Submitted:
26 August 2023
Posted:
29 August 2023
You are already at the latest version
Abstract
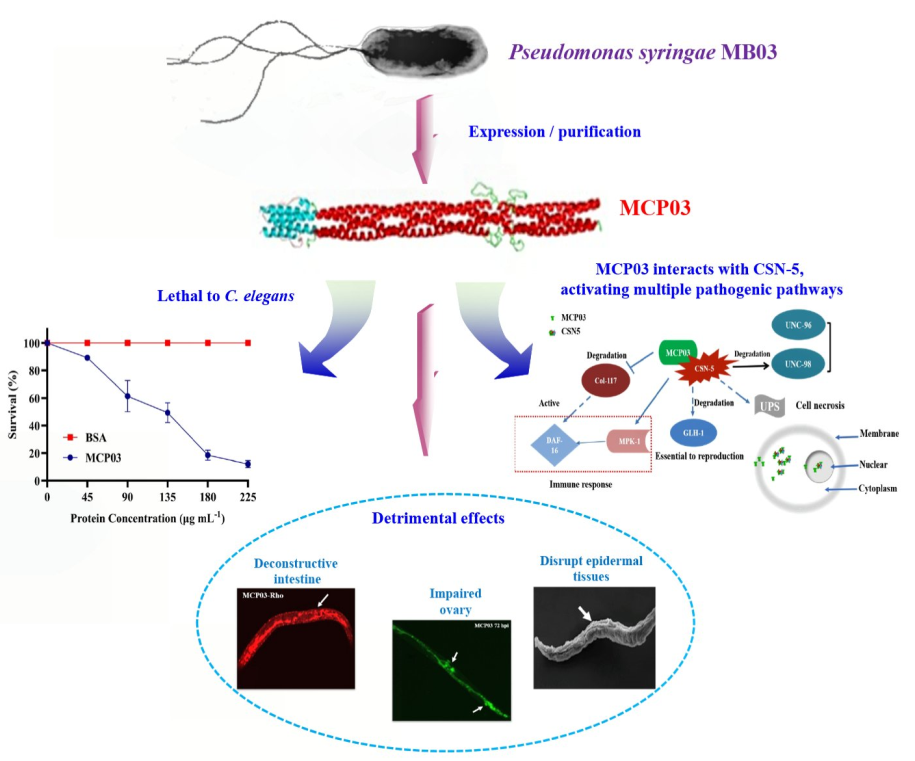
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial and Nematode Strains, Plasmids, and Culture Conditions
2.2. Cloning, Expression, Purification, and Labelling of MCP03 Protein
2.3. Nematicidal Bioassay
2.4. Analysis of C. elegans Intestinal Pathology
2.5. Yeast Two-Hybrid Assay
2.6. Expression and Purification of Recombinant Proteins from C. elegans
2.7. Scanning Electron Microscope (SEM)
2.8. Pull-Down Assay
2.9. Western Blot Analysis
2.10. Construction of csn-5 RNAi Strain
2.11. Co-Localization
2.12. Real-Time Quantitative PCR (RT-qPCR)
2.13. Data Analysis
3. Results
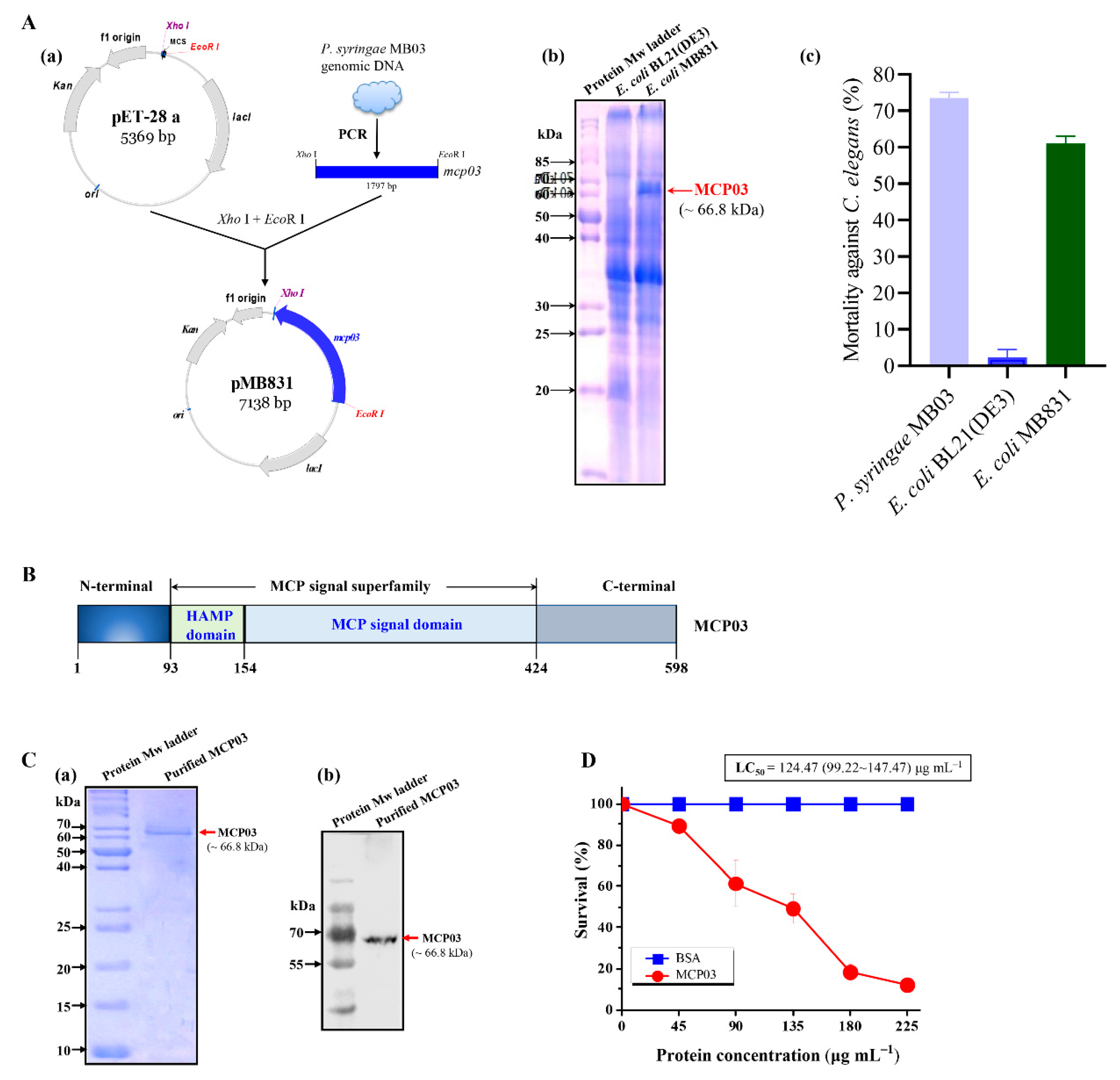
3.1. Molecular Characterization, Expression and Purification, and Nematicidal Activity of MCP03
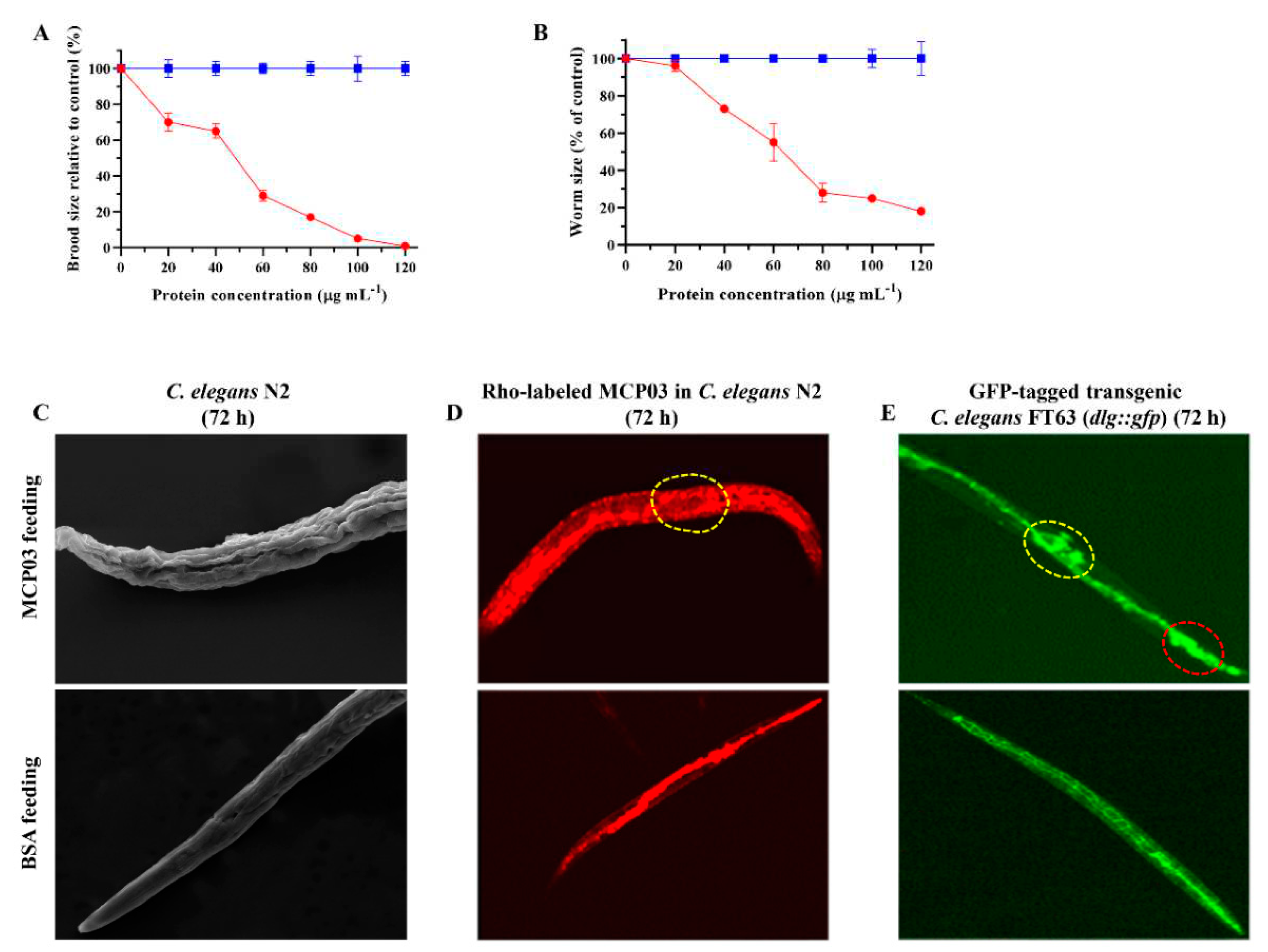
3.2. MCP03 Affects Different Phenotypes of C. elegans
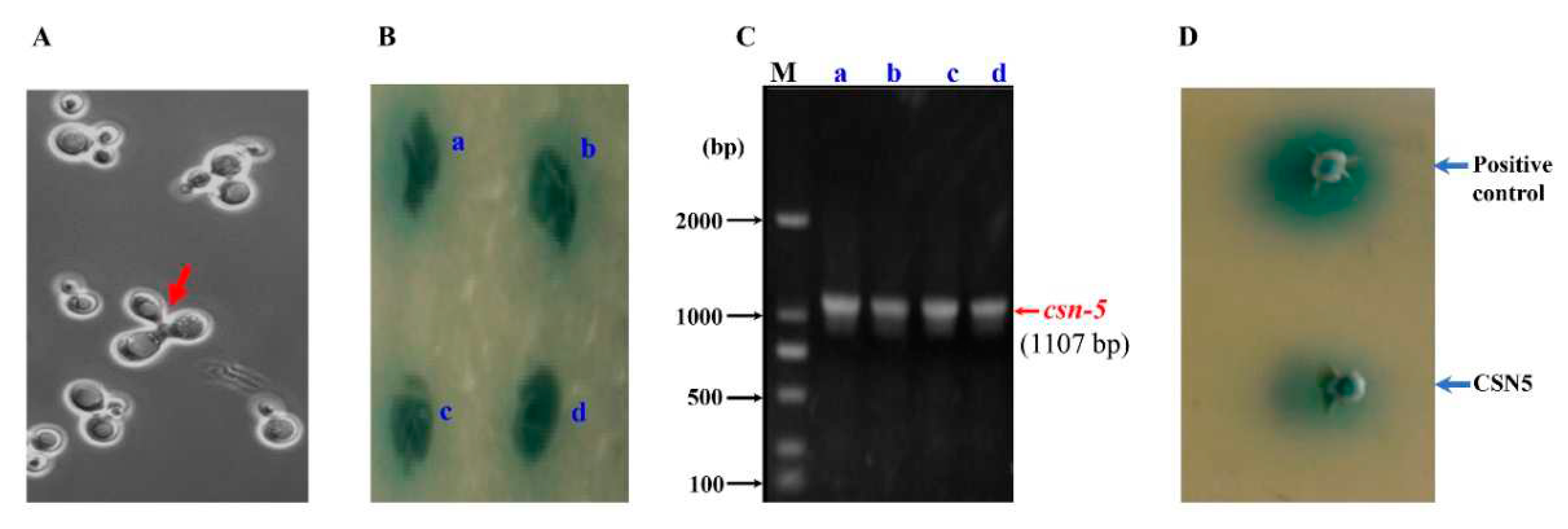
3.3. Identification of CSN-5 As A Putative Receptor of MCP03
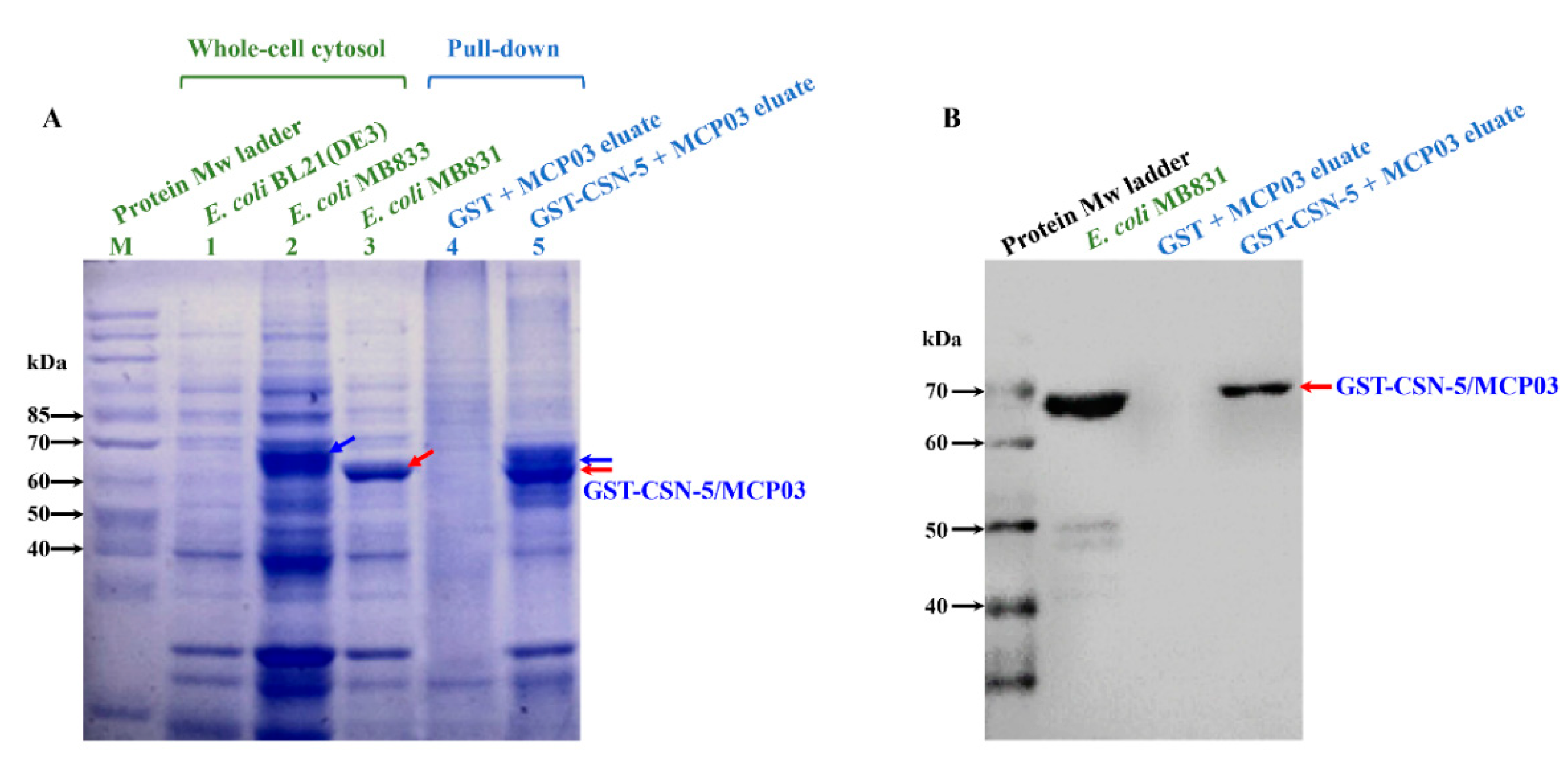
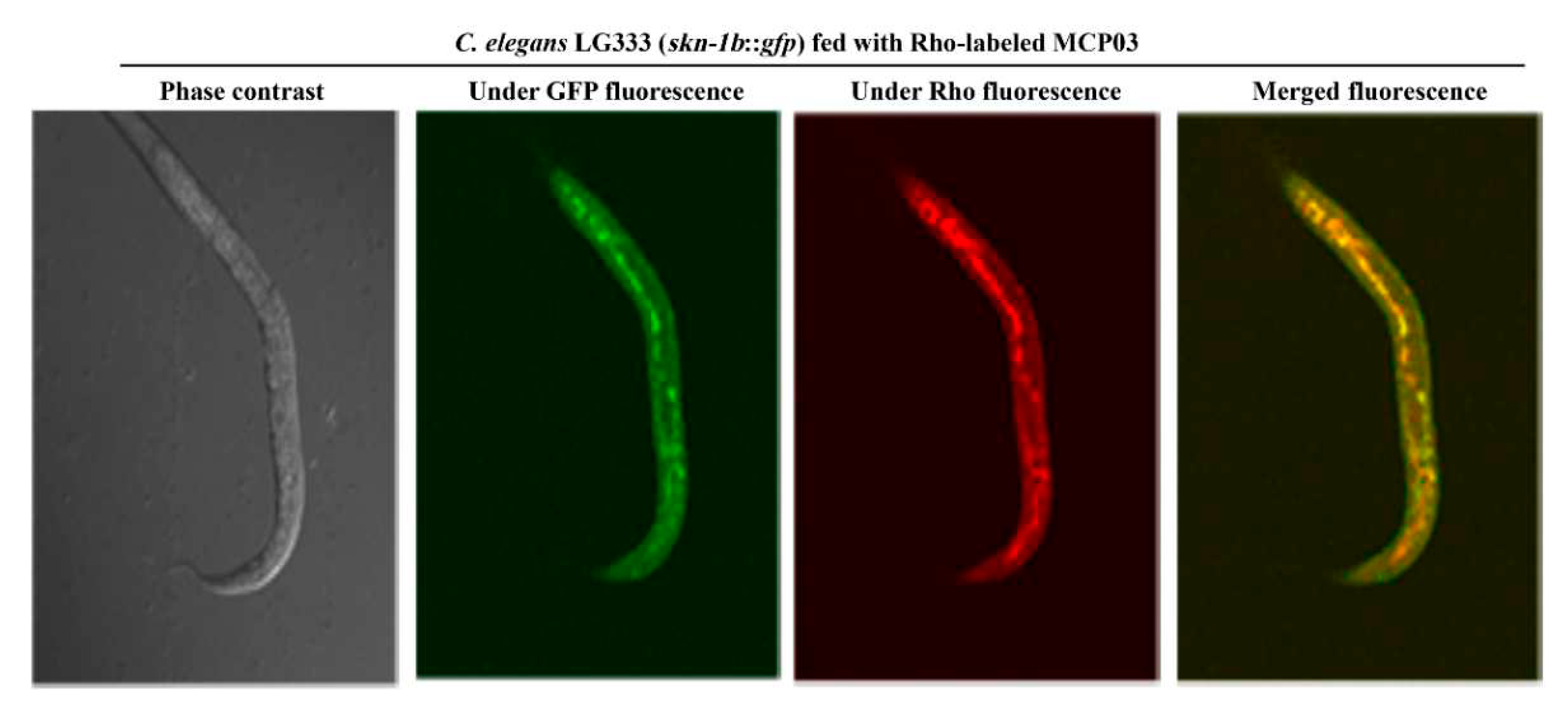
3.4. In Vivo and In Vitro Binding Interaction of MCP03 with CSN-5
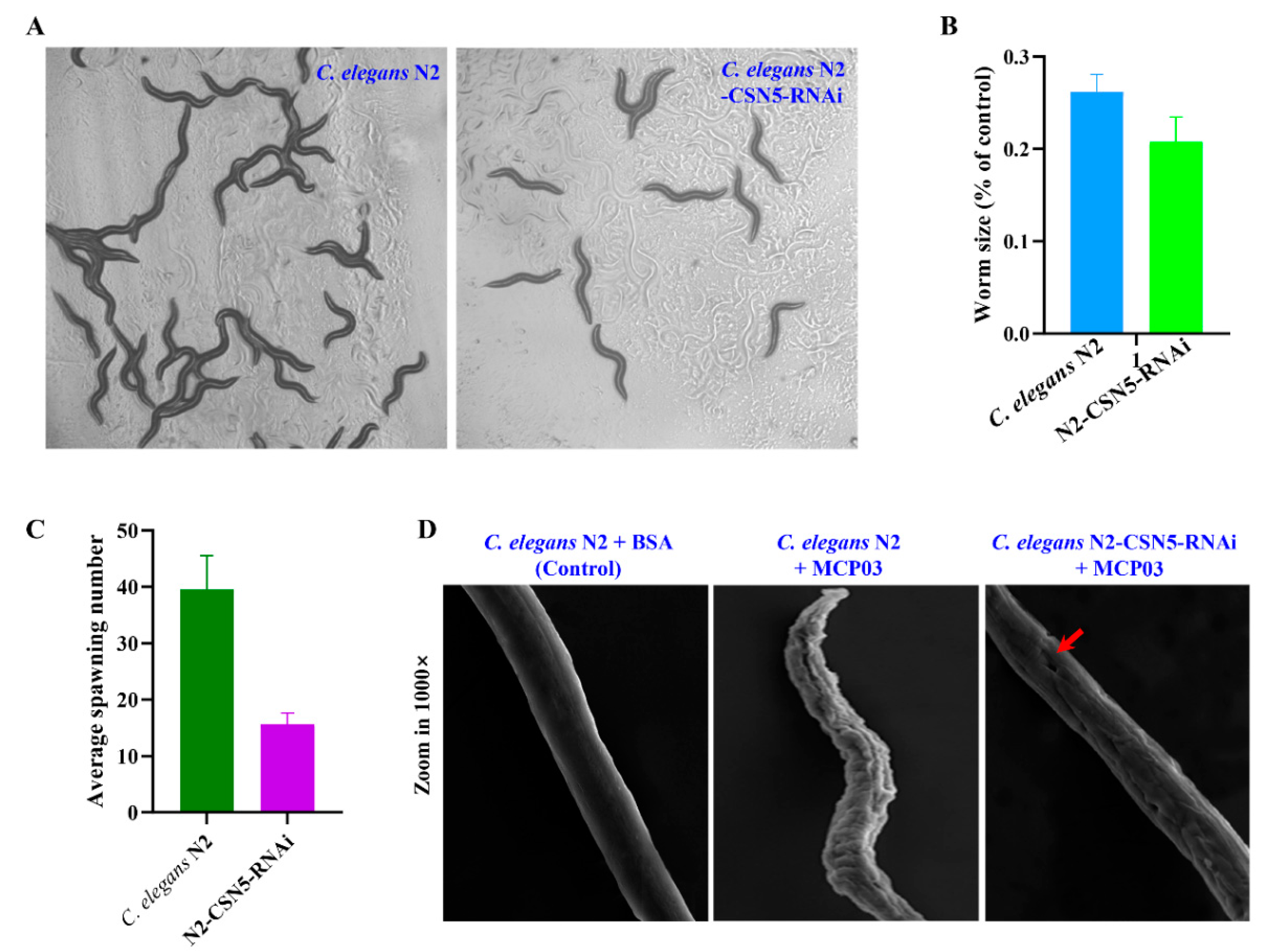
3.5. Effects of Silencing csn-5 by RNAi on Brood Size, Growth Size, and Cuticle Integrity
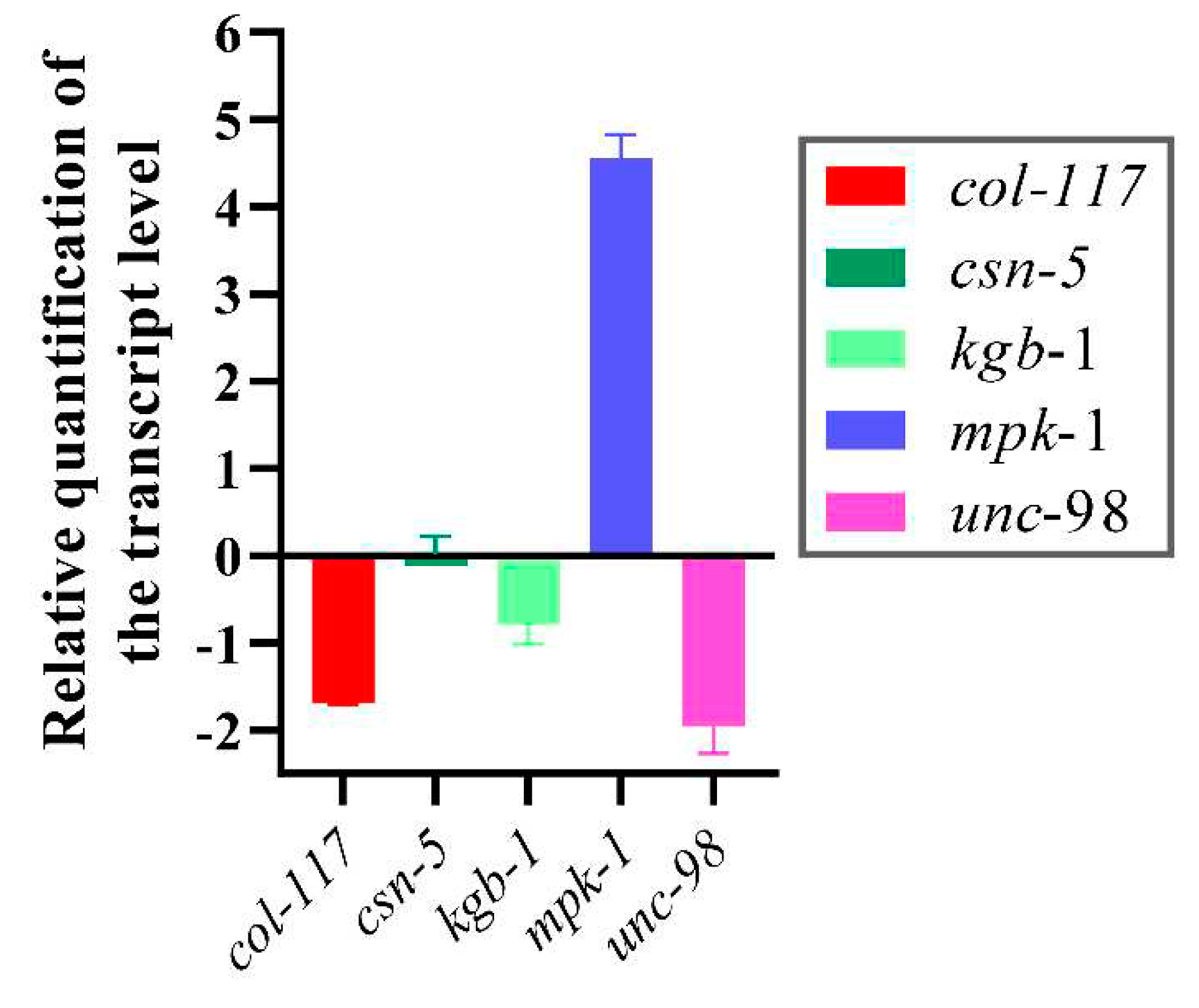
3.6. RT-qPCR Analysis of Selected Genes of N2
4. Discussion
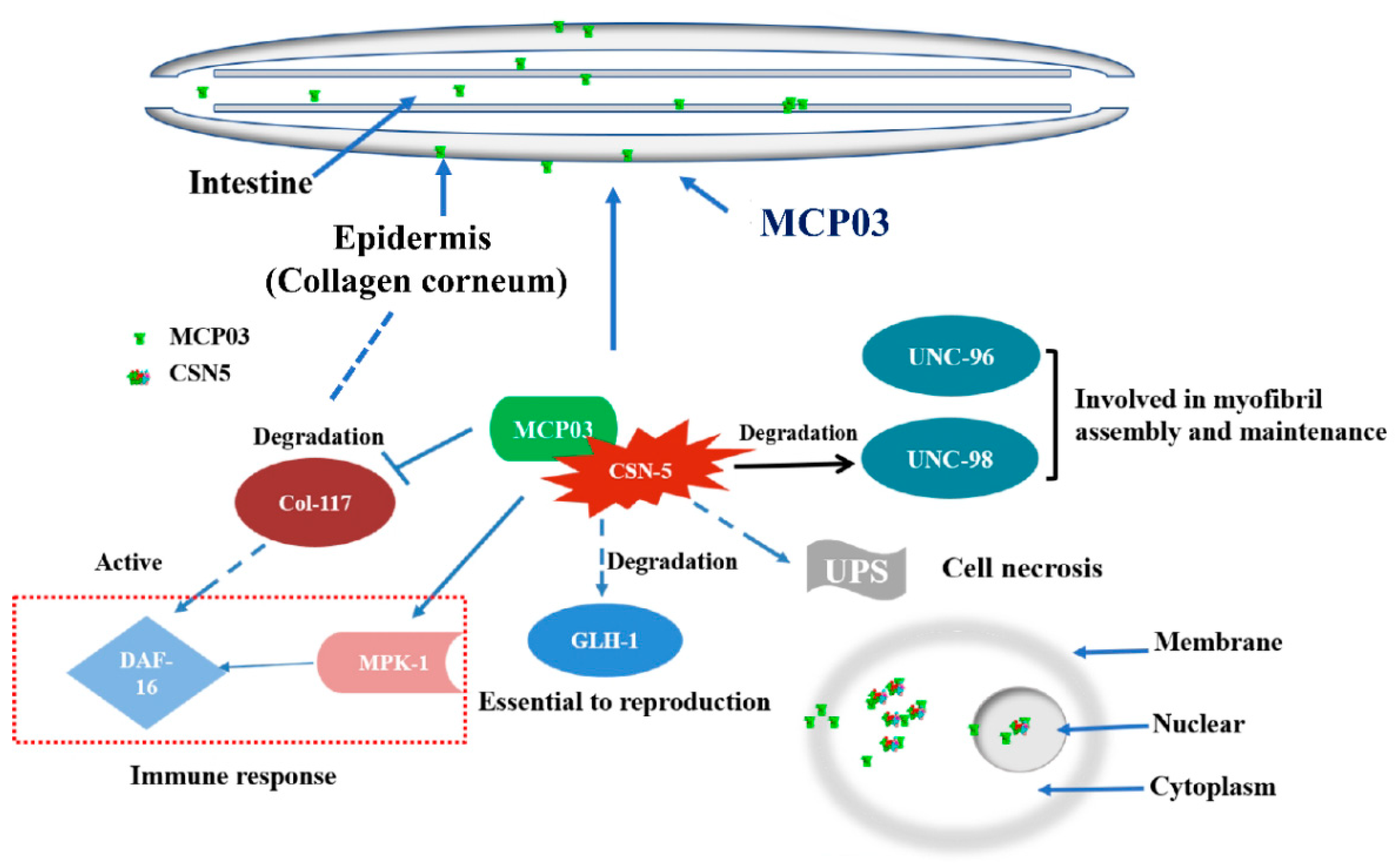
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; Caillaud, M.C.; Coutinho, P.M.; Dasilva, C.; De Luca, F.; Deau, F.; Esquibet, M.; Flutre, T.; Goldstone, J.V.; Hamamouch, N.; Hewezi, T.; Jaillon, O.; Jubin, C.; Leonetti, P.; Magliano, M.; Maier, T.R.; Markov, G.V.; McVeigh, P.; Pesole, G.; Poulain, J.; Robinson-Rechavi, M.; Sallet, E.; Ségurens, B.; Steinbach, D.; Tytgat, T.; Ugarte, E.; van Ghelder, C.; Veronico, P.; Baum, T.J.; Blaxter, M.; Bleve-Zacheo, T.; Davis, E.L.; Ewbank, J.J.; Favery, B.; Grenier, E.; Henrissat, B.; Jones, J.T.; Laudet, V.; Maule, A.G.; Quesneville, H.; Rosso, M.N.; Schiex, T.; Smant, G.; Weissenbach, J.; Wincker, P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol 2008, 26, 909–915. [Google Scholar] [CrossRef]
- Topalović, O.; Geisen, S. Nematodes as suppressors and facilitators of plant performance. New Phytol 2023, 238, 2305–2312. [Google Scholar] [CrossRef]
- Castillo, P.; Navas-Cortés, J.A.; Landa, B.B.; Jiménez-Díaz, R.M.; Vovlas, N. Plant-parasitic nematodes attacking chickpea and their in planta interactions with rhizobia and phytopathogenic fungi. Plant Dis 2008, 92, 840–853. [Google Scholar] [CrossRef]
- Sabbahi, R.; Hock, V.; Azzaoui, K.; Saoiabi, S.; Hammouti, B. A global perspective of entomopathogens as microbial biocontrol agents of insect pests. J Agric Food Res 2022, 10, 100376. [Google Scholar] [CrossRef]
- Luo, H.; Xiong, J.; Zhou, Q.; Xia, L.; Yu, Z. The effects of Bacillus thuringiensis Cry6A on the survival, growth, reproduction, locomotion, and behavioral response of Caenorhabditis elegans. Appl Microbiol Biotechnol 2013, 97, 10135–10142. [Google Scholar] [CrossRef]
- Peng, D.; Lin, J.; Huang, Q.; Zheng, W.; Liu, G.; Zheng, J.; Zhu, L.; Sun, M. A novel metalloproteinase virulence factor is involved in Bacillus thuringiensis pathogenesis in nematodes and insects. Environ Microbiol 2016, 18, 846–862. [Google Scholar] [CrossRef]
- Dubern, J.F.; Cigana, C.; De Simone, M.; Lazenby, J.; Juhas, M.; Schwager, S.; Bianconi, I.; Döring, G.; Eberl, L.; Williams, P. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ Microbiol 2015, 17, 4379–4393. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, R.; Ding, M.; Liu, Y.; Li, L. Biocontrol of the root-knot nematode Meloidogyne incognita by a nematicidal bacterium Pseudomonas simiae MB751 with cyclic dipeptide. Pest Manag Sci 2021, 77, 4365–4374. [Google Scholar] [CrossRef]
- Phani, V.; Shivakumara, T.N.; Davies, K.G.; Rao, U. Meloidogyne incognita fatty acid- and retinol-binding protein (Mi-FAR-1) affects nematode infection of plant roots and the attachment of Pasteuria penetrans endospores. Front Microbiol 2017, 8, 2122. [Google Scholar] [CrossRef]
- Tedesco, P.; Di Schiavi, E.; Esposito, F.P.; de Pascale, D. Evaluation of Burkholderia cepacia complex bacteria pathogenicity using Caenorhabditis elegans. Bio Protoc 2016, 6, e1964. [Google Scholar] [CrossRef]
- Huang, X.; Tian, B.; Niu, Q.; Yang, J.; Zhang, L.; Zhang, K. An extracellular protease from Brevibacillus laterosporus G4 without parasporal crystals can serve as a pathogenic factor in infection of nematodes. Res Microbiol 2005, 156, 719–727. [Google Scholar] [CrossRef]
- Crickmore, N. Using worms to better understand how Bacillus thuringiensis kills insects. Trends Microbiol 2005, 13, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Wadhams, G.H.; Armitage, J.P. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol 2004, 5, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Salah Ud-Din, A.I. M.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: A core sensing element in prokaryotes and archaea. Cell Mol Life Sci 2017, 74, 3293–3303. [Google Scholar] [CrossRef] [PubMed]
- Flack, C.E.; Parkinson, J.S. A zipped-helix cap potentiates HAMP domain control of chemoreceptor signaling. Proc Natl Acad Sci USA 2018, 115, E3519–E3528. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, S.; Suzuki, D.; Itoh, Y.; Suzuki, K.; Tajima, H.; Hyakutake, A.; Homma, M.; Butler-Wu, S.M.; Camilli, A.; Kawagishi, I. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect Immun 2012, 80, 3170–3178. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, S.; Hwang, H.; Kim, K.P.; Kang, D.H.; Ryu, S. Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect Immun 2015, 83, 197–204. [Google Scholar] [CrossRef]
- McLaughlin, H.P.; Caly, D.L.; McCarthy, Y.; Ryan, R.P.; Dow, J.M. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS One 2012, 7, e42205. [Google Scholar] [CrossRef]
- Yu, X.; Lund, S.P.; Scott, R.A.; Greenwald, J.W.; Records, A.H.; Nettleton, D.; Lindow, S.E.; Gross, D.C.; Beattie, G.A. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci USA 2013, 110, E425–434. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 2018, 42. [Google Scholar] [CrossRef]
- Ali, M.; Sun, Y.; Xie, L.; Yu, H.; Bashir, A.; Li, L. The pathogenicity of Pseudomonas syringae MB03 against Caenorhabditis elegans and the transcriptional response of nematicidal genes upon different nutritional conditions. Front Microbiol 2016, 7, 805. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.; Sun, Y.; Yu, X.; Sun, X.; Li, L. Nematicidal effects of 2-methyl-aconitate isomerase from the phytopathogen Pseudomonas syringae MB03 on the model nematode Caenorhabditis elegans. J Invertebr Pathol 2021, 185, 107669. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Gu, T.; Yu, X.; Bashir, A.; Wang, Z.; Sun, X.; Ashraf, N.M.; Li, L. Identification of the genes of the plant pathogen Pseudomonas syringae MB03 required for the nematicidal activity against Caenorhabditis elegans through an integrated approach. Front Microbiol 2022, 13, 826962. [Google Scholar] [CrossRef] [PubMed]
- Kaletta, T.; Hengartner, M.O. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 2006, 5, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yan, Q.; Chen, J.; He, Y.; Wang, J.; Zhang, H.; Yu, Z.; Li, L. Molecular characterization of an ice nucleation protein variant (InaQ) from Pseudomonas syringae and the analysis of its transmembrane transport activity in Escherichia coli. Int J Biol Sci 2012, 8, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. 2001.
- Porta-de-la-Riva, M.; Fontrodona, L.; Villanueva, A.; Cerón, J. Basic Caenorhabditis elegans methods: Synchronization and observation. J Vis Exp 2012, 64, e4019. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Griffitts, J.S.; Whitacre, J.L.; Stevens, D.E.; Aroian, R.V. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science 2001, 293, 860–864. [Google Scholar] [CrossRef]
- Manan, A.; Bazai, Z.A.; Fan, J.; Yu, H.; Li, L. The Nif3-family protein YqfO03 from Pseudomonas syringae MB03 has multiple nematicidal activities against Caenorhabditis elegans and Meloidogyne incognita. Int J Mol Sci 2018, 19, 3915. [Google Scholar] [CrossRef]
- Paiano, A.; Margiotta, A.; De Luca, M.; Bucci, C. Yeast two-hybrid assay to identify interacting proteins. Curr Protoc Protein Sci 2019, 95, e70. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Total RNA extraction from Caenorhabditis elegans. Cold Spring Harb Protoc 2020, 2020, 101683. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Cai, X.Q.; Harris, T.L.; Gooyit, M.; Wood, M.; Lardy, M.; Janda, K.D. Disarming Pseudomonas aeruginosa virulence factor LasB by leveraging a Caenorhabditis elegans infection model. Chem Biol 2015, 22, 483–491. [Google Scholar] [CrossRef]
- Conte Jr, D.; MacNeil, L.T.; Walhout, A.J.; Mello, C.C. RNA interference in Caenorhabditis elegans. Curr Protoc Mol Biol 2015, 109, 26.3.1–26.3.30. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Qin, N.; Xu, D.; Li, J.; Deng, X.W. COP9 signalosome: Discovery, conservation, activity, and function. J Integr Plant Biol 2020, 62, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Pintard, L.; Kurz, T.; Glaser, S.; Willis, J.H.; Peter, M.; Bowerman, B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol 2003, 13, 911–921. [Google Scholar] [CrossRef]
- Inoue, H.; Hisamoto, N.; An, J.H.; Oliveira, R.P.; Nishida, E.; Blackwell, T.K.; Matsumoto, K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 2005, 19, 2278–2283. [Google Scholar] [CrossRef]
- Broday, L.; Hauser, C.A.; Kolotuev, I.; Ronai, Z. Muscle-epidermis interactions affect exoskeleton patterning in Caenorhabditis elegans. Dev Dyn 2007, 236, 3129–3136. [Google Scholar] [CrossRef]
- Gerke, P.; Keshet, A.; Mertenskötter, A.; Paul, R.J. The JNK-like MAPK KGB-1 of Caenorhabditis elegans promotes reproduction, lifespan, and gene expressions for protein biosynthesis and germline homeostasis but interferes with hyperosmotic stress tolerance. Cell Physiol Biochem 2014, 34, 1951–1973. [Google Scholar] [CrossRef]
- Gieseler, K.; Qadota, H.; Benian, G. Development, structure, and maintenance of C. elegans body wall muscle. WormBook 2018, 2017. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.; Leung-Chiu, W.M.; Montgomery, R.; Orsborn, A.; Kuznicki, K.; Gressman-Coberly, E.; Mutapcic, L.; Bennett, K. The GLH proteins, Caenorhabditis elegans P granule components, associate with CSN-5 and KGB-1, proteins necessary for fertility, and with ZYX-1, a predicted cytoskeletal protein. Dev Biol 2002, 251, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.S.; Cha, D.S.; Choi, Y.; Lee, J.W.; Lee, M.H. MPK-1/ERK is required for the full activity of resveratrol in extended lifespan and reproduction. Aging Cell 2019, 18, e12867. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.K.; Qadota, H.; Stark, T.J.; Mercer, K.B.; Wortham, T.S.; Anyanful, A.; Benian, G.M. CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle. Mol Biol Cell 2015, 20, 3608–3616. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Liu, Y.; Li, M.; Tang, Z.; Muhammad, S.; Zheng, J.; Wan, D.; Peng, D.; Ruan, L.; Sun, M. Dissimilar crystal proteins Cry5Ca1 and Cry5Da1 synergistically act against Meloidogyne incognita and delay Cry5Ba-based nematode resistance. Appl Environ Microbiol 2017, 83, e03505–e03516. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Hofmann, K.; von Arnim, A.G.; Chamovitz, D.A. PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci 2001, 6, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.N.; Pei, D.S.; Zheng, J.N. The COP9 signalosome subunit 6 (CSN6): a potential oncogene. Cell Div 2013, 8, 14. [Google Scholar] [CrossRef]
- Braus, G.H.; Irniger, S.; Bayram, Ö. Fungal development and the COP9 signalosome. Curr Opin Microbiol 2010, 13, 672–676. [Google Scholar] [CrossRef]
- Sandhu, A.; Badal, D.; Sheokand, R.; Tyagi, S.; Singh, V. Specific collagens maintain the cuticle permeability barrier in Caenorhabditis elegans. Genetics 2021, 217, iyaa047. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




