Submitted:
22 May 2025
Posted:
26 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Yeast Strain and Culture Media
2.2. Determination of Rapamycin Concentration
2.3. Growth Assays in Microtiter Plates
2.4. Fermentation Tests
2.5. Quantification of Extracellular Metabolites
2.6. Analysis of Relative Gene Expression
3. Results
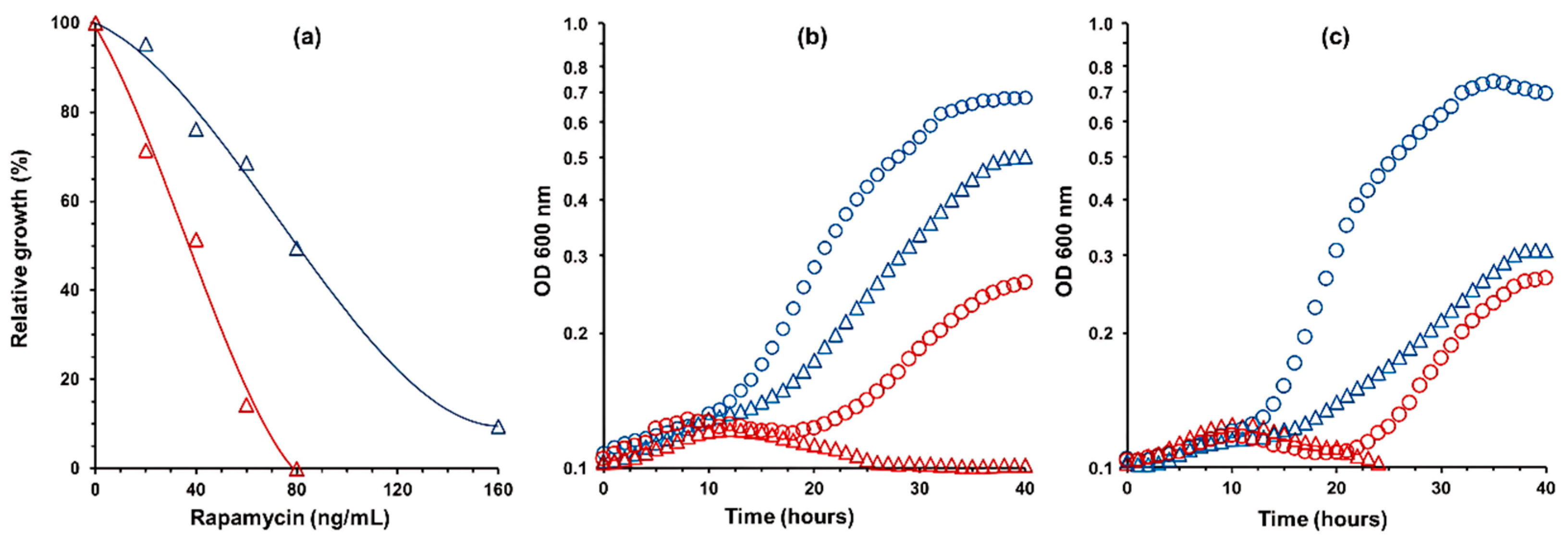
3.1. Rapamycin Acts as an Inhibitor of the TORC1 in D. bruxellensis
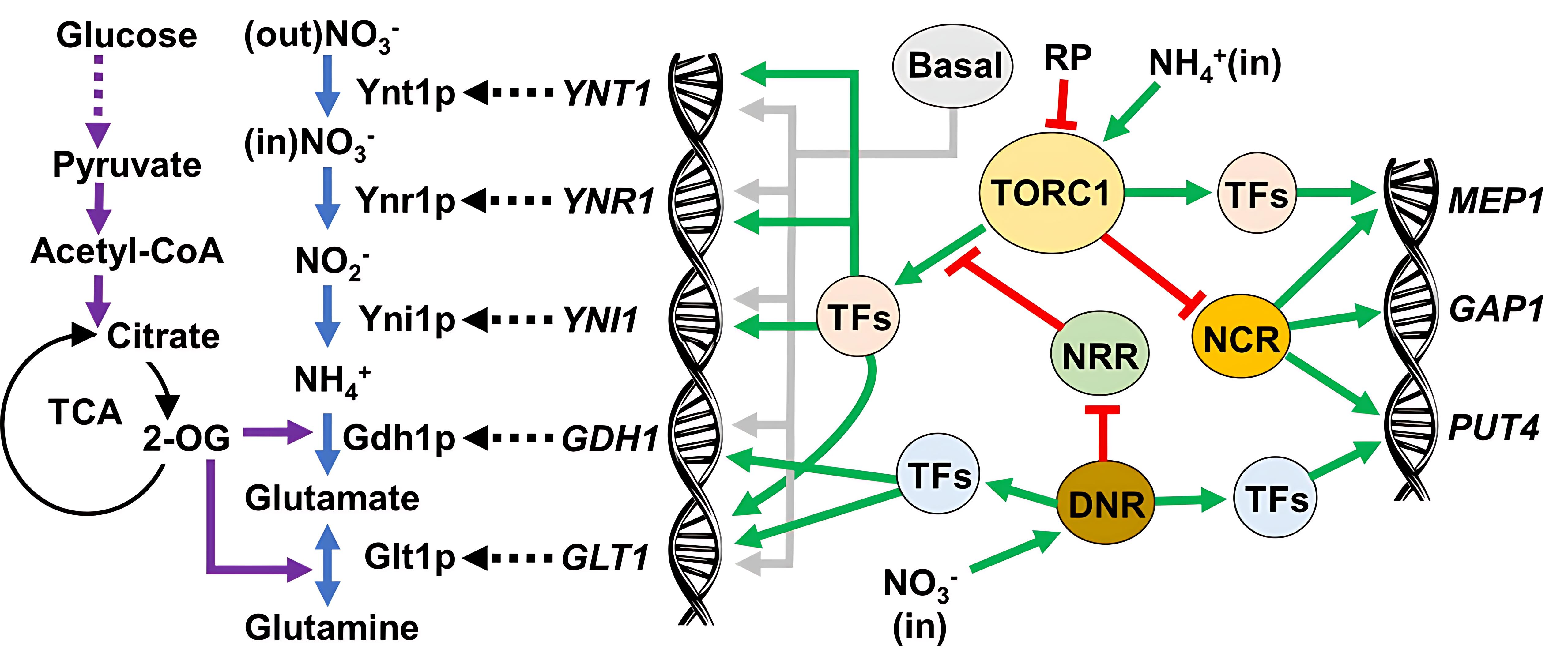
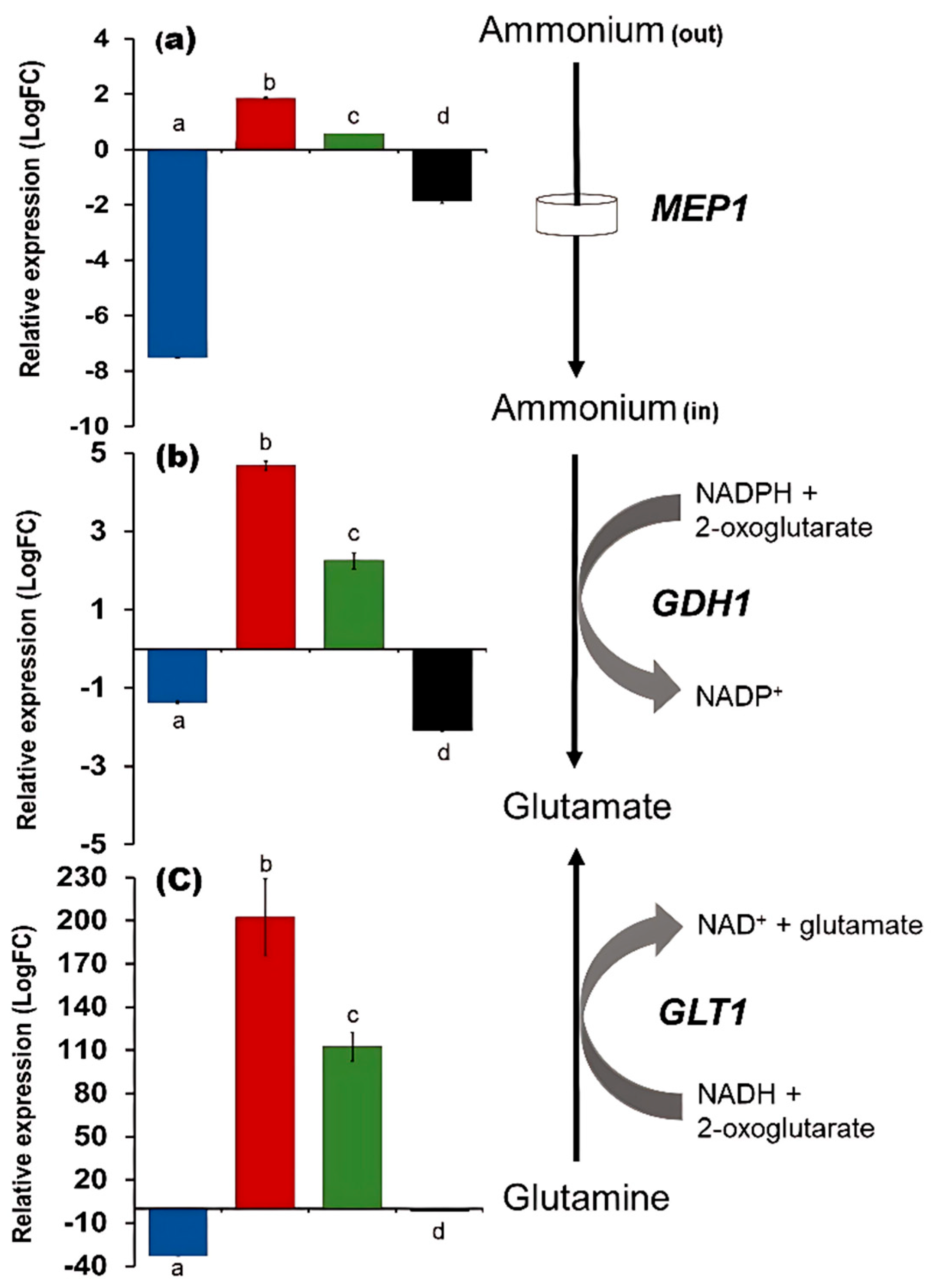
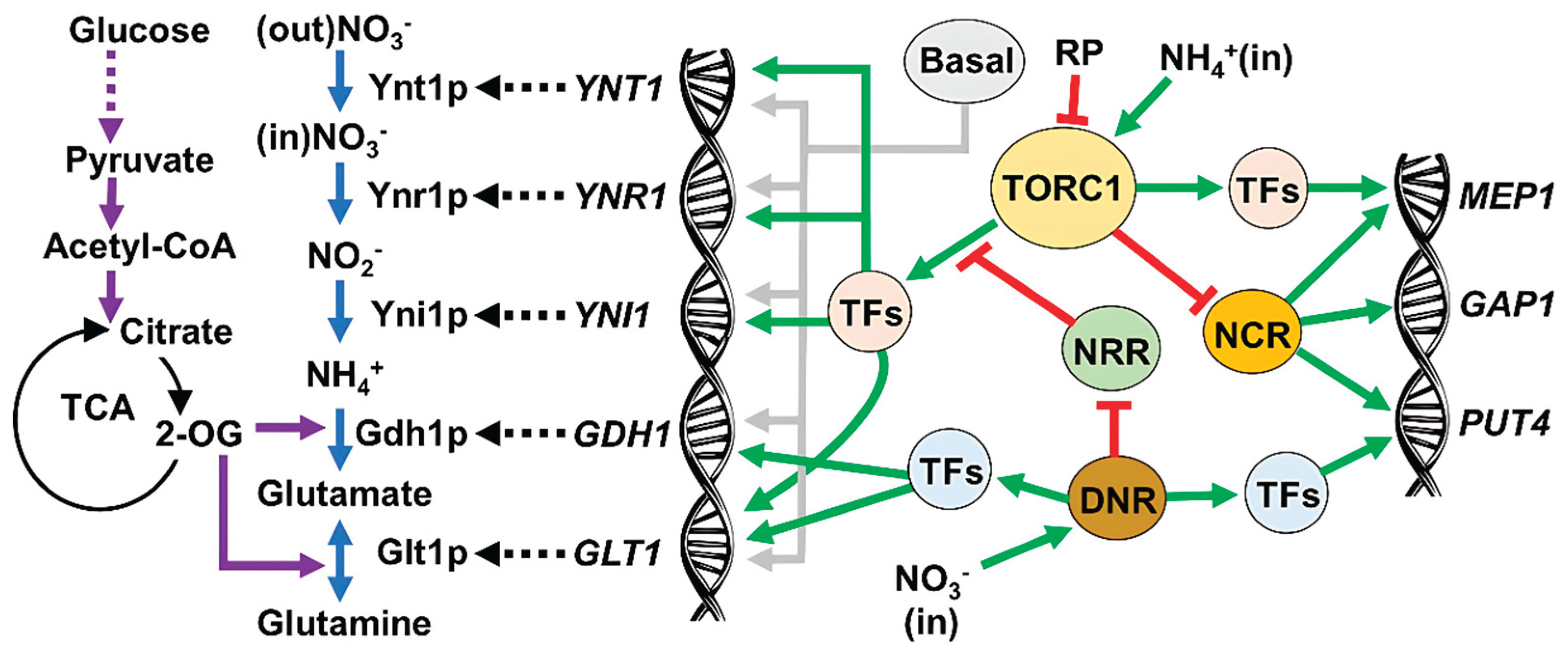
3.2. TOR Mechanism Controls the Nitrogen Metabolism at the Glutamine Level
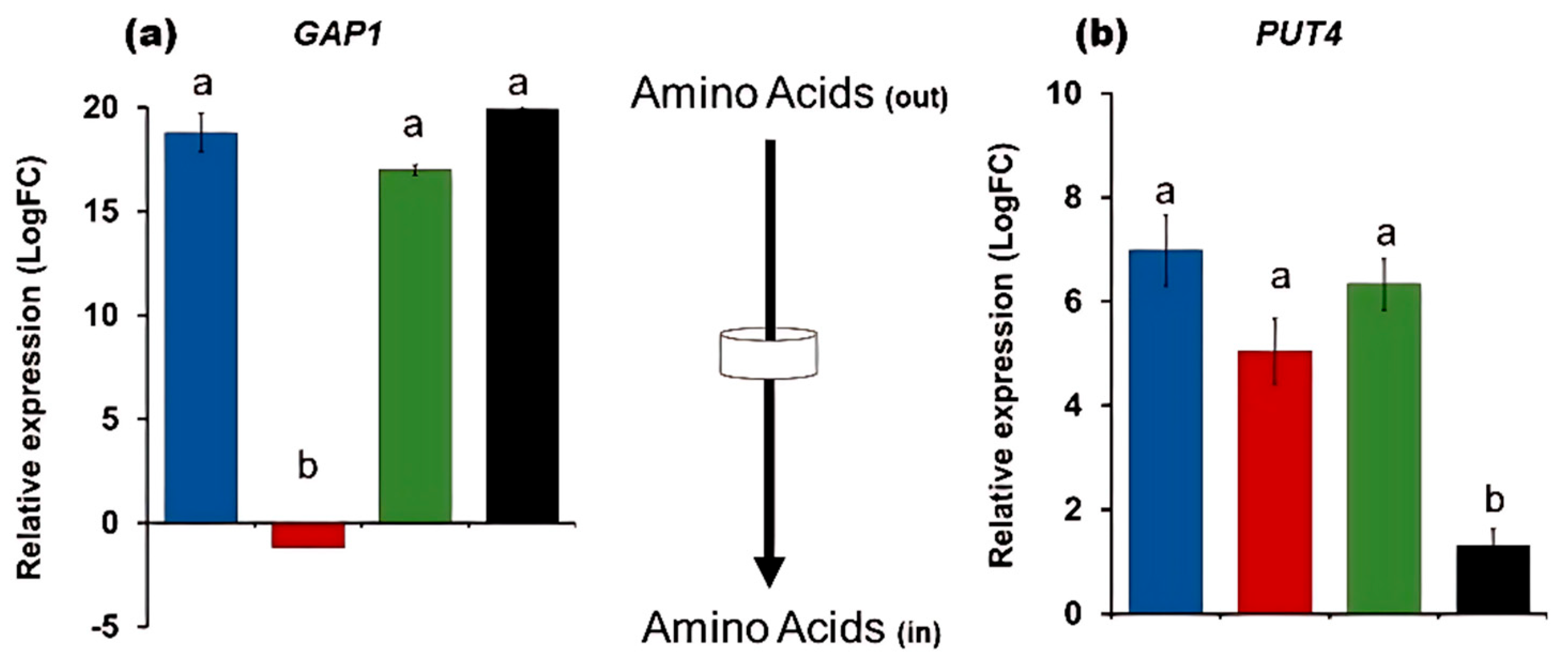
3.3. The Expression of NIT Genes Is Under the Control of the TORC1
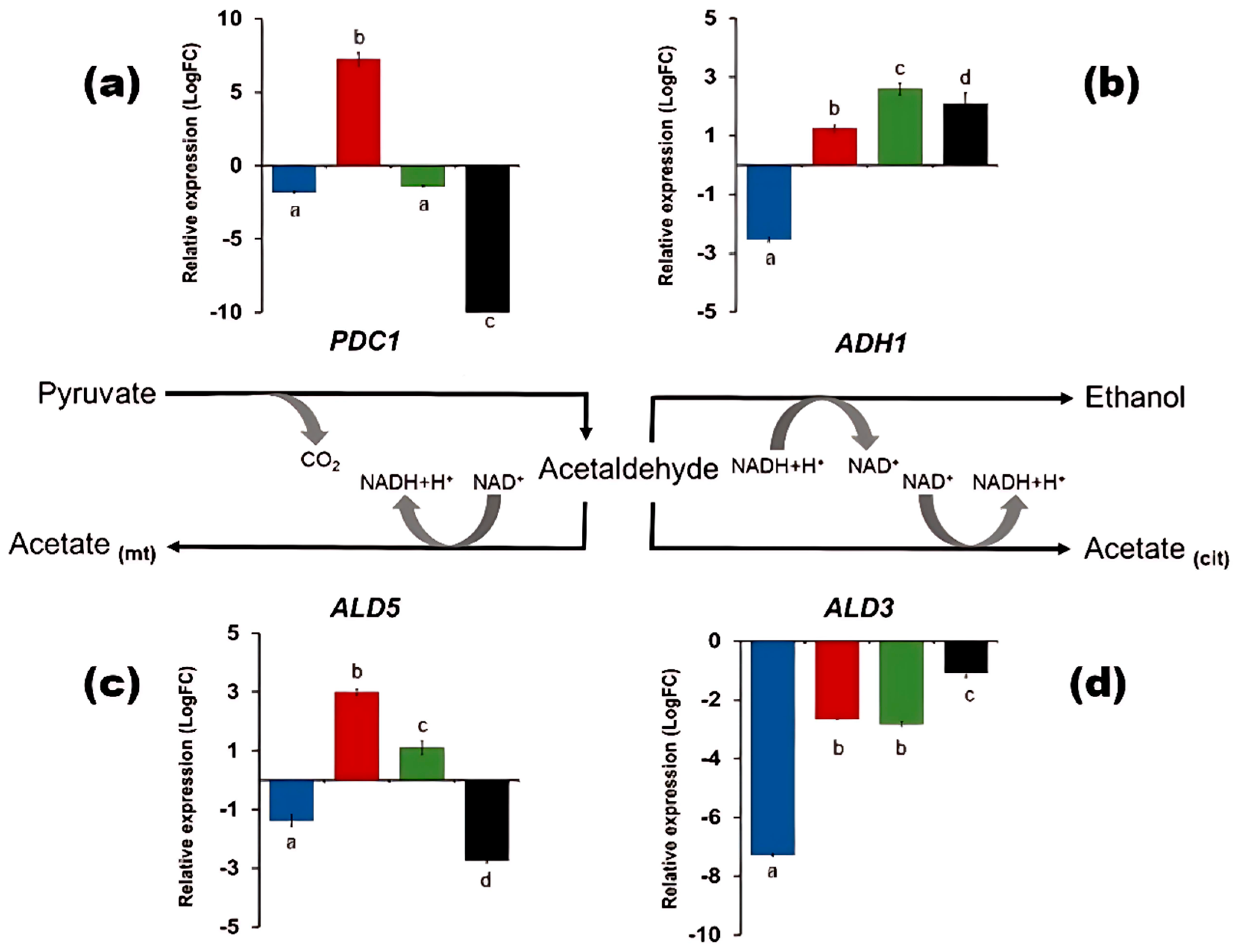
3.4. Partial Inactivation of TORC1 Hampered the Fermentation Metabolism in Nitrate
4. Discussion
4.1. TORC1 and the Control of the Nitrogen Central Metabolism
4.2. NCR Is Not a Player in the Nitrate Metabolism
4.3. TORC1 and Its Influence on the Physiology of D. bruxellensis in Nitrate
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Souza Liberal, A.T.; Basílio, A.C.; Do Monte Resende, A.; et al. Identification of Dekkera bruxellensis as a major contaminant yeast in continuous fuel ethanol fermentation. J. Appl. Microbiol. 2007, 102, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Basilio, A.C.M.; Araujo; P. R.L.; Morais, J.O.F.; et al. Detection and identification of wild yeast contaminants of the industrial fuel ethanol fermentation process. Curr. Microbiol. 2008, 56, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Renouf, V.; Falcou, M.; Miot-Setier, C.; et al. Interactions between Brettanomyces bruxellensis and other yeast species during the initial stages of winemaking. J. Appl. Microbiol. 2006, 100, 1208–1219. [Google Scholar] [CrossRef]
- De Barros Pita, W.; Teles, G.H.; Peña-Moreno, I.C.; et al. The biotechnological potential of the yeast Dekkera bruxellensis. World J. Microbiol. Biotechnol. 2019, 35, 103. [Google Scholar] [CrossRef]
- Harrouard, J.; Eberlein, C.; Ballestra, P.; et al. Brettanomyces bruxellensis: Overview of the genetic and phenotypic diversity of an anthropized yeast . Mol. Ecol. 2023, 32, 2374–2395. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, J.; Eberhard, T.; Schnürer, J.; Passoth, V. Fermentation characteristics of Dekkera bruxellensis strains. Appl. Microbiol. Biotechnol. 2010, 87, 1487–1497. [Google Scholar] [CrossRef]
- De Barros Pita, W.; Leite, F.C.B; . De Souza Liberal, A.T.; Simões, D.A.; De Morais Jr, M.A. The ability to use nitrate confers advantage to Dekkera bruxellensis over S. cerevisiae and can explain its adaptation to industrial fermentation processes. Antonie van Leeuwenhoek 2011, 1, 1–9. [Google Scholar] [CrossRef]
- Leite, F.C.B.; Basso, T.O.; De Pita Barros, W.; et al. Quantitative aerobic physiology of the yeast Dekkera bruxellensis, a major contaminant in bioethanol production plants. FEMS Yeast Res. 2013, 1, 34–43. [Google Scholar] [CrossRef]
- Parente, D.C.; Vidal, E.E.; Leite, F.C.; et al. Production of sensory compounds by means of the yeast Dekkera bruxellensis in different nitrogen sources with the prospect of producing cachaça. Yeast 2015, 32, 77–87. [Google Scholar] [CrossRef]
- Reis, A.L.S.; Damilano, L.D.; Menezes, R.S.C.; De Morais Jr, M.A. Second-generation ethanol from sugarcane and sweet sorghum bagasses using the yeast Dekkera bruxellensis. Ind. Crops Prod. 2016, 92, 255–262. [Google Scholar] [CrossRef]
- De Barros Pita, W.; Teles, G.H.; Peña-Moreno, I.C.; et al. The biotechnological potential of the yeast Dekkera bruxellensis. World J. Microbiol. Biotechnol. 2019, 35, 103. [Google Scholar] [CrossRef] [PubMed]
- Peña-Moreno, I.C.; Parente, D.C.; Da Silva; K. M.; et al. Comparative proteomic analyses reveal the metabolic aspects and biotechnological potential of nitrate assimilation in the yeast Dekkera bruxellensis. Appl. Microbiol. Biotechnol. 2021, 105, 1585–1600. [Google Scholar] [CrossRef] [PubMed]
- Piškur, J.; Rozpedowska, E.; Polakova, S.; et al. How did Saccharomyces evolve to become a good brewer? Trends in Genet. 2006, 22, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.M.; Teles, G.H.; Parente, D.C.; et al. Biological diversity of carbon assimilation among isolates of the yeast Brettanomyces bruxellensis from wine and fuel-ethanol industrial processes. FEMS Yeast Res. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Canonico, L.; Agarbati, A.; Comitini, F.; Ciani, M. Relationships among origin, genotype, and oenological traits of Brettanomyces yeasts. Int. J. Mol. Sci. 2024, 25, 11781. [Google Scholar] [CrossRef]
- Pereira, L.F.; Bassi, A.P.G.; Avansini, S.H.; et al. The physiological characteristics of the yeast Dekkera bruxellensis in fully fermentative conditions with cell recycling and in mixed cultures with Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 2012, 101, 529–539. [Google Scholar] [CrossRef]
- Bassi, A.P.; Paraluppi, A.L.; Reis, V.R.; Ceccato-Antonini, S.R. Potassium metabisulphite as a potential biocide against Dekkera bruxellensis in fuel ethanol fermentations. Lett. Appl. Microbiol. 2015, 60, 248–258. [Google Scholar] [CrossRef]
- Blomqvist, J.; Nogué, V.S.; Gorwa-Grauslund, M.; Passoth, V. Physiological requirements for growth and competitiveness of Dekkera bruxellensis under oxygen-limited or anaerobic conditions. Yeast 2012, 29, 265–274. [Google Scholar] [CrossRef]
- Galafassi, S.; Capusoni, C.; Moktaduzzaman, M.; Compagno, C. Utilization of nitrate abolishes the “Custers effect” in Dekkera bruxellensis and determines a different pattern of fermentation products. J. Ind. Microbiol. Biotechnol. 2013, 40, 297–303. [Google Scholar] [CrossRef]
- Parente, D.C.; Cajueiro, D.B.B.; Peña-Moreno, I.C.; et al. On the catabolism of amino acids in the yeast Dekkera bruxellensis and the implications for industrial fermentation processes. Yeast 2017, 35, 299–309. [Google Scholar] [CrossRef]
- Cajueiro, D.B.B.; Parente, D.C.; Leite, F.C.B.; et al. Glutamine: a major player in nitrogen catabolite repression in the yeast Dekkera bruxellensis. Antonie Van Leeuwenhoek 2017, 110, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Peña-Moreno, I.C.; Parente, D.C.; Da Silva, J.M.; et al. Nitrate boosts anaerobic ethanol production in an acetate-dependent manner in the yeast Dekkera bruxellensis. J. Ind. Microbiol. Biotechnol. 2019, 46, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.R.; Teles, G.H.; De Carvalho, J.M.; et al. The antioxidant defense of Dekkera bruxellensis against hydrogen peroxide and its relationship to nitrate metabolism. J. Appl. Microbiol. 2023, 134, lxad065. [Google Scholar] [CrossRef]
- De Barros Pita, W.; Castro-Silva, D.; Simões, D.A.; et al. Physiology and gene expression profiles of Dekkera bruxellensis in response to carbon and nitrogen availability. Antonie Van Leeuwenhoek 2013, 105, 855–868. [Google Scholar] [CrossRef]
- De Barros Pita, W.; Tiukova, I.; Leite, F.C.B.; et al. The influence of nitrate on the physiology of the yeast Dekkera bruxellensis grown under oxygen limitation. Yeast 2013, 30, 111–117. [Google Scholar] [CrossRef]
- Magasanik, B.; Kaiser, C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef]
- Siverio, J.M. Assimilation of nitrate by yeasts . FEMS Microbiol. Rev. 2002, 26, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Georis, I.; Tate, J.J.; Cooper, T.G.; Dubois, E. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J. Biol. Chem. 2011, 286, 44897–44912. [Google Scholar] [CrossRef]
- Crespo, J.L.; Hall, M.N. Elucidating TOR signaling and rapamycin action: Lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002, 66, 579–591. [Google Scholar] [CrossRef]
- Teles, G.H; Da Silva, J.M.; Mendonça, A.A.; De Morais Jr, M.A.; De Barros Pita. First aspects on acetate metabolism in the yeast Dekkera bruxellensis: a few keys for improving ethanol fermentation. Yeast 2018, 35, 577–584. [Google Scholar] [CrossRef]
- Teles, G.H. , Xavier, M.R.; Da Silva, J.M.; et al. The metabolism of respiring carbon sources by Dekkera bruxellensis and its relation with the production of acetate. Appl. Biochem. Biotechnol. 2023, 195, 6369–6391. [Google Scholar] [CrossRef] [PubMed]
- De Barros Pita, W.; Leite, F.C.B.; De Souza Liberal, A.T.; et al. A new set of reference genes for RT-qPCR assays in the yeast Dekkera bruxellensis. Can. J. Microbiol. 2012, 58, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Teles, G.H.; Da Silva, J.M.; Xavier, M.R.; et al. Metabolic and biotechnological insights on the analysis of the Pdh bypass and acetate production in the yeast Dekkera bruxellensis. J. Biotechnol. 2022, 355, 42–52. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Francis, B.R.; White, K.H.; Thorsness, P.E. Mutations in the ATP1p and Atp3p subunits of yeast ATP synthase differentially affect respiration and fermentation in Saccharomyces cerevisiae. J. Bioenerg. Biomembr. 2007, 39, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Betram, P.G.; Choi, J.H.; Carvalho, J.; et al. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 2000, 275, 35727–35733. [Google Scholar] [CrossRef]
- Ter Schure, E.G.; Van Riel, N.A.W.; Verrips, C.T. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2000, 24, 67–83. [Google Scholar] [CrossRef]
- Valenzuela, L.; Ballario, P.; Aranda, C.; Filetici, P.; González, A. Regulation of expression of GLT1, the gene encoding glutamate synthase in Saccharomyces cerevisiae. J. Bacteriol. 1998, 180, 3533–3540. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Siewers, V.; Nielsen, J. Ach1 is involved in shuttling mitochondrial acetyl units for cytosolic C2 provision in Saccharomyces cerevisiae lacking pyruvate decarboxylase. FEMS Yeast Res. 2015, 15, fov015. [Google Scholar] [CrossRef]

| YNB-NH4 | YNB-NO3 | ||||
| REF | iTOR | REF | iTOR | ||
| Mass balance* | |||||
| Sugar consumption (g L−1) | 134.5 | 134.9 | 138.9 | 16.1 | |
| Biomass yield (g g−1) | 0.07 | 0.05 | 0.02 | 0.06 | |
| Ethanol yield (g g−1) | 0.43 | 0.26 | 0.31 | 0.00 | |
| Acetate yield (g g−1) | 0.00 | 0.00 | 0.05 | 0.07 | |
| CO2** yield (g g−1) | 0.43 | 0.27 | 0.35 | 0.07 | |
| Mass recovery (%) | 93.3 | 58.3 | 73.3 | 20.7 | |
| Carbon balance* | |||||
| Carbon from substrate (mmol) | 4,483 | 4,497 | 4,631 | 537 | |
| Carbon to biomass (mmol) | 377 | 270 | 111 | 38 | |
| Carbon to ethanol (mmol) | 2,519 | 1,551 | 1,892 | 0.0 | |
| Carbon to acetate (mmol) | 0.0 | 0.0 | 253 | 39 | |
| Carbon to CO2** (mmol) | 1,329 | 826 | 1,094 | 23 | |
| Carbon recovery (%) | 94.2 | 58.8 | 72.4 | 18.6 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





