Submitted:
08 June 2025
Posted:
09 June 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Sample Collection
DNA Extraction and Amplification of Cytb and D-loop Genes
DNA Sequence Data Processing
Results
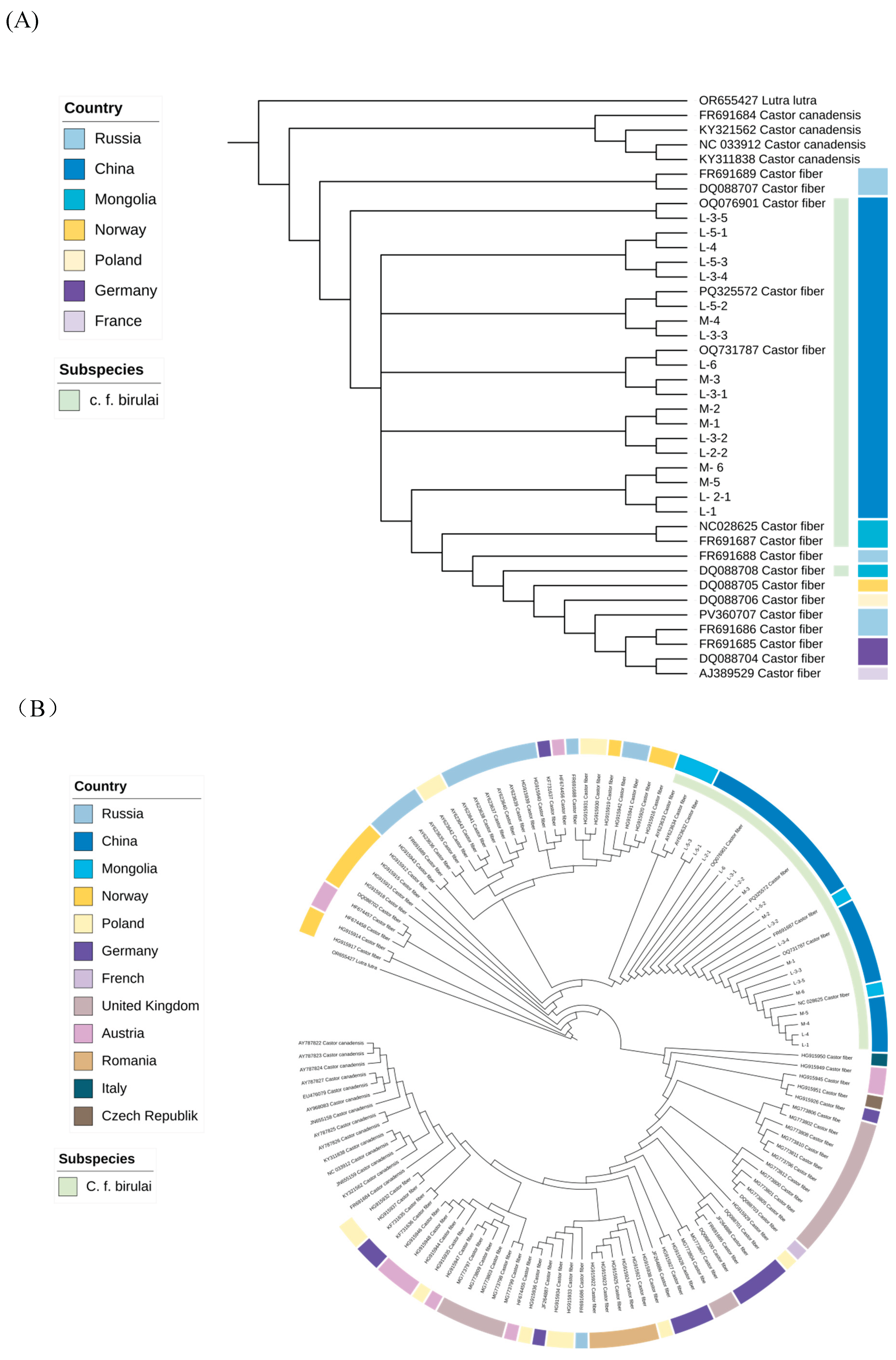
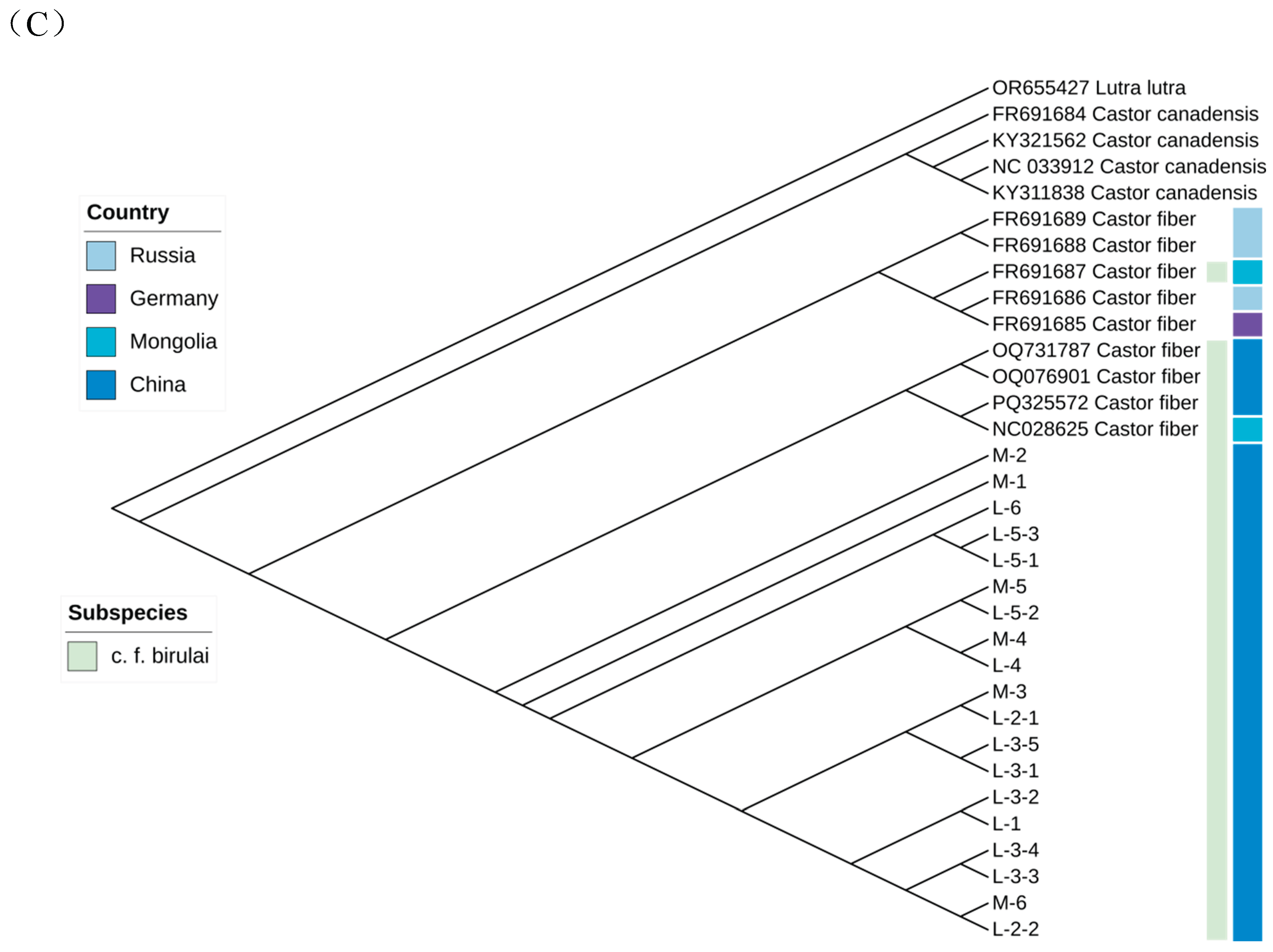
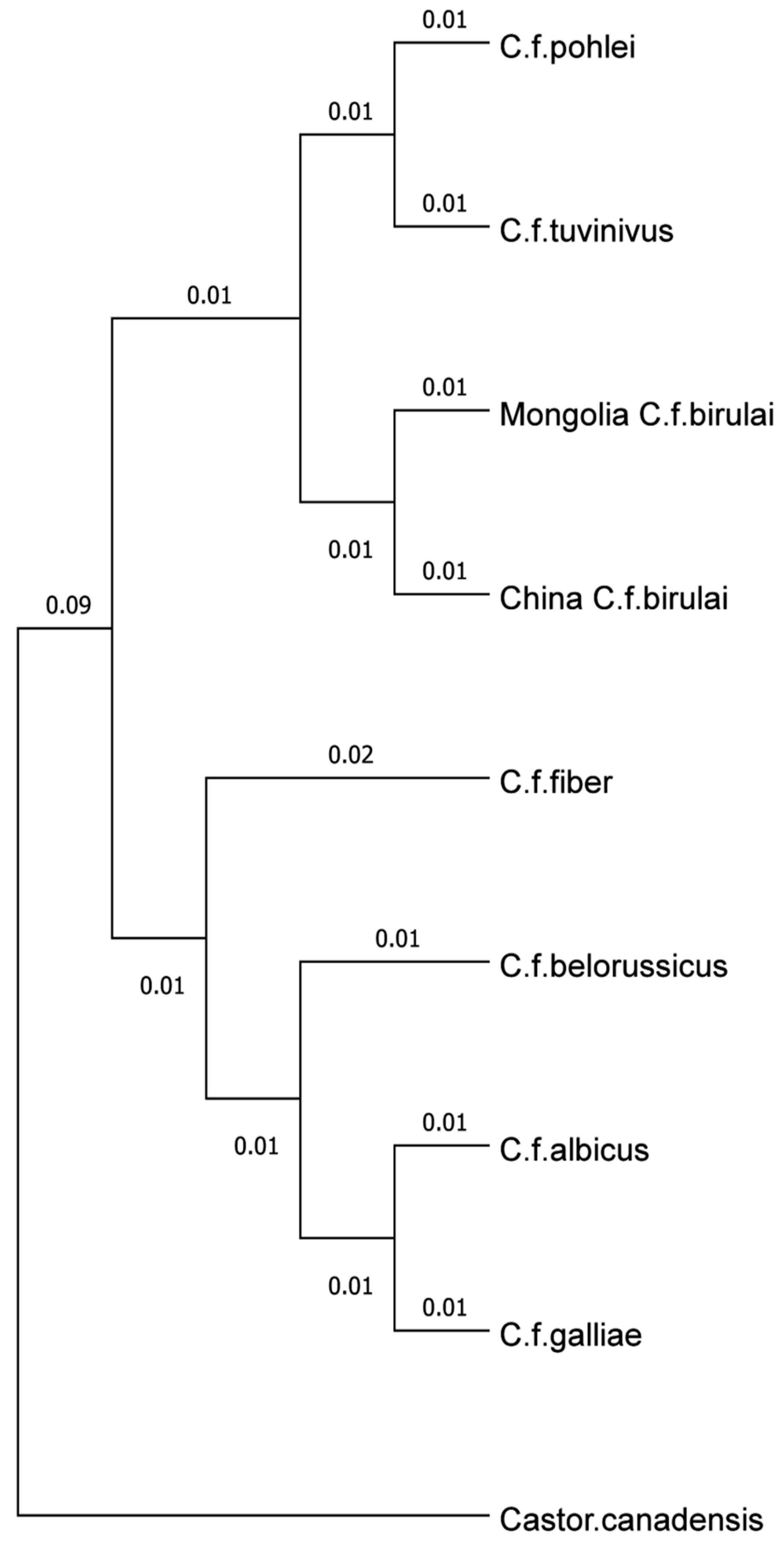
Mitochondrial Sequence Analysis
Genetic Diversity of C.f. birulai
Genetic Structure of Xinjiang C.f. birulai
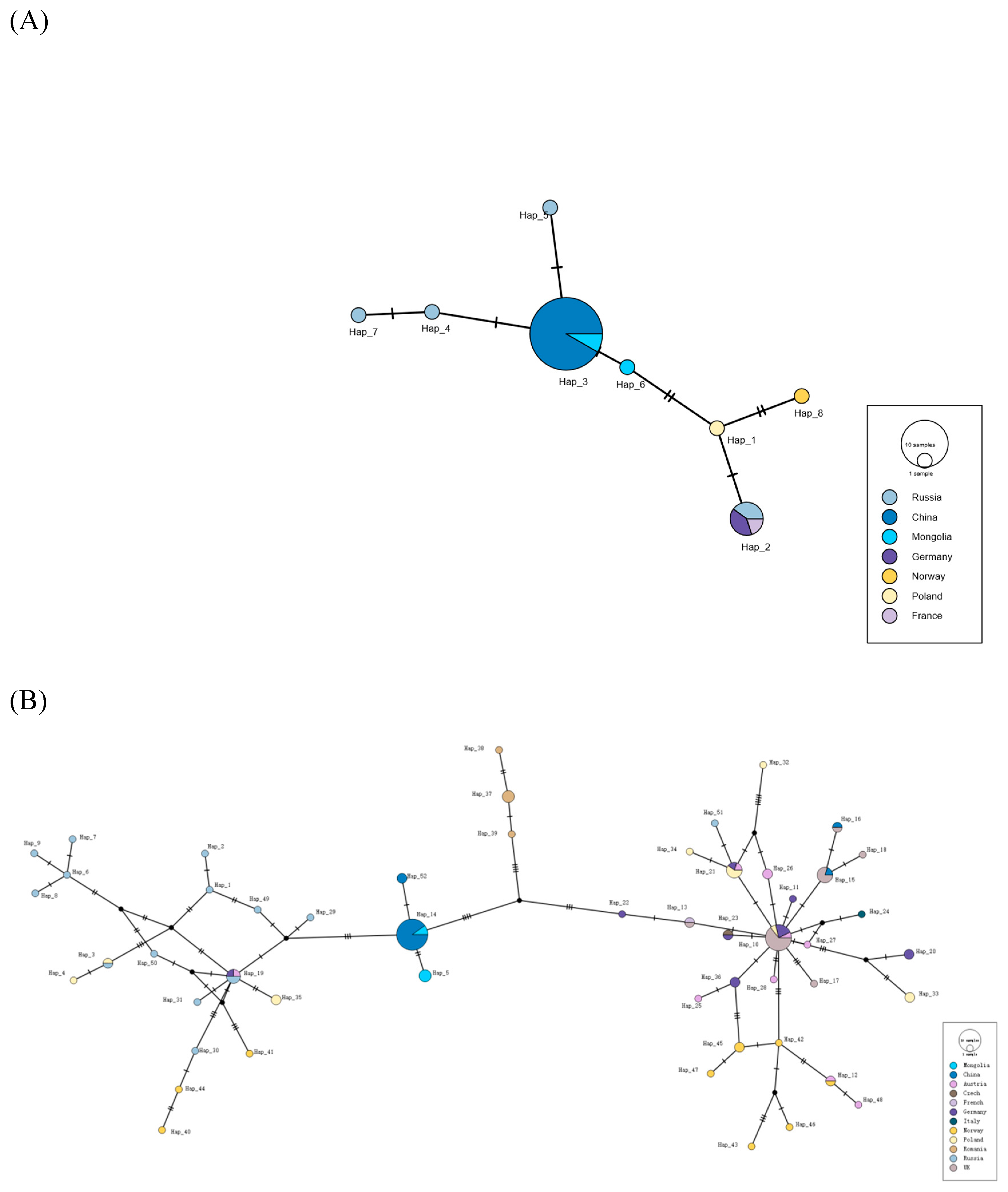
Haplotype Network Analysis
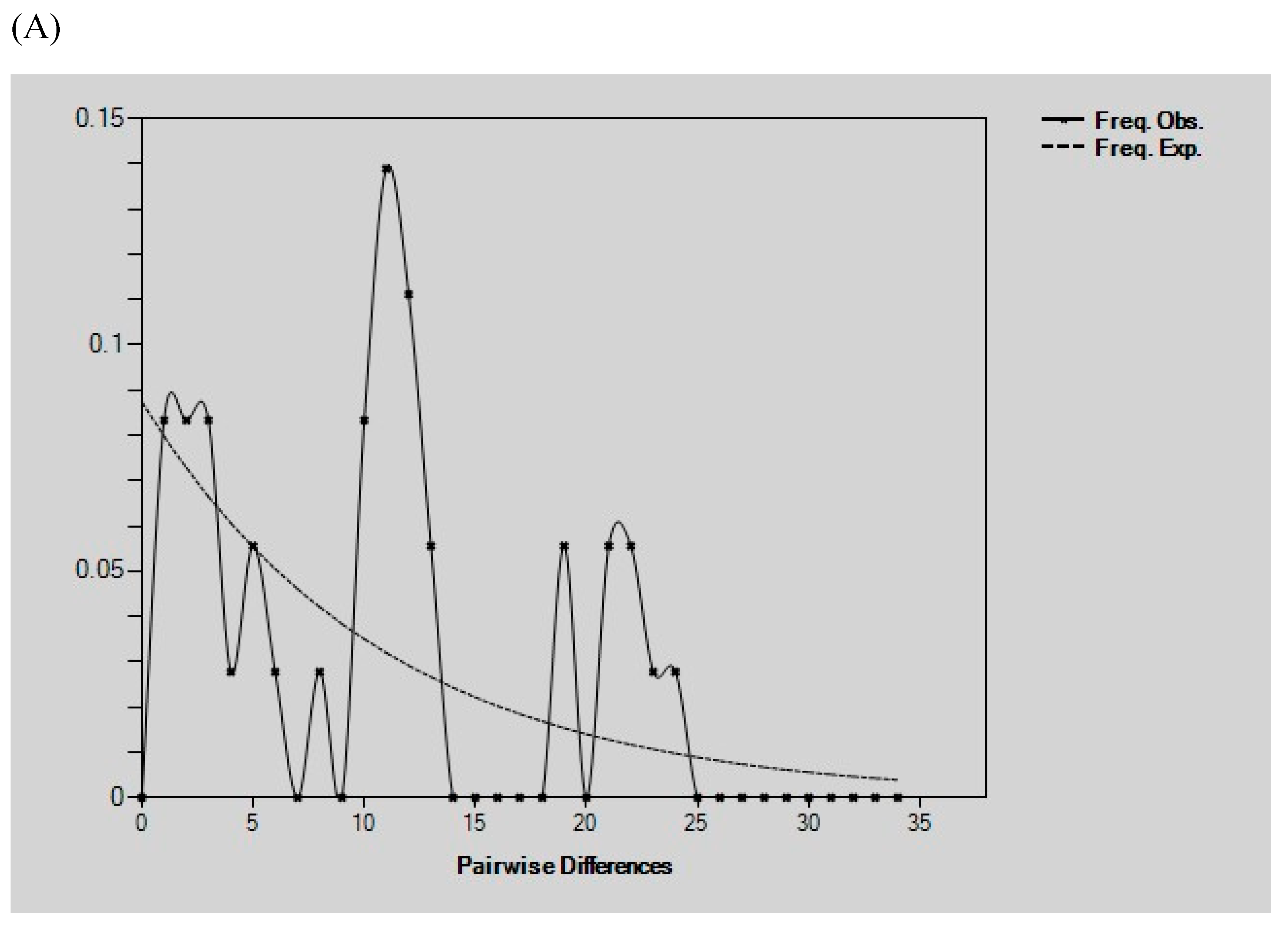
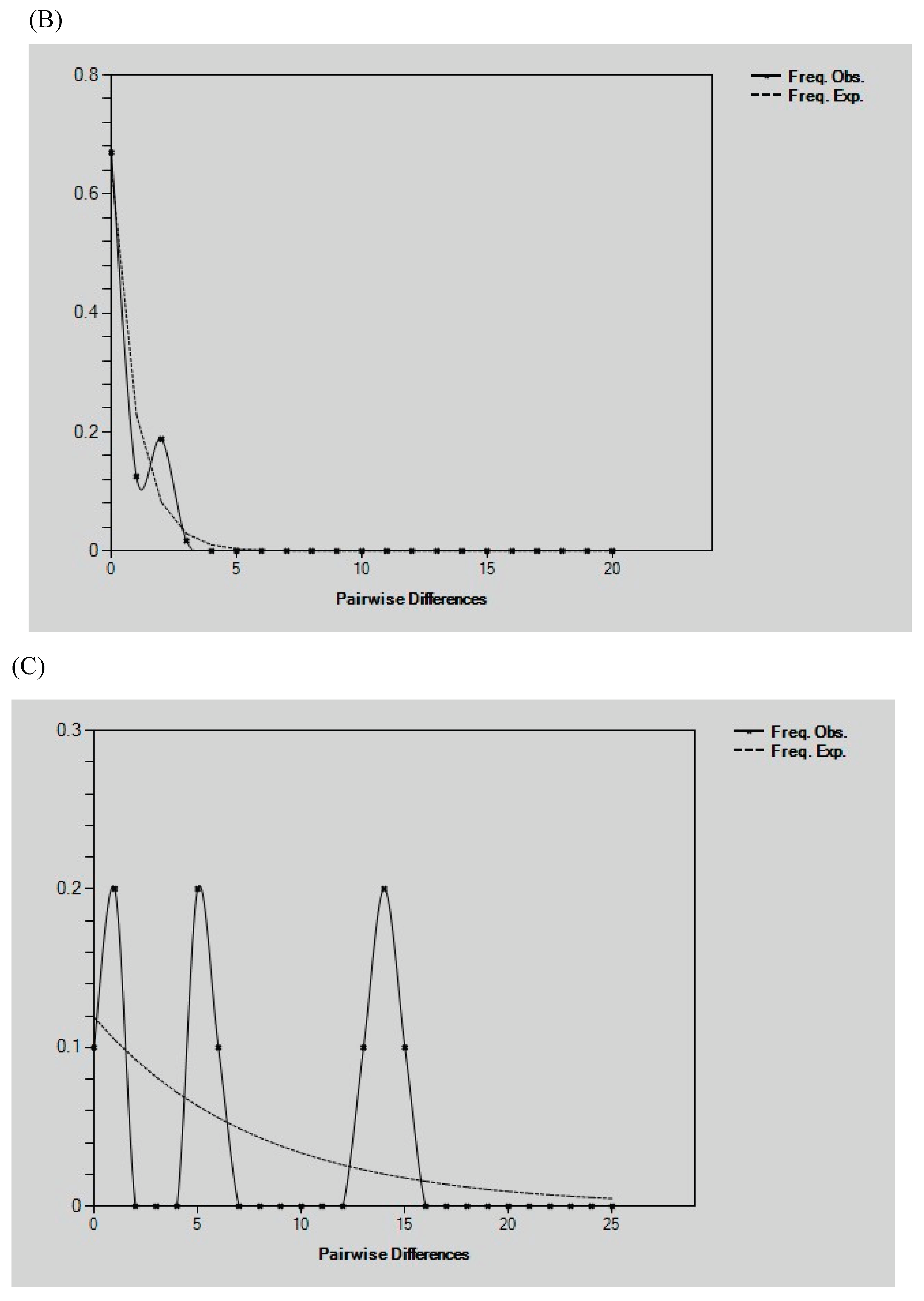
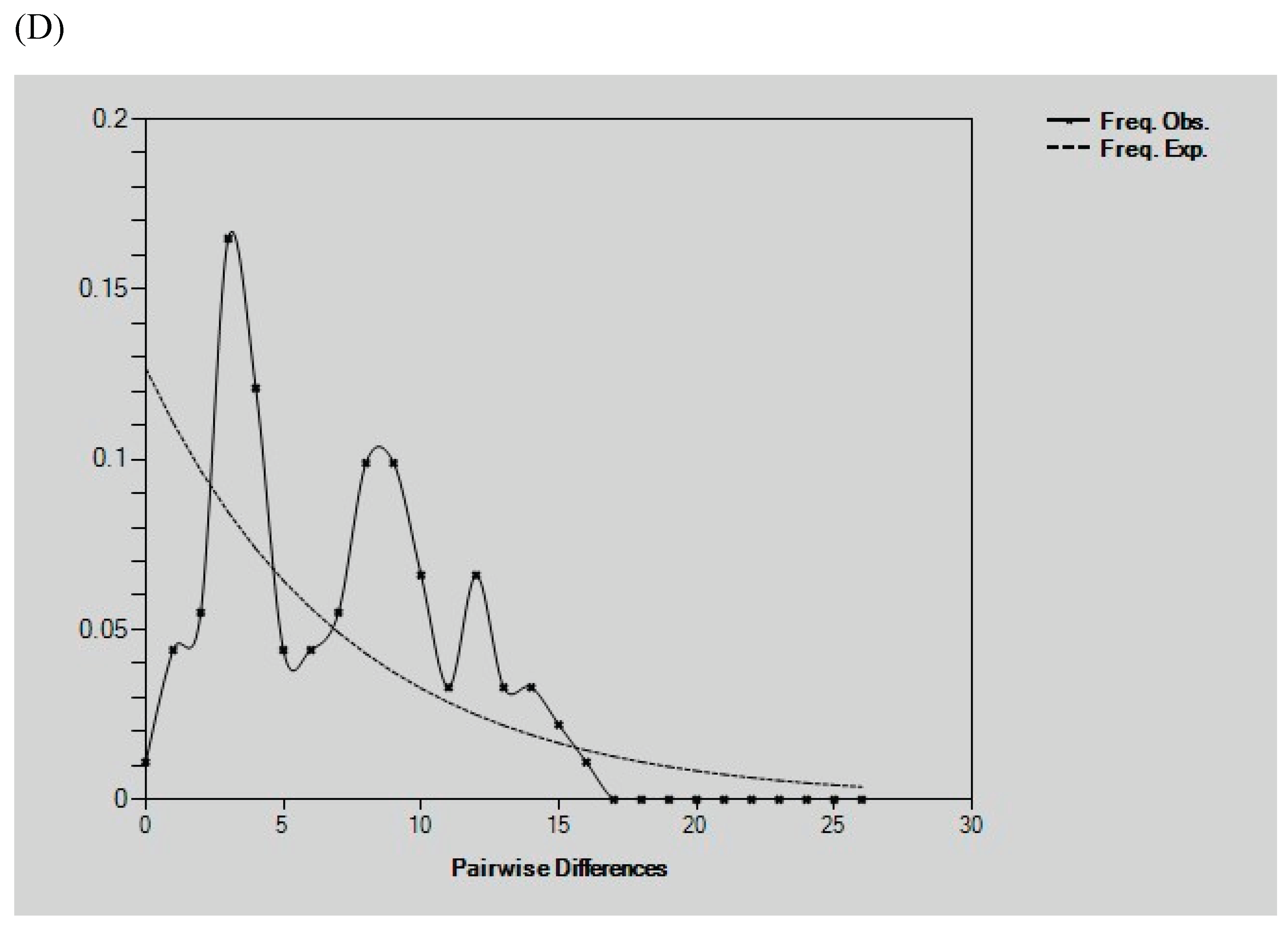
Demographic History Analysis
Discussion
Acknowledgments
References
- Rosell, F.; BozsÉR, O.; Collen, P.; Parker, H. Ecological impact of beavers Castor fiber and Castor canadensisand their ability to modify ecosystems. Mammal Review 2005, 35, 248-276. [CrossRef]
- Biedrzycka, A.; Konior, M.; Babik, W.; Świsłocka, M.; Ratkiewicz, M. Admixture of two phylogeographic lineages of the Eurasian beaver in Poland. Mammalian Biology 2014, 79, 287-296. [CrossRef]
- Halley, D.J.; Saveljev, A.P.; Rosell, F.J.M.r. Population and distribution of beavers Castor fiber and Castor canadensis in Eurasia. 2021, 51, 1-24.
- Falaschi, M.; Ficetola, G.F.; Viviano, A.; Mazza, G.; Mori, E. Environmental suitability and potential range expansion of the Eurasian beaver in Italy. Animal Conservation 2023, 27, 324-337. [CrossRef]
- Gaywood, M.J. Reintroducing the Eurasian beaver Castor fiber to Scotland. Mammal Review 2017, 48, 48-61. [CrossRef]
- Halley, D.J. Sourcing Eurasian beaver Castor fiber stock for reintroductions in Great Britain and Western Europe. Mammal Review 2010, 41, 40-53. [CrossRef]
- Marr, M.M.; Brace, S.; Schreve, D.C.; Barnes, I. Identifying source populations for the reintroduction of the Eurasian beaver, Castor fiber L. 1758, into Britain: evidence from ancient DNA. Scientific Reports 2018, 8. [CrossRef]
- Campbell-Palmer, R.; Rosell, F.; Naylor, A.; Cole, G.; Mota, S.; Brown, D.; Fraser, M.; Pizzi, R.; Elliott, M.; Wilson, K.; et al. Eurasian beaver (Castor fiber) health surveillance in Britain: Assessing a disjunctive reintroduced population. Veterinary Record 2021, 188. [CrossRef]
- Senn, H.; Ogden, R.; Frosch, C.; Syrůčková, A.; Campbell-Palmer, R.; Munclinger, P.; Durka, W.; Kraus, R.H.S.; Saveljev, A.P.; Nowak, C.; et al. Nuclear and mitochondrial genetic structure in the Eurasian beaver (Castor fiber) – implications for future reintroductions. Evolutionary Applications 2014, 7, 645-662. [CrossRef]
- Ducroz, J.-F.; Stubbe, M.; Saveljev, A.P.; Heidecke, D.; Samjaa, R.; Ulevičius, A.; Stubbe, A.; Durka, W. Genetic Variation and Population Structure of the Eurasian Beaver Castor fiber in Eastern Europe and Asia. Journal of Mammalogy 2005, 86, 1059-1067. [CrossRef]
- Halley, D.J.; Saveljev, A.P.; Rosell, F. Population and distribution of beaversCastor fiberand Castor canadensisin Eurasia. Mammal Review 2020, 51, 1-24. [CrossRef]
- Chu, H.; Jiang, Z. Distribution and conservation of the Sino-Mongolian beaver Castor fiber birulai in China. Oryx 2009, 43. [CrossRef]
- Saveljev, A.P.; Niamosor, B.; Shaariybuu, B.; Batseren, D. Self-eating in beavers — trophic opportunism or reaction on stress Extreme case from Mongolia. Russian Journal of Theriology 2016, 15, 68-74. [CrossRef]
- Halley, D.; Rosell, F.; Saveljev, A.J.B.F. Population and distribution of Eurasian beaver (Castor fiber). 2012, 18, 168-175.
- Horn, S.; Durka, W.; Wolf, R.; Ermala, A.; Stubbe, A.; Stubbe, M.; Hofreiter, M.J.P.o. Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species. 2011, 6, e14622.
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer Premier: Program for Design of Degenerate Primers from a Protein Sequence. BioTechniques 2018, 24, 318-319. [CrossRef]
- Li, L.; Wang, X.; Gai, J.; Yu, D.J.D.S. Isolation and characterization of a seed-specific isoform of microsomal omega-6 fatty acid desaturase gene (FAD2-1B) from soybean: Full Length Research Article. 2008, 19, 28-36.
- Burland, T.G.J.B.m.; protocols. DNASTAR’s Lasergene sequence analysis software. 1999, 71-91.
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 2021, 38, 3022-3027. [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Molecular Biology and Evolution 2017, 34, 3299-3302. [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S.J.E.b. Arlequin (version 3.0): an integrated software package for population genetics data analysis. 2005, 1, 117693430500100003.
- Tajima, F.J.G. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. 1989, 123, 585-595.
- Fu, Y.-X.J.G. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. 1997, 147, 915-925.
- Weir, B.S.; Cockerham, C.C. Estimatingf-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358-1370. [CrossRef]
- Exeoeffr, L.; Smouse, P.; Quattro, J.J.G. Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. 1992, 131, 479-491.
- Moritz, C.J.T.i.e.; evolution. Defining ‘evolutionarily significant units’ for conservation. 1994, 9, 373-375.
- Hoban, S.; Bruford, M.W.; da Silva, J.M.; Funk, W.C.; Frankham, R.; Gill, M.J.; Grueber, C.E.; Heuertz, M.; Hunter, M.E.; Kershaw, F.; et al. Genetic diversity goals and targets have improved, but remain insufficient for clear implementation of the post-2020 global biodiversity framework. Conservation Genetics 2023, 24, 181-191. [CrossRef]
- Zhang, Y.M.; Sheikh, S.I.; Ward, A.K.G.; Forbes, A.A.; Prior, K.M.; Stone, G.N.; Gates, M.W.; Egan, S.P.; Zhang, L.; Davis, C.; et al. Delimiting the cryptic diversity and host preferences of Sycophila parasitoid wasps associated with oak galls using phylogenomic data. Molecular Ecology 2022, 31, 4417-4433. [CrossRef]
- Frankham, R. Genetics and extinction. Biological Conservation 2005, 126, 131-140. [CrossRef]
- Loire, E.; Chiari, Y.; Bernard, A.; Cahais, V.; Romiguier, J.; Nabholz, B.; Lourenço, J.M.; Galtier, N.J.G.B. Population genomics of the endangered giant Galápagos tortoise. 2013, 14, 1-11.
- Guo, Y.; Chang, J.; Han, L.; Liu, T.; Li, G.; Garber, P.A.; Xiao, N.; Zhou, J. The Genetic Status of the Critically Endangered Hainan Gibbon (Nomascus hainanus): A Species Moving Toward Extinction. Frontiers in Genetics 2020, 11. [CrossRef]
- Kuang, W.; Zinner, D.; Li, Y.; Yao, X.; Roos, C.; Yu, L. Recent Advances in Genetics and Genomics of Snub-Nosed Monkeys (Rhinopithecus) and Their Implications for Phylogeny, Conservation, and Adaptation. Genes 2023, 14. [CrossRef]
- Fernández-Morán, J.; Saavedra, D.; Manteca-Vilanova, X.J.J.o.Z.; Medicine, W. Reintroduction of the Eurasian otter (Lutra lutra) in northeastern Spain: trapping, handling, and medical management. 2002, 33, 222-227.
- Canestrelli, D.; Frosch, C.; Kraus, R.H.S.; Angst, C.; Allgöwer, R.; Michaux, J.; Teubner, J.; Nowak, C. The Genetic Legacy of Multiple Beaver Reintroductions in Central Europe. PLoS ONE 2014, 9. [CrossRef]
- Tigano, A.; Friesen, V.L. Genomics of local adaptation with gene flow. Molecular Ecology 2016, 25, 2144-2164. [CrossRef]
- Gouskov, A.; Vorburger, C. Postglacial recolonizations, watershed crossings and human translocations shape the distribution of chub lineages around the Swiss Alps. BMC Evolutionary Biology 2016, 16. [CrossRef]
- Caramelli, D.; Elsner, J.; Hofreiter, M.; Schibler, J.; Schlumbaum, A. Ancient mtDNA diversity reveals specific population development of wild horses in Switzerland after the Last Glacial Maximum. Plos One 2017, 12. [CrossRef]
- Groombridge, J.J.; Raisin, C.; Bristol, R.; Richardson, D.S.J.R.b.I.s.; management. Genetic consequences of reintroductions and insights from population history. 2012, 395-440.
- Preatoni, D.; Mustoni, A.; Martinoli, A.; Carlini, E.; Chiarenzi, B.; Chiozzini, S.; Van Dongen, S.; Wauters, L.A.; Tosi, G. Conservation of brown bear in the Alps: space use and settlement behavior of reintroduced bears. Acta Oecologica 2005, 28, 189-197. [CrossRef]
- Swenson, J.E.; Taberlet, P.; Bellemain, E. Genetics and conservation of European brown bears Ursus arctos. Mammal Review 2011, 41, 87-98. [CrossRef]
- Yang, S.; Zhang, T.; Li, Y.; Xu, S.; Zhang, M.; Hu, X.; Liu, S.; Hu, D.; Wronski, T. Identifying personality traits and their potential application to the management of captive forest musk deer (Moschus berezovskii). Applied Animal Behaviour Science 2021, 234. [CrossRef]
- Tarav, M.; Tokunaga, M.; Kondo, T.; Kato-Mori, Y.; Hoshino, B.; Dorj, U.; Hagiwara, K. Problems in the Protection of Reintroduced Przewalski's Horses (Equus ferus przewalskii) Caused by Piroplasmosis. Journal of Wildlife Diseases 2017, 53, 911-915. [CrossRef]
- Crandall, K.A.; Bininda-Emonds, O.R.; Mace, G.M.; Wayne, R.K.J.T.i.e.; evolution. Considering evolutionary processes in conservation biology. 2000, 15, 290-295.
- Stock, S.E.; Klop-Toker, K.; Wallace, S.; Kelly, O.; Callen, A.; Seeto, R.; Mahony, S.V.; Hayward, M.W.; Mahony, M.J. Uncovering inbreeding, small populations, and strong genetic isolation in an Australian threatened frog, Litoria littlejohni. Conservation Genetics 2023, 24, 575-588. [CrossRef]
- Perrin, A.; Khimoun, A.; Faivre, B.; Ollivier, A.; de Pracontal, N.; Théron, F.; Loubon, M.; Leblond, G.; Duron, O.; Garnier, S. Habitat fragmentation differentially shapes neutral and immune gene variation in a tropical bird species. Heredity 2020, 126, 148-162. [CrossRef]
- Perrin, A.; Khimoun, A.; Faivre, B.; Ollivier, A.; de Pracontal, N.; Théron, F.; Loubon, M.; Leblond, G.; Duron, O.; Garnier, S.J.H. Habitat fragmentation differentially shapes neutral and immune gene variation in a tropical bird species. 2021, 126, 148-162.
- Lan, T.; Yang, S.; Li, H.; Zhang, Y.; Li, R.; Sahu, S.K.; Deng, W.; Liu, B.; Shi, M.; Wang, S.; et al. Large-scale genome sequencing of giant pandas improves the understanding of population structure and future conservation initiatives. Proceedings of the National Academy of Sciences 2024, 121. [CrossRef]
- Wang, P.; Burley, J.T.; Liu, Y.; Chang, J.; Chen, D.; Lu, Q.; Li, S.-H.; Zhou, X.; Edwards, S.; Zhang, Z.; et al. Genomic Consequences of Long-Term Population Decline in Brown Eared Pheasant. Molecular Biology and Evolution 2021, 38, 263-273. [CrossRef]
- Hodgson, J.A.; Moilanen, A.; Wintle, B.A.; Thomas, C.D. Habitat area, quality and connectivity: striking the balance for efficient conservation. Journal of Applied Ecology 2010, 48, 148-152. [CrossRef]
| Genes | Bp | Haplotypes | Sample |
| Cytb | 800 | \ | \ |
| D-loop | 592 | Hap_1 | L_1、L_2_1、L_2_2、L_3_1、L_3_2、L_3_3、L_3_4、L_3_5、L_4、L_5_2、M_2、M_3、M_4、M_5、M_6 |
| Hap_2 | L_5_1、L_5_3 | ||
| Hap_3 | L_6、M_1、M_2 | ||
| Cytb+D-loop | 1258 | Hap_1 | L_1、L_2_1、L_2_2、L_3_1、L_3_2、L_3_3、L_3_4、L_3_5、L_4、L_5_2、M_2、M_3、M_4、M_5、M_6 |
| Hap_2 | L_5_1、L_5_3 | ||
| Hap_3 | L_6、M_1、M_2 |
| Population | H | π | K | |||
| Cytb | D-LOOP | Cytb | D-loop | Cytb | D-loop | |
| China-C. f. birulai | 1 | 0.576 | \ | 0.00515 | \ | 2.41558 |
| Mongolia-C. f. birulai | 2 | 0.600 | 0.00083 | 0.00248 | 0.667 | 1.20 |
| Castor fiber | 7 | 0.967 | 0.00411 | 0.02123 | 3.289 | 8.15398 |
| Castor canadensis | 3 | 0.989 | 0.00440 | 0.01472 | 3.500 | 6.89011 |
| country | Subspecies | C.f. pohlei |
mogolin_ C.f. briulai |
C.f. tuvinivus | C.f. albicus | C.f. fiber | C.f. galliae |
China_ C.f. briulai |
C.f. belorussicus | Castor canadensis |
| Russia | C.f. pohlei | - | 0.86900 | 0.95059 | 0.94203 | 0.89286 | 0.89831 | 0.97056 | 0.90323 | 0.93333 |
| mogolin |
mogolin_ C.f. briulai |
0.02389 | - | 0.83434 | 0.95033 | 0.94059 | 0.93750 | 0.67055 | 0.93407 | 0.93627 |
| Russia | C.f. tuvinivus | 0.02357 | 0.02826 | - | 0.90330 | 0.84158 | 0.85965 | 0.95059 | 0.85321 | 0.93368 |
| Germany | C.f. albicus | 0.05007 | 0.04446 | 0.05262 | - | 0.95349 | 0.87500 | 0.98880 | 0.84615 | 0.93267 |
| Norway | C.f. fiber | 0.04022 | 0.04370 | 0.04367 | 0.03063 | - | NA | 0.99093 | NA | 0.92314 |
| French | C.f. galliae | 0.04248 | 0.04147 | 0.04958 | 0.01118 | 0.02770 | - | 0.99043 | NA | 0.92350 |
| China |
China_ C.f. briulai |
0.02494 | 0.00269 | 0.03002 | 0.04200 | 0.04125 | 0.03902 | - | 0.98987 | 0.96743 |
| Russia | C.f. belorussicus | 0.04474 | 0.03919 | 0.04727 | 0.00906 | 0.02990 | 0.01047 | 0.03676 | - | 0.91914 |
| Canada | Castor canadensis | 0.24048 | 0.21939 | 0.23211 | 0.22851 | 0.23189 | 0.23341 | 0.21630 | 0.21754 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





