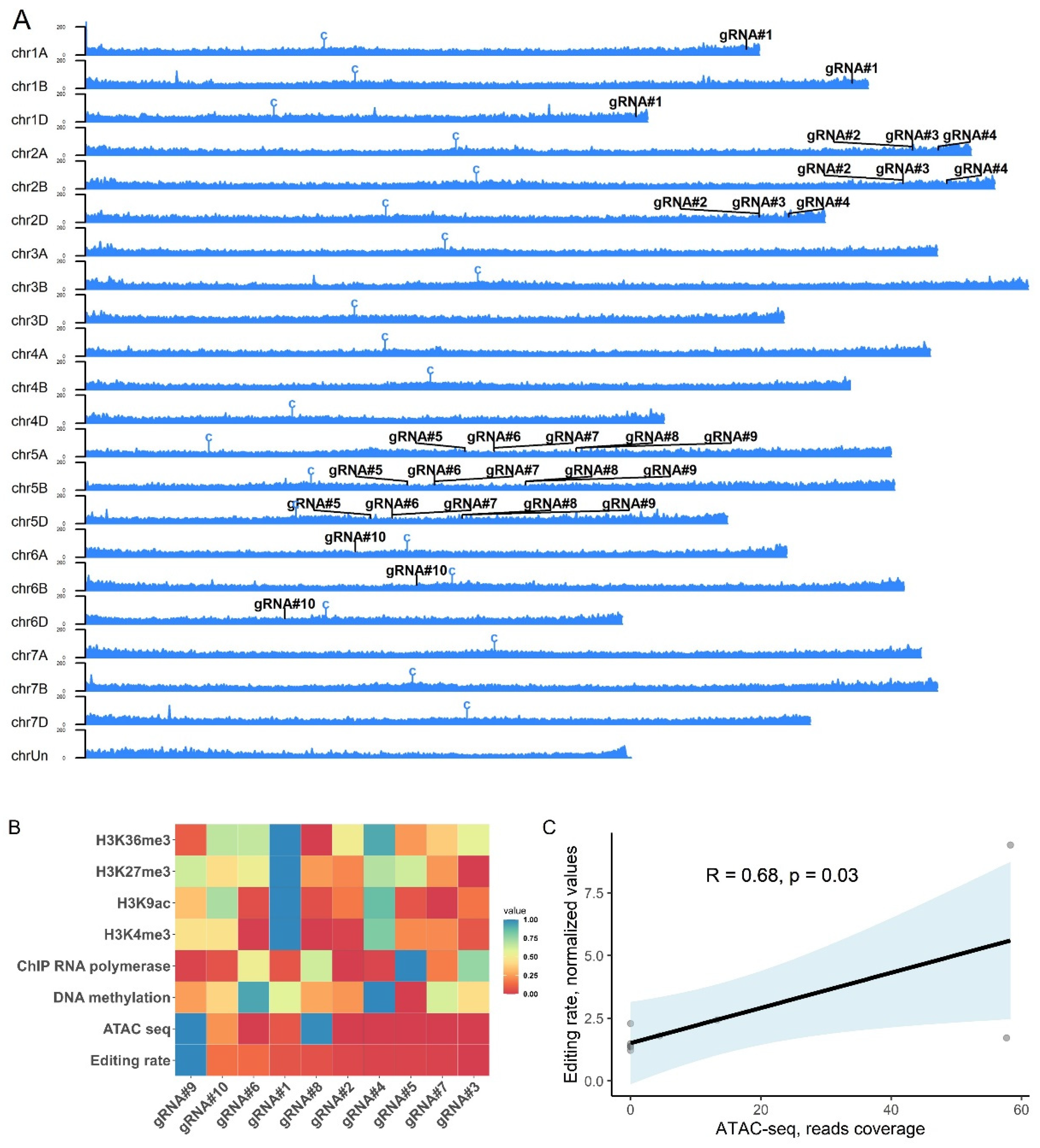
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walkowiak, S., et al., Multiple wheat genomes reveal global variation in modern breeding. Nature, 2020. 588(7837): p. 277-283. [CrossRef]
- Davière, J.M. and P. Achard, A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol Plant, 2016. 9(1): p. 10-20. [CrossRef]
- Rebetzke, G.J., et al., Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crops Research, 2007. 100(1): p. 10-23. [CrossRef]
- Chen, H., M. Neubauer, and J.P. Wang, Enhancing HR Frequency for Precise Genome Editing in Plants. Frontiers in Plant Science, 2022. 13. [CrossRef]
- Lazar, S., et al., RECAS9: Recombining wild species introgression via mitotic gene editing in barley. bioRxiv, 2020: p. 2020.01.07.897280.
- Wang, K., et al., The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat Plants, 2022. 8(2): p. 110-117. [CrossRef]
- Johnson, K., et al., Rapid and highly efficient morphogenic gene-mediated hexaploid wheat transformation. Frontiers in Plant Science, 2023. 14. [CrossRef]
- Debernardi, J.M., et al., A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol, 2020. 38(11): p. 1274-1279. [CrossRef]
- Allen, F., et al., Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol, 2018. [CrossRef]
- Concordet, J.P. and M. Haeussler, CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res, 2018. 46(W1): p. W242-W245. [CrossRef]
- Xiang, X., et al., Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat Commun, 2021. 12(1): p. 3238. [CrossRef]
- Naim, F., et al., Are the current gRNA ranking prediction algorithms useful for genome editing in plants? PLoS One, 2020. 15(1): p. e0227994.
- Weiss, T., et al., Epigenetic features drastically impact CRISPR–Cas9 efficacy in plants. Plant Physiology, 2022. 190(2): p. 1153-1164. [CrossRef]
- Concia, L., et al., Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biol, 2020. 21(1): p. 104. [CrossRef]
- Hayta, S., et al., An Efficient Agrobacterium-Mediated Transformation Protocol for Hexaploid and Tetraploid Wheat. Curr Protoc, 2021. 1(3): p. e58. [CrossRef]
- An, G., et al., Binary vectors. pp. 1988, A3/1-19, In: Gelvin, SB and. Schilperoort, RA (eds.), Plant molecular ….
- Xu, W., et al., Evaluation of Factors Affecting In Planta Gene Editing Efficiency in Wheat (Triticum aestivum L.). ACS Agricultural Science & Technology, 2022. 2(2): p. 222-231. [CrossRef]
- Collier, R., et al., Accurate measurement of transgene copy number in crop plants using droplet digital PCR. 2017. 90(5): p. 1014-1025. [CrossRef]
- Warner, P., et al., An investigation of a rapid DNA extraction method for routine MAS in the S.A. Barley Improvement Program. 2002.
- Jouanin, A., et al., Optimisation of droplet digital PCR for determining copy number variation of α-gliadin genes in mutant and gene-edited polyploid bread wheat. Journal of Cereal Science, 2020. 92: p. 102903. [CrossRef]
- Garrido, J., M. Aguilar, and P. Prieto, Identification and validation of reference genes for RT-qPCR normalization in wheat meiosis. Sci Rep, 2020. 10(1): p. 2726.
- Coulter, S.J., Mitigation of the effect of variability in digital PCR assays through use of duplexed reference assays for normalization. BioTechniques, 2018. 65(2): p. 86-91. [CrossRef]
- Jia, J., et al., Homology-mediated inter-chromosomal interactions in hexaploid wheat lead to specific subgenome territories following polyploidization and introgression. Genome Biology, 2021. 22(1): p. 26. [CrossRef]
- International Wheat Genome Sequencing, C., Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 2018. 361(6403).
- Gel, B. and E. Serra, karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics, 2017. 33(19): p. 3088-3090. [CrossRef]
- Luo, J., et al., Pyramiding favorable alleles in an elite wheat variety in one generation by CRISPR-Cas9-mediated multiplex gene editing. Molecular Plant, 2021. 14(6): p. 847-850. [CrossRef]
- Abdelrahman, M., et al., Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Frontiers in Plant Science, 2021. 12. [CrossRef]
- Gelvin, S.B., Integration of Agrobacterium T-DNA into the Plant Genome. Annual Review of Genetics, 2017. 51(1): p. 195-217. [CrossRef]
- Li, J., et al., Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol J, 2021. 19(3): p. 427-429. [CrossRef]
- Wang, W., et al., Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. Crispr j, 2018. 1(1): p. 65-74. [CrossRef]
- Song, X.J., et al., A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet, 2007. 39(5): p. 623-30. [CrossRef]
- Cram, D., et al., WheatCRISPR: a web-based guide RNA design tool for CRISPR/Cas9-mediated genome editing in wheat. BMC Plant Biology, 2019. 19(1): p. 474. [CrossRef]
- Doench, J.G., et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 2016. 34(2): p. 184-191. [CrossRef]
- Concia, L., et al., Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biology, 2020. 21(1): p. 104. [CrossRef]
- Richardson, T., et al., Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue and Organ Culture (PCTOC), 2014. 119(3): p. 647-659. [CrossRef]
- Ishida, Y., et al., Wheat (Triticum aestivum L.) Transformation Using Immature Embryos, in Agrobacterium Protocols: Volume 1, K. Wang, Editor. 2015, Springer New York: New York, NY. p. 189-198.
- Wang, K., et al., Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol J, 2017. 15(5): p. 614-623. [CrossRef]
- Gelvin, S.B., Agrobacterium-mediated plant transformation: the biology behind the "gene-jockeying" tool. Microbiol Mol Biol Rev, 2003. 67(1): p. 16-37, table of contents.
- Che, P., et al., Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Communications Biology, 2022. 5(1): p. 344. [CrossRef]
- Meng, X., et al., Construction of a Genome-Wide Mutant Library in Rice Using CRISPR/Cas9. Molecular Plant, 2017. 10(9): p. 1238-1241. [CrossRef]
- Wu, X., et al., Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol, 2014. 32(7): p. 670-6. [CrossRef]
- Daer, R.M., et al., The Impact of Chromatin Dynamics on Cas9-Mediated Genome Editing in Human Cells. ACS Synth Biol, 2017. 6(3): p. 428-438. [CrossRef]
- Yarrington, R.M., et al., Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc Natl Acad Sci U S A, 2018. 115(38): p. 9351-9358. [CrossRef]
- Liu, G., et al., Modulating chromatin accessibility by transactivation and targeting proximal dsgRNAs enhances Cas9 editing efficiency in vivo. Genome Biol, 2019. 20(1): p. 145. [CrossRef]
- Miguel, C. and L. Marum, An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. Journal of Experimental Botany, 2011. 62(11): p. 3713-3725. [CrossRef]
- Lee, K. and P.J. Seo, Dynamic Epigenetic Changes during Plant Regeneration. Trends in Plant Science, 2018. 23(3): p. 235-247. [CrossRef]
- Buchholzer, M. and W.B. Frommer, An increasing number of countries regulate genome editing in crops. New Phytol, 2023. 237(1): p. 12-15. [CrossRef]

| Replicate | Cultivar | Agrobacterium strain/helper plasmid | Number of embryos transformed | Number of PCR-confirmed transgenics recovered | Transformation efficiency, % |
|---|---|---|---|---|---|
| #1 | Fielder | AGL1/pSOUP | 192 | 44 | 22.9 |
| #2 | Fielder | AGL1/pSOUP | 145 | 70 | 48.3 |
| #3 | Fielder | AGL1/pSOUP | 190 | 70 | 36.8 |
| #4 | Fielder | AGL1/pSOUP | 221 | 151 | 68.3 |
| Average | 187 | 84 | 44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





