Submitted:
30 January 2025
Posted:
01 February 2025
You are already at the latest version
Abstract
Bursaphelenchus xylophilus, the causative agent of pine wilt disease (PWD), poses a severe global threat to coniferous forests. Esteya vermicola, an endoparasitic nematophagous fungus, exhibits promising biocontrol potential against this pinewood nematode. The vesicular transport system, evolutionarily conserved in eukaryotes, is essential for fungal pathogenicity. Based on our genome sequence of E. vermicola CBS115803, we identified EvSec22, a gene encoding a SNARE protein implicated in vesicular transport process. This study investigates the role of EvSec22 in E. vermicola during nematode infection, utilizing our optimized gene knockout methodology. Infection assays revealed that EvSec22 deletion significantly impaired the pathogenicity of E. vermicola against B. xylophilus. Phenotypic analyses revealed that the ΔEvSec22 mutant exhibited suppressed hyphal growth, reduced conidiation, and abnormal septal spacing. Furthermore, the mutant showed significantly diminished tolerance to osmotic stress (sorbitol) and oxidative stress (hydrogen peroxide). Overall, the EvSec22 gene is associated with the virulence of E. vermicola CBS115803 against B. xylophilus, and its deletion also impacts the normal growth of E. vermicola and its tolerance to abiotic stress. This study providing new insights into SNARE protein functions in fungal biocontrol agents.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Vector Construction and Fungal Transformation
2.2. Infectivity Assay of B. xylophilus
2.3. Conidia Count
2.4. Growth Assay on Solid Medium
2.5. Hyphal Septal Distance Measurement
2.6. Abiotic Stress Assay
2.7. Statistical Analysis
3. Result
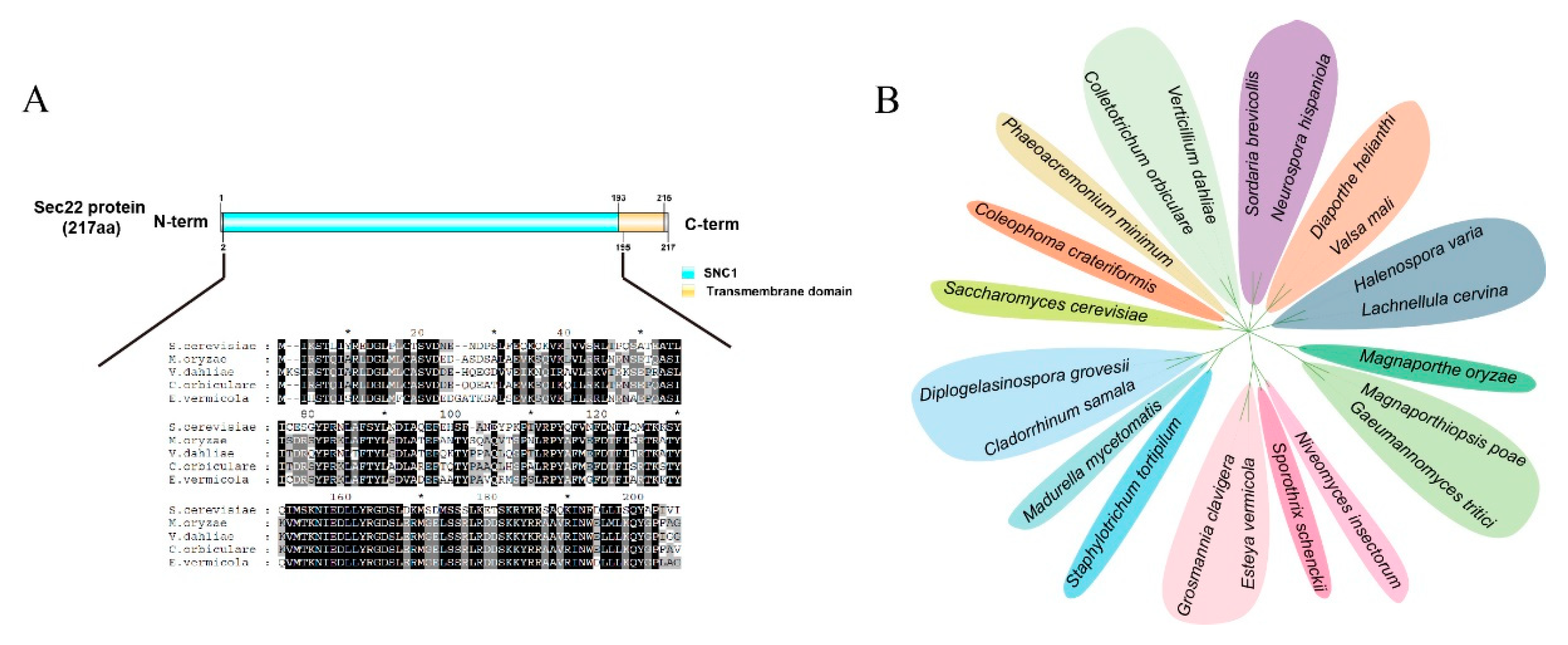
3.1. Sequence Analysis of EvSec22 in E. vermicola CBS115803
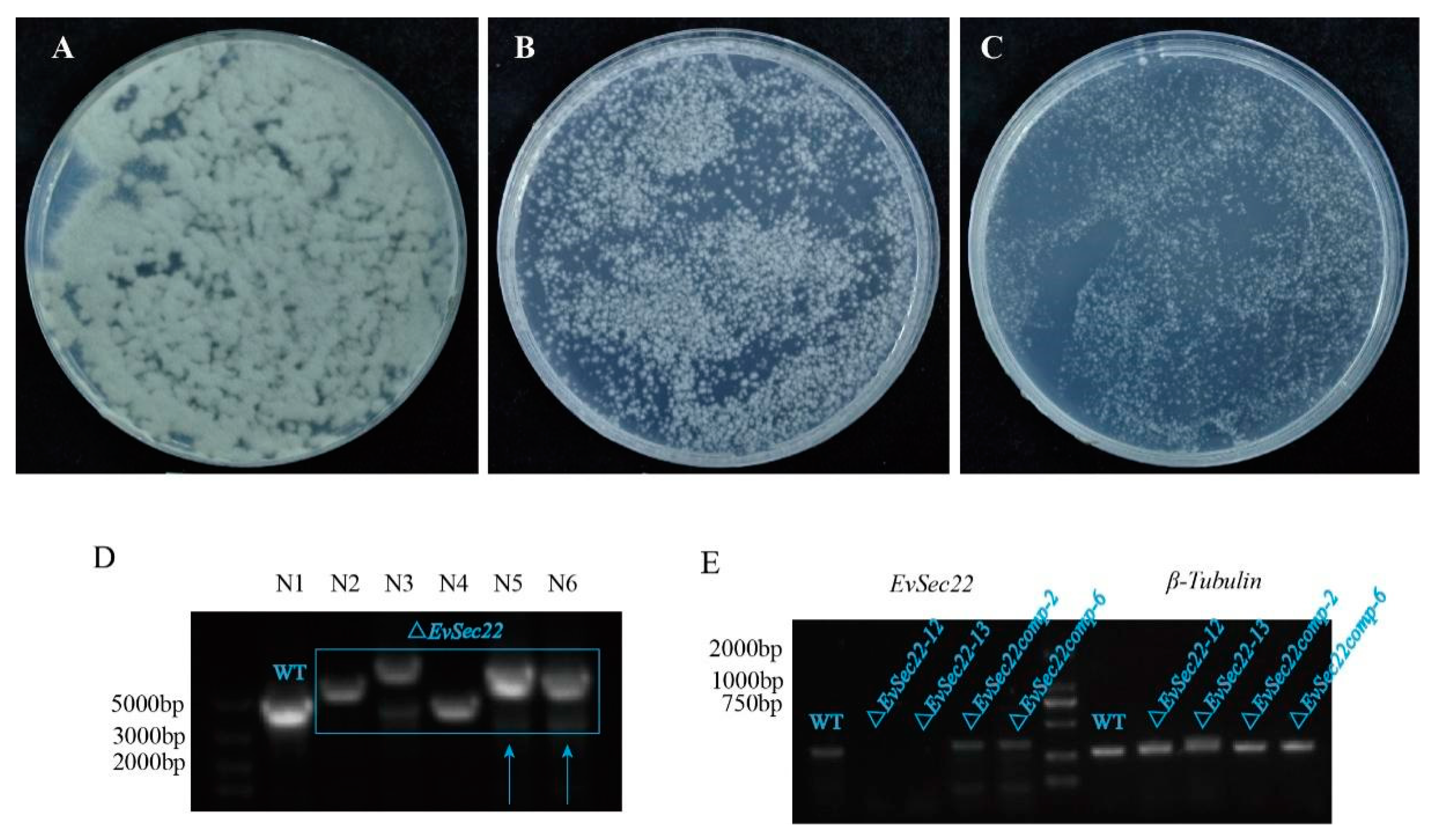
3.2. Optimization of E. vermicola Transformation Methods and Construction of EvSec22 Mutants and Complementary Strains
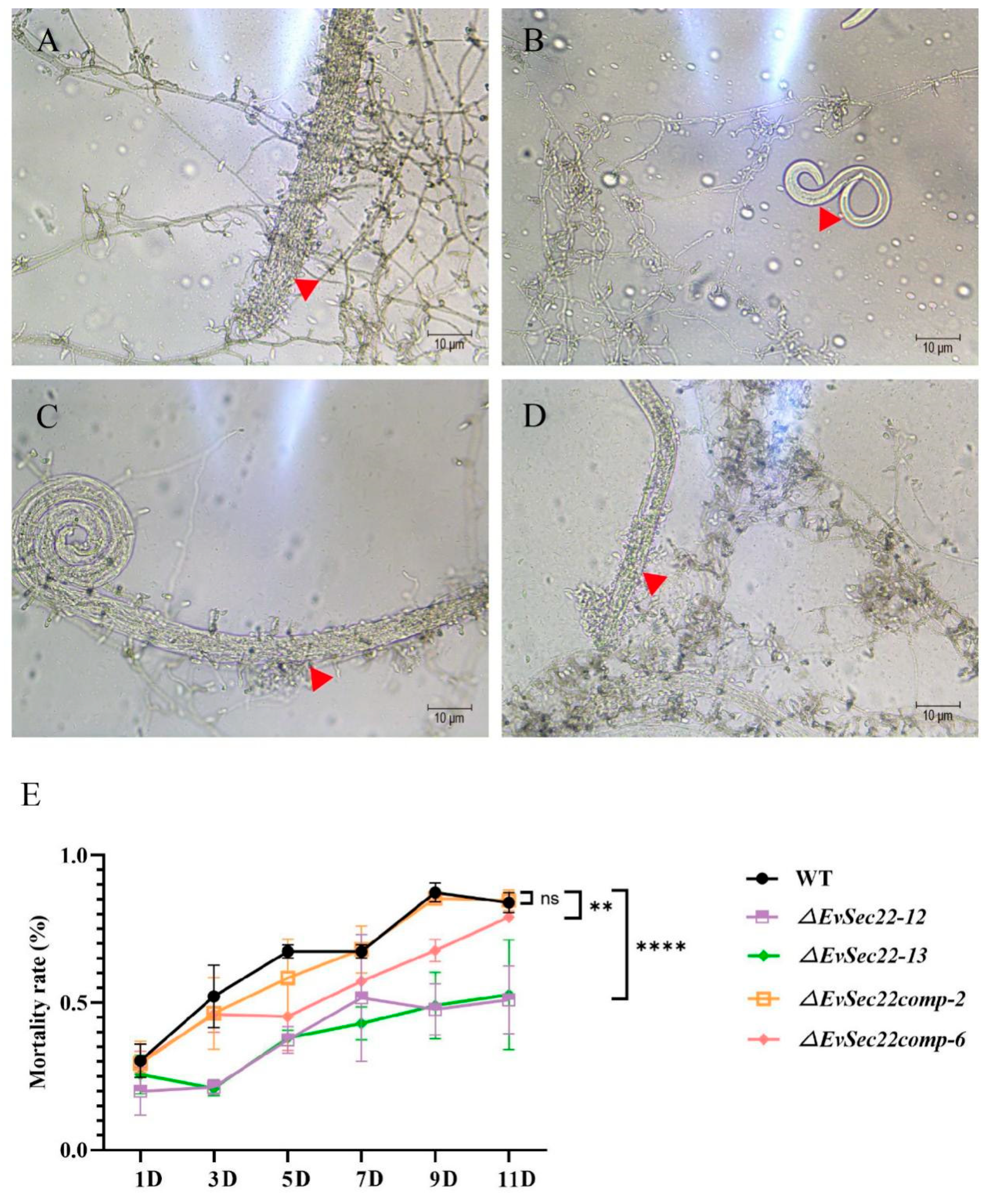
3.3. EvSec22 Mutants Impaired the Infectivity of E. vermicola Against B. xylophilus
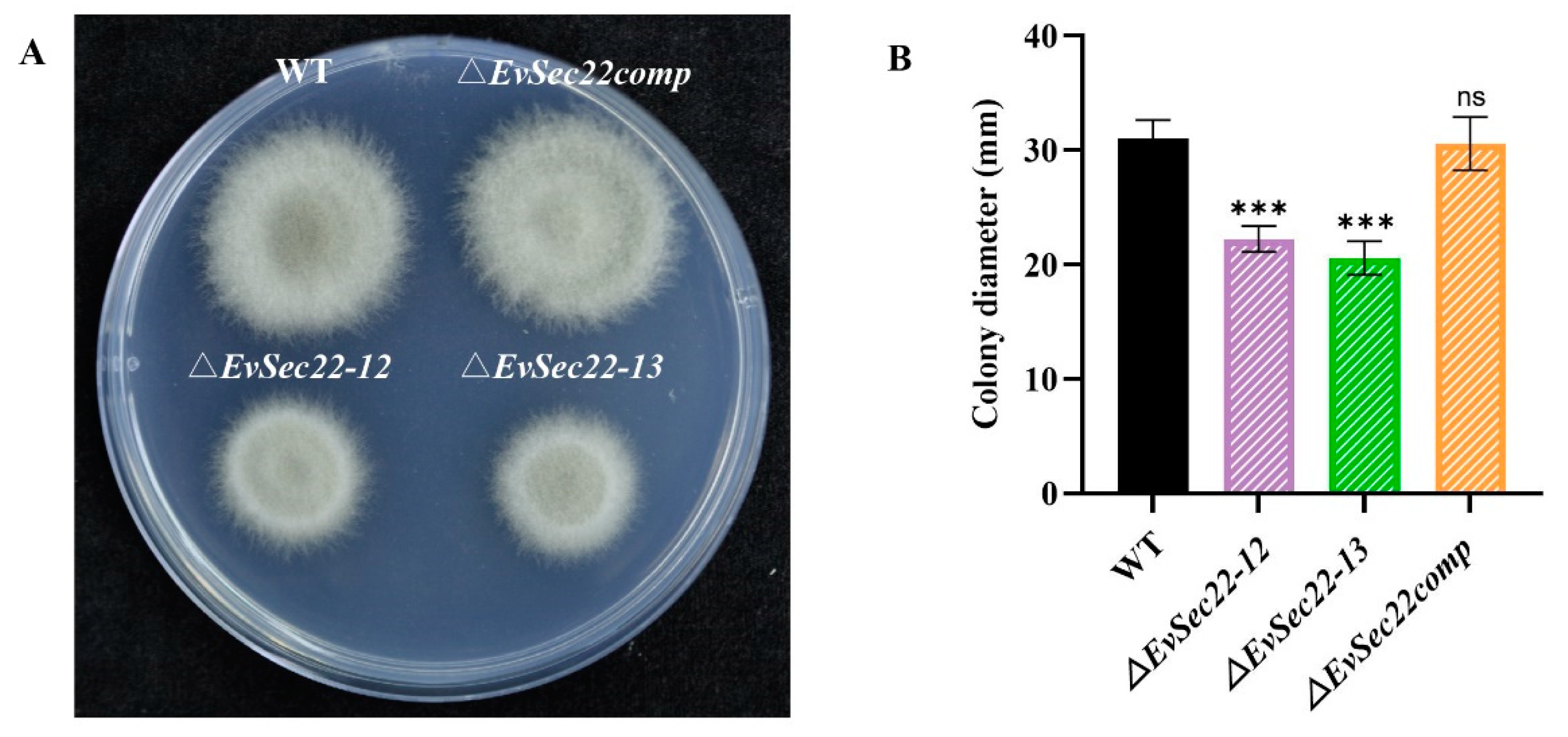
3.4. Loss of EvSec22 Leads to Slower Hyphal Growth and Hyphal Septal Spacing in E. vermicola
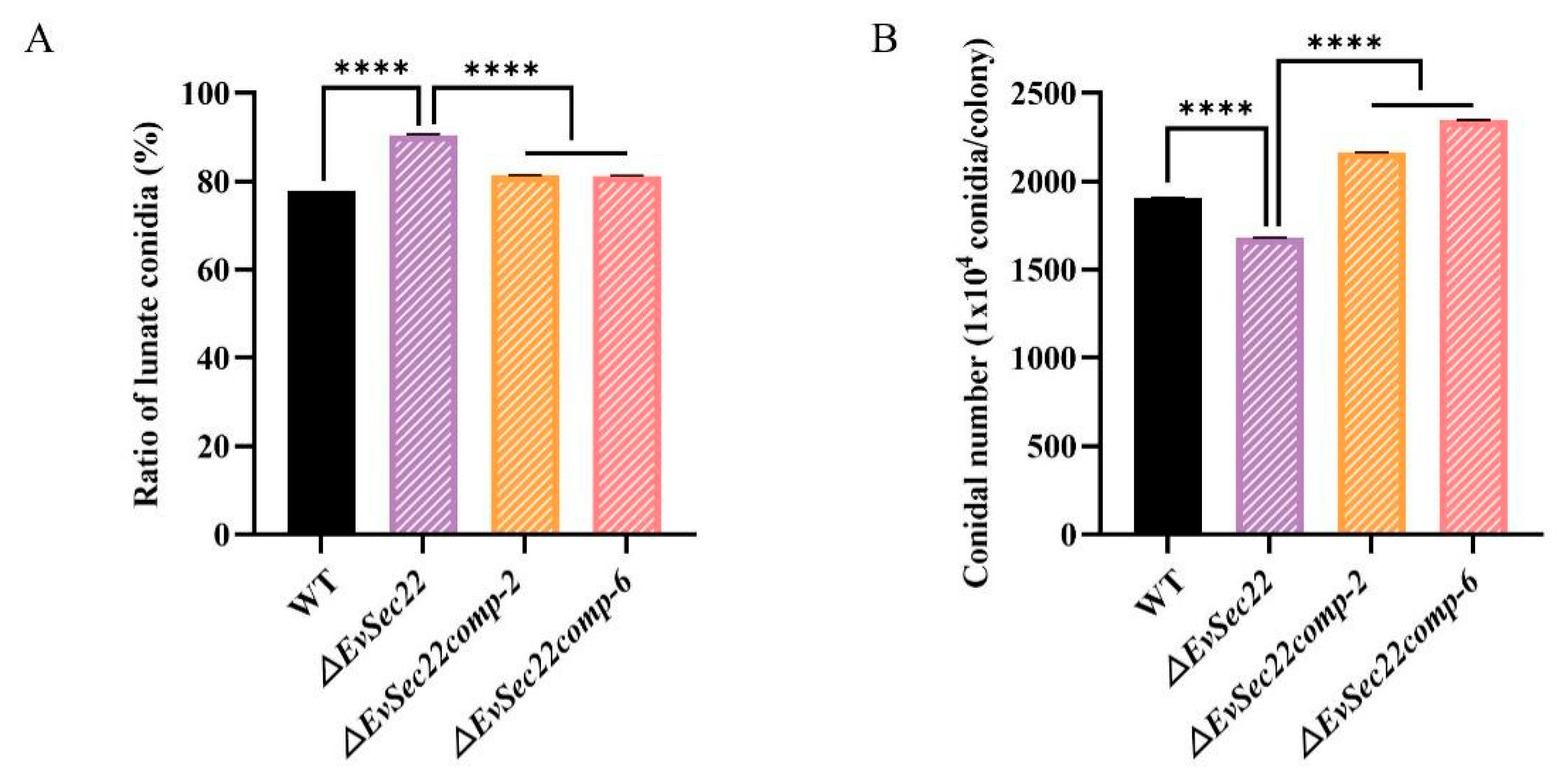
3.5. EvSec22 Deletion Reduces Total Conidia Count but Increases the Proportion of Lunate-Shaped Conidia
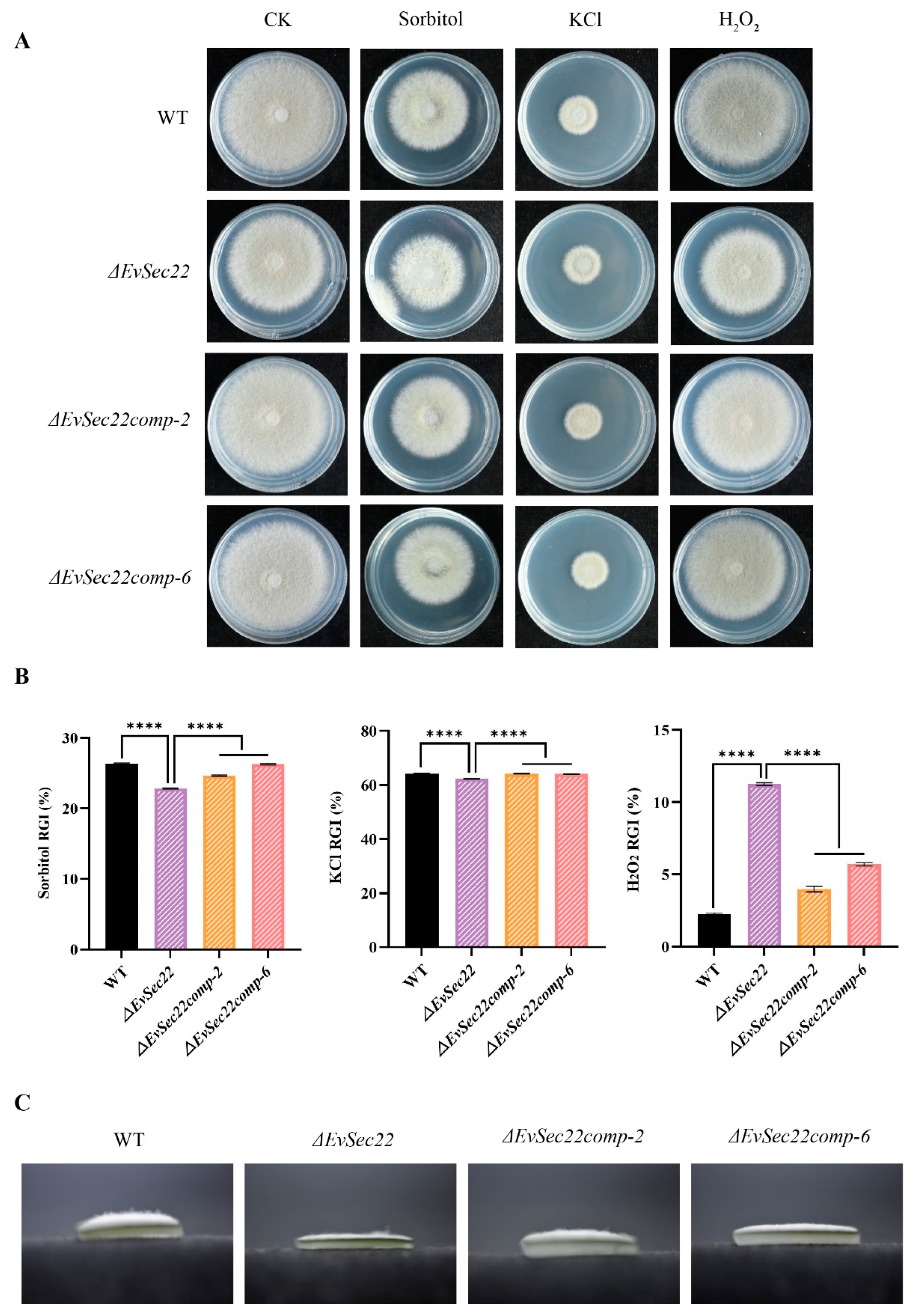
3.6. Deletion of EvSec22 Affects E. vermicola's Tolerance to Abiotic Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Nickle, W.R.; Golden, A.M.; Mamiya, Y.; Wergin, W.P. On the Taxonomy and Morphology of the Pine Wood Nematode, Bursaphelenchus xylophilus (Steiner &Buhrer 1934) Nickle 1970. J Nematol. 1981, 13, 385-92.
- Mamiya, Y. Pine wood nematode, Bursaphelenchus lignicolus Mamiya and Kiyohara, as a causal agent of pine wilting disease. Environmental Science, Biology, Agricultural and Food Sciences. 1972.
- Jones J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.; Perry, R.N. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 2013, 14, 946-61. [CrossRef]
- Hunt, D., Pine wilt disease: a worldwide threat to forest ecosystems. Nematology, 2009. [CrossRef]
- Robertson, L.; Arcos, S.C.; Escuer, M.; Merino, R.S.; Espárrago, G.; Abelleira, A.; Navas, A. Incidence of the pinewood nematode Bursaphelenchus xylophlius Steiner & Buhrer, 1934 (Nickle, 1970) in Spain. Nematology. 2011, 13, 755-757.
- Zhao, L.L.; Mota, M.; Vieira, P.; Butcher, R.A.; Sun, J.H. Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends Parasitol. 2014, 30, 299-308. [CrossRef]
- Chen, J.X.; Li, Q.X.; Song, B.A. Chemical Nematicides: Recent Research Progress and Outlook. J Agric Food Chem. 2020, 68, 12175-12188. [CrossRef]
- Liang, W.L.; Yu, A.X.; Wang, G.D.; Zheng, F.; Jia, J.L.; Xu, H.H. Chitosan-based nanoparticles of avermectin to control pine wood nematodes. Int J Biol Macromol. 2018, 112, 258-263. [CrossRef]
- Affokpon, A.; Coyne, D.L.; Htay, C.C.; Agbèdè, R.D.; Lawouin, L.; Coosemans, J. Biocontrol potential of native Trichoderma isolates against root-knot nematodes in West African vegetable production systems. Soil Biology & Biochemistry, 2011, 43, 600-608. [CrossRef]
- Kerry, B.R. Rhizosphere Interactions and the Exploitation of Microbial Agents for the Biological Control of Plant-Parasitic Nematodes. Annu Rev Phytopathol. 2000, 38:423-441. [CrossRef]
- Liou, J.Y.; Shih, J.Y.; Tzean, S.S. Esteya, a new nematophagous genus from Taiwan, attacking the pinewood nematode (Bursaphelenchus xylophilus). Mycological Research, 1999, 103, 242-248. [CrossRef]
- Wang, C.Y.; Fang, Z.M.; Sun, B.S.; Gu, L.J.; Zhang, K.Q.; Sung, C.K. High infectivity of an endoparasitic fungus strain, Esteya vermicola, against nematodes. J Microbiol. 2008, 46, 380-9. [CrossRef]
- Lin, F.; Ye, J.L.; Wang, H.G.; Zhang, A.J.; Zhao, B.G. Host deception: predaceous fungus, Esteya vermicola, entices pine wood nematode by mimicking the scent of pine tree for nutrient. PLoS One. 2013, 8, e71676. [CrossRef]
- Wang, C.Y.; Fang, Z.M.; Wang, Z.; Zhang, D.L.; Gu, L.J.; Lee, M.R.; Liu, L.; Sung, C.K. Biological control of the pinewood nematode Bursaphelenchus xylophilus by application of the endoparasitic fungus Esteya vermicola. J BioControl. 2011. 56, 91-100. [CrossRef]
- Wang, X.; Wang, T.T.; Wang, J.C.; Guan, T.L.; Li, H.M. Morphological, molecular and biological characterization of Esteya vermicola, a nematophagous fungus isolated from intercepted wood packing materials exported from Brazil. J Mycoscience. 2014, 55, 367-377. [CrossRef]
- Bonifacino, J.S.; Glick, B.S. The mechanisms of vesicle budding and fusion. Cell. 2004, 116, 153-66. [CrossRef]
- Zhang, Y.; Yang, J. Securing SNAREs for assembly. J Biol Chem. 2020, 295, 10136-10137. [CrossRef]
- Jahn, R.; Scheller, R.H. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006, 7, 631-43. [CrossRef]
- Fasshauer, D.; Sutton, R.B.; Brunger, A.T.; Jahn, R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998, 95, 15781-6. [CrossRef]
- Shao, K.K.; Li, F.; Yang, Y.; Wang, N.; Gao, X.D.; Nakanishi, H. Characteristics of SNARE proteins are defined by distinctive properties of SNARE motifs. Biochim Biophys Acta Gen Subj. 2020, 1864, 129658. [CrossRef]
- Aalto, M.K.; Ronne, H.; Keränen, S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993, 12, 4095-104. [CrossRef]
- Adnan, M.; Islam, W.; Waheed, A.; Hussain, Q.; Shen, L.; Wang, J.; Liu, G. SNARE Protein Snc1 Is Essential for Vesicle Trafficking, Membrane Fusion and Protein Secretion in Fungi. Cells. 2023, 12, 1547. [CrossRef]
- Ballensiefen, W.; Ossipov, D.; Schmitt, H.D. Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J Cell Sci. 1998, 111, 1507-20. [CrossRef]
- Chen, L.; Lau, M.S.Y.; Banfield, D.K. Multiple ER-Golgi SNARE transmembrane domains are dispensable for trafficking but required for SNARE recycling. Mol Biol Cell. 2016, 27, 2633-41. [CrossRef]
- Wang, J.; Tian, L.; Zhang, D.D.; Short, D.P.G.; Zhou, L.; Song, S.S.; Liu, Y.; Wang, D.; Kong, Z.Q.; Cui, W.Y.; Ma, X.F.; Klosterman, S.J.; Subbarao, K.V.; Chen, J.Y.; Dai, X.F. SNARE-Encoding Genes VdSec22 and VdSso1 Mediate Protein Secretion Required for Full Virulence in Verticillium dahliae. Mol Plant Microbe Interact. 2018, 31, 651-664. [CrossRef]
- Irieda, H.; Maeda, H.; Akiyama, K.; Hagiwara, A.; Saitoh, H.; Uemura, A.; Terauchi, R.; Takano, Y. Colletotrichum orbiculare Secretes Virulence Effectors to a Biotrophic Interface at the Primary Hyphal Neck via Exocytosis Coupled with SEC22-Mediated Traffic. Plant Cell. 2014, 26, 2265-2281. [CrossRef]
- Song, W.W.; Dou, X.Y.; Qi, Z.Q.; Wang, Q.; Zhang, X.; Zhang, H.F.; Guo, M.; Dong, S.M.; Zhang, Z.G.; Wang, P.; Zheng X.B. R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS One. 2010, 5, e13193. [CrossRef]
- Zhou, S.Z.; Liu, S.; Guo, C.C.; Wei, H.W.; He, Z.H.; Liu, Z.Q.; Li, X.Y. The C2H2 Transcription Factor Con7 Regulates Vegetative Growth, Cell Wall Integrity, Oxidative Stress, Asexual Sporulation, Appressorium and Hyphopodium Formation, and Pathogenicity in Colletotrichum graminicola and Colletotrichum siamense. J Fungi (Basel). 2024, 10, 495. [CrossRef]
- Scott, B.; Eaton, C.J. Role of reactive oxygen species in fungal cellular differentiations. Curr Opin Microbiol. 2008, 11, 488-93. [CrossRef]
- Xie, C.J.; Li, Q.L.; Yang, X.Y. Characterization of VdASP F2 Secretory Factor from Verticillium dahliae by a Fast and Easy Gene Knockout System. Mol Plant Microbe Interact. 2017, 30, 444-454. [CrossRef]
- Wang, H.H.; Wang, Y.B.; Yin, C.; Gao, J.; Tao, R.; Sun, Y.L.; Wang, C.Y.; Wang, Z., Li, Y.X.; Sung, C.K. In vivo infection of Bursaphelenchus xylophilus by the fungus Esteya vermicola. Pest Manag Sci. 2020, 76, 2854-2864. [CrossRef]
- Chen, C.; Hu, Z.J.; Zheng, X.Y.; Yuan, J.J.; Zou, R.; Wang, Y.L.; Peng, X.; Xie C.J. The essential role of arginine biosynthetic genes in lunate conidia formation, conidiation, mycelial growth, and virulence of nematophagous fungus, Esteya vermicola CBS115803. Pest Manag Sci. 2024, 80, 786-796. [CrossRef]
- Li, N., Wang, B.J.; Cui, X.Y.; Hou, J.; Zhang, N. Biocontrol activities of grey mould of grapes with the volatile organic compounds generated by yeast HXMG-1 isolated from grapes. Journal of Plant Diseases and Protection, 2024, 131, 1387-1397. [CrossRef]
- Guo, C.M.; Yang, X.; Shi, H.L.; Chen, C.; Hu, Z.J.; Zheng, X.Y.; Yang, X.Y.; Xie, C.J. Identification of VdASP F2-interacting protein as a regulator of microsclerotial formation in Verticillium dahliae. Microb Biotechnol. 2022, 15, 2040-2054. [CrossRef]
- Liu, J.; Wang, Z.K.; Sun, H.H.; Ying, S.H.; Feng, M.G. Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol. 2017, 19,1808-1821. [CrossRef]
- Adnan, M.; Fang, W.; Sun, P.; Zheng, Y.; Abubakar, Y.S.; Zhang, J.; Lou, Y.; Zheng, W.; Lu, G.D. R-SNARE FgSec22 is essential for growth, pathogenicity and DON production of Fusarium graminearum. Curr Genet. 2020, 66, 421-435. [CrossRef]
- Zhu, Y.M.; Zhou, D.X.; Bai, N., Liu, Q.Q.; Zhao, N, Yang, J.K.;. SNARE Protein AoSec22 Orchestrates Mycelial Growth, Vacuole Assembly, Trap Formation, Stress Response, and Secondary Metabolism in Arthrobotrys oligospora. J Fungi (Basel). 2023, 9, 75. [CrossRef]
- Traeger, S.; Nowrousian, M. Functional Analysis of Developmentally Regulated Genes chs7 and sec22 in the Ascomycete Sordaria macrospora. G3 (Bethesda). 2015, 5, 1233-45. [CrossRef]
- Angelova, M.B.; Pashova, S.B.; Spasova, B.K.; Vassilev, S.V.; Slokoska, L.S. Oxidative stress response of filamentous fungi induced by hydrogen peroxide and paraquat. Mycol Res. 2005, 109, 150-8. [CrossRef]
- Ferrigo, D.; Scarpino, V.; Vanara, F.; Causin, R.; Raiola, A.; Blandino, M. Influence of H2O2-Induced Oxidative Stress on In Vitro Growth and Moniliformin and Fumonisins Accumulation by Fusarium proliferatum and Fusarium subglutinans. Toxins (Basel). 2021 ,13, 653. [CrossRef]
- Liu, Y.W.; Gong, X.D.; Li, M.X.; Si, H.L.; Zhou, Q.H.; Liu, X.C.; Fan, Y.; Zhang, X.Y.; Han, J.M.; Gu, S.Q.; Dong, J.G. Effect of Osmotic Stress on the Growth, Development and Pathogenicity of Setosphaeria turcica. Front Microbiol. 2021, 12, 706349. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



