Submitted:
05 May 2025
Posted:
08 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Preparation and Phenotyping
2.3. QTL Mapping for Growth Traits
2.4. Screening of Candidate Genes for Growth Traits QTL and Functional Enrichment Analysis
2.5. Transcriptome Library Creation and Raw Data Processing
2.6. Screening of DEGs and Functional Enrichment Analysis
2.7. QTL and Transcriptome Association Analysis
2.8. Real-Time Fluorescence Quantitative PCR (qRT-PCR) Validation
3. Results
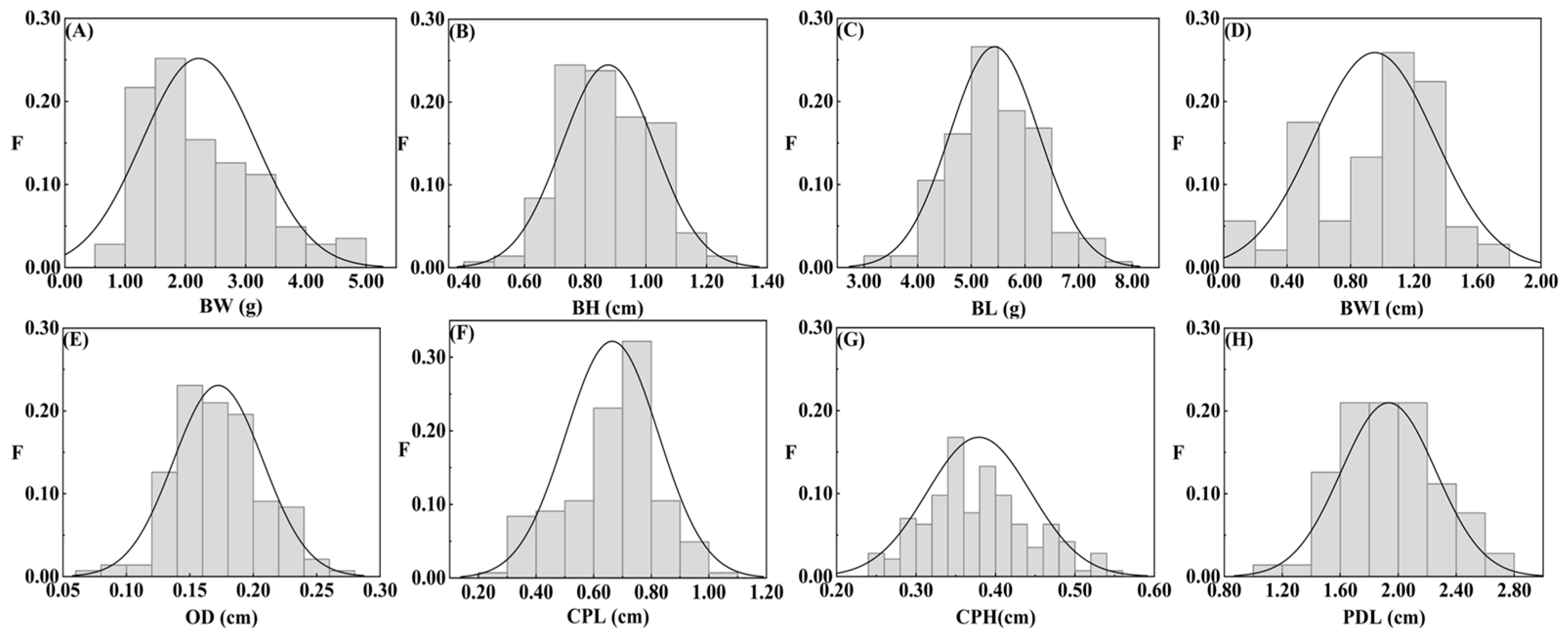
3.1. Morphological Statistics
3.2. QTL Analysis and Candidate Gene Identification
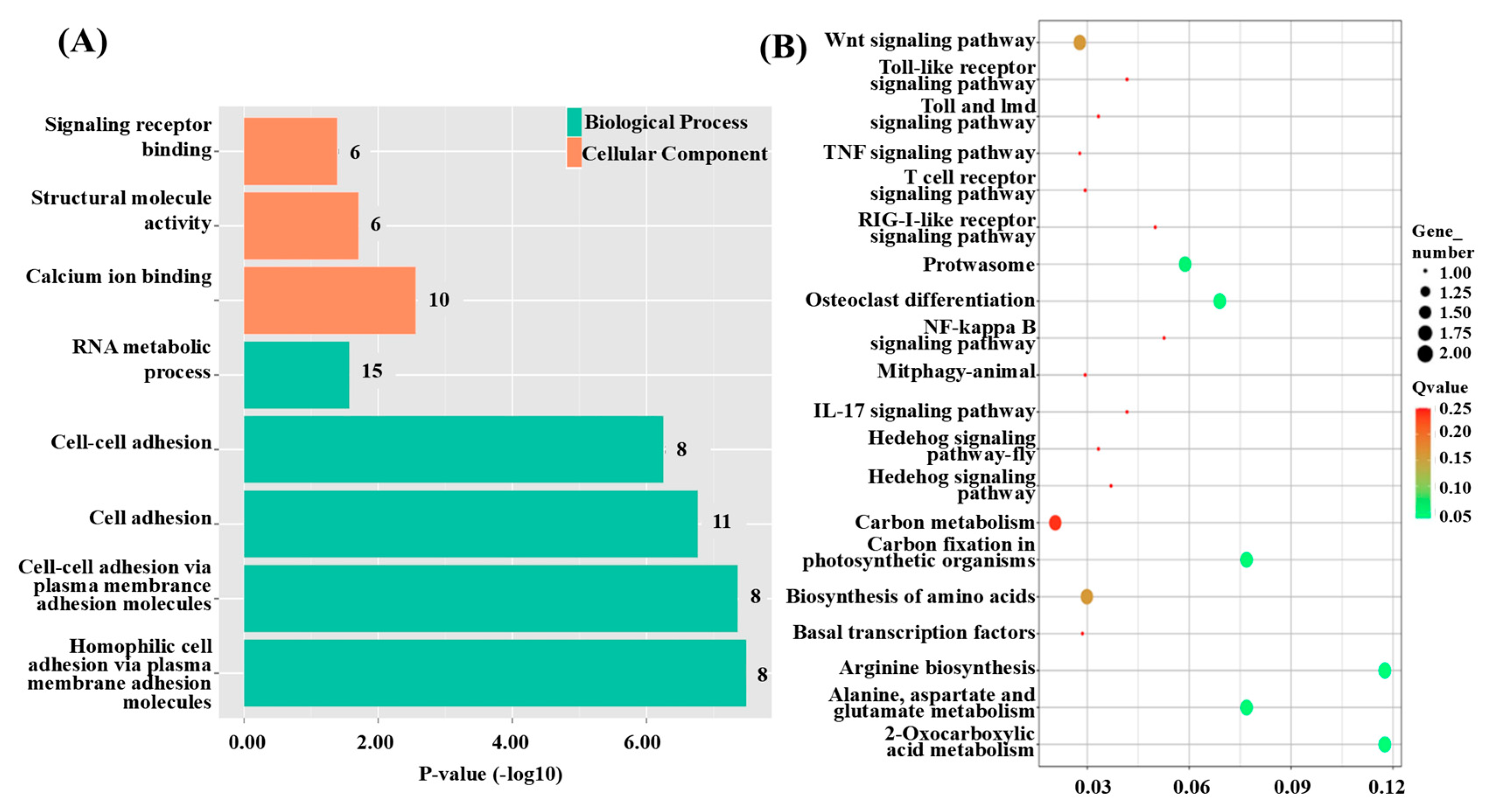
3.3. Functional Annotation of QTL Candidate Genes
3.4. Quality Control of Transcriptome Sequencing Data
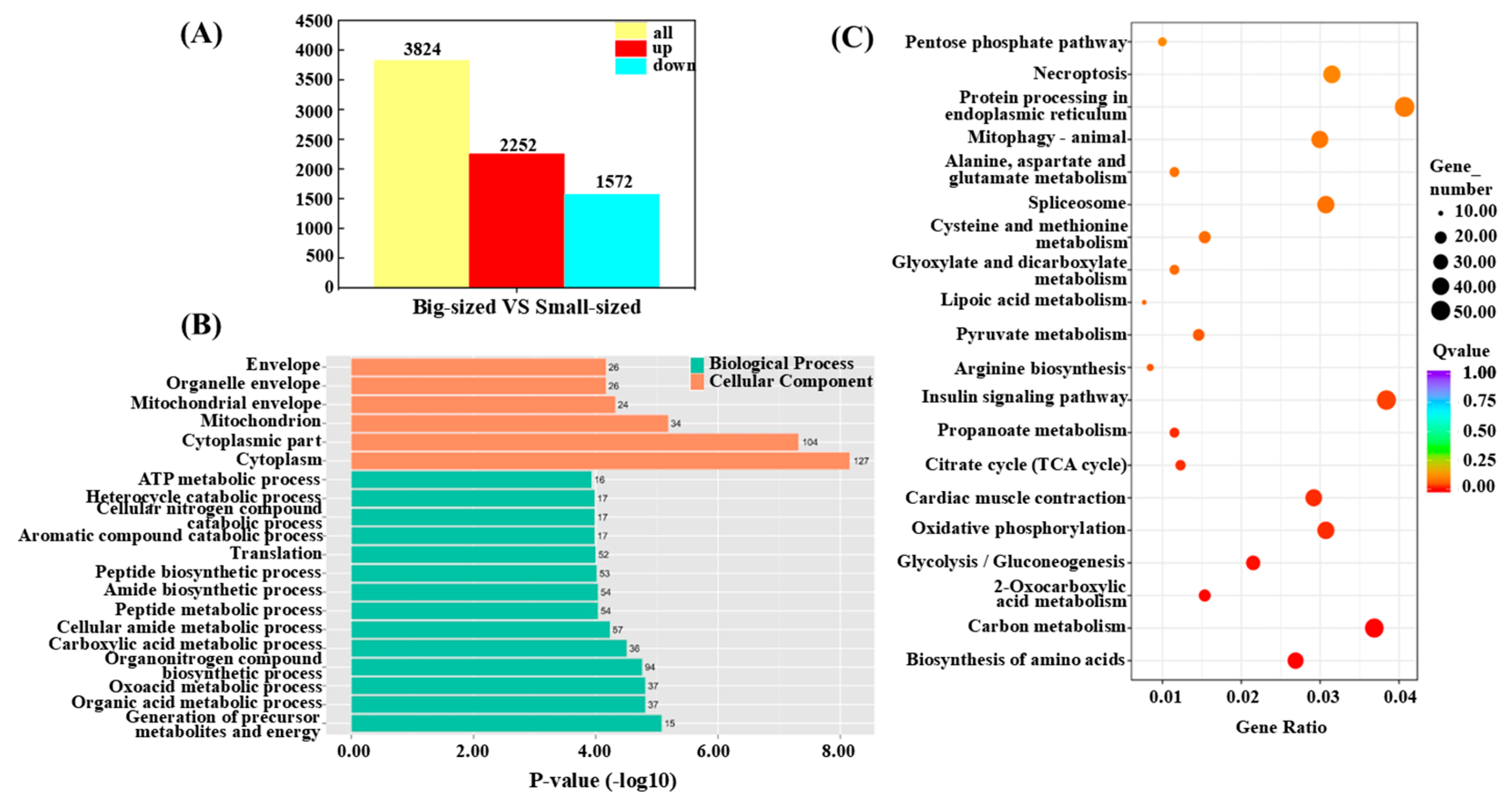
3.5. Differential Gene Expression Analysis Related to Body Size
3.6. Functional Classification of DEGs
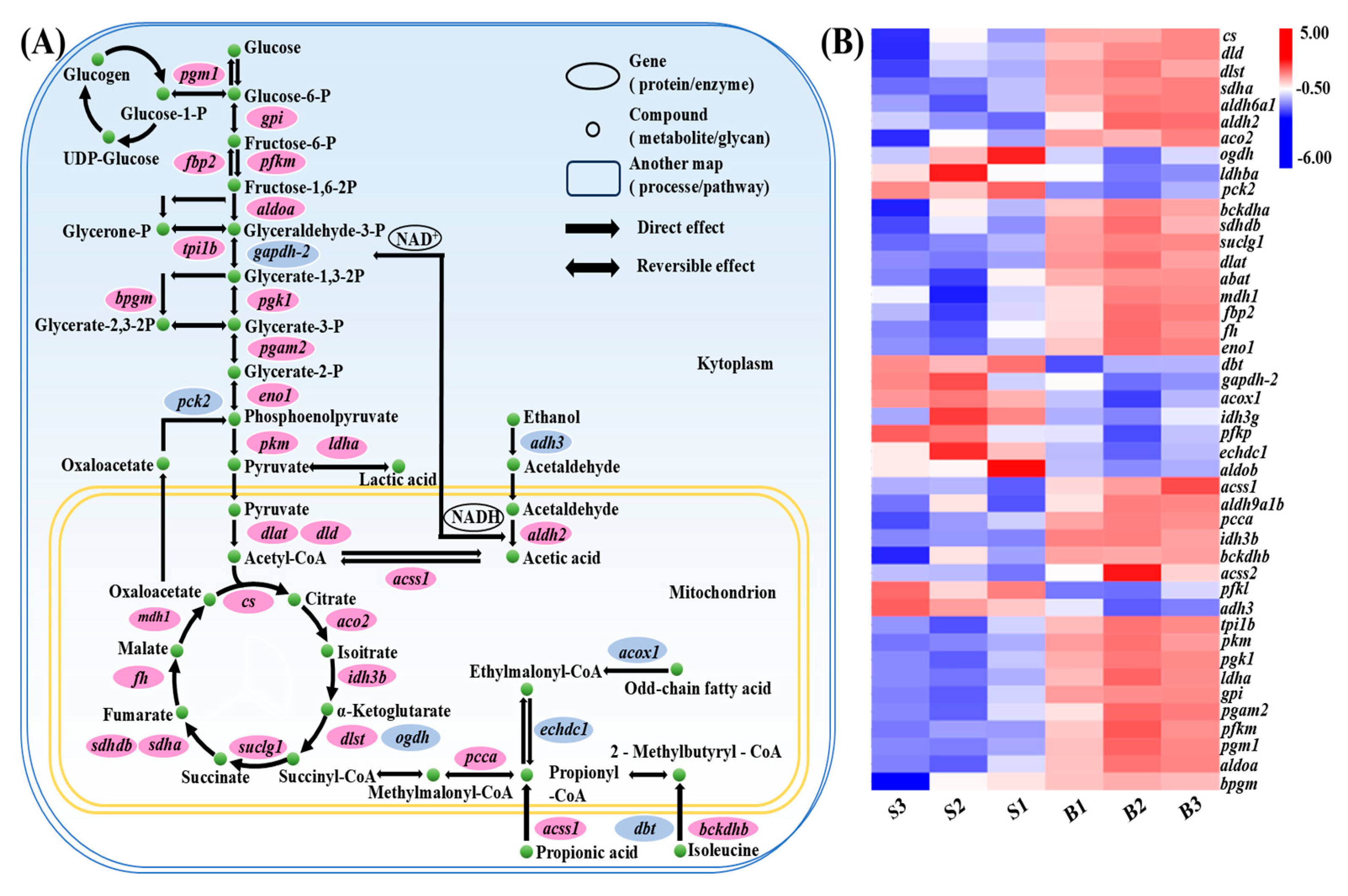
3.7. Carbohydrate Metabolism Pathway Analysis
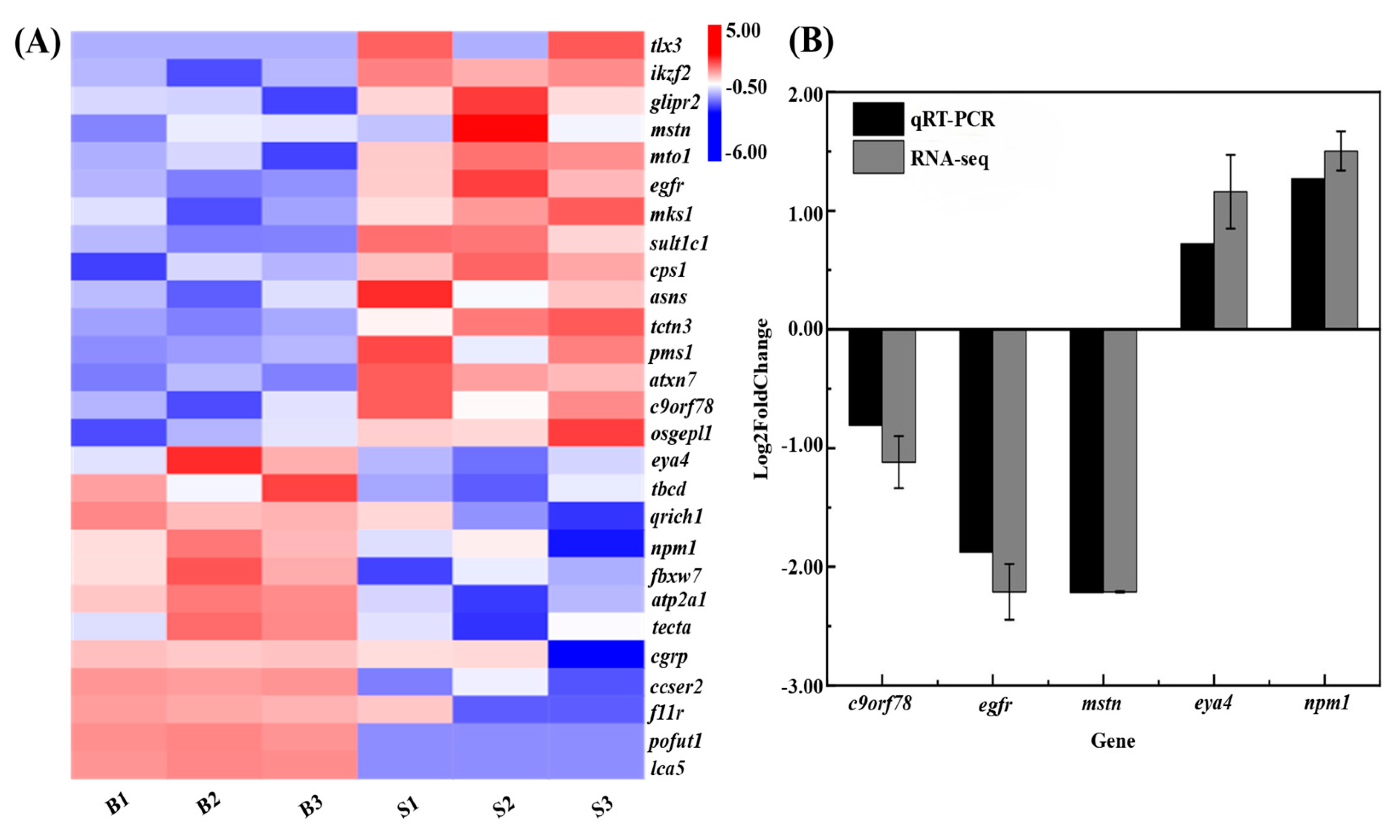
3.8. Integration of QTL Mapping and Transcriptome Data
4. Discussion
4.1. QTL Mapping of Growth Traits in C. fuscus
4.2. Transcriptome Enrichment Pathways Associated with body Size in C. fuscus
4.3. Identification and Characterization of Growth-Associated Genes in in C. fuscus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, J.; Wang, A.; Mao, S.; Xu, X.; Li, J.; Shen, Y. Construction of High-Density Genetic Linkage Map and QTL Mapping for Growth Performance in Black Carp (Mylopharyngodon Piceus). Aquaculture 2022, 549, 737799. [Google Scholar] [CrossRef]
- Ashton, D.T.; Ritchie, P.A.; Wellenreuther, M. Fifteen Years of Quantitative Trait Loci Studies in Fish: Challenges and Future Directions. Molecular Ecology 2017, 26, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Ndandala, C.B.; Dai, M.; Mustapha, U.F.; Li, X.; Liu, J.; Huang, H.; Li, G.; Chen, H. Current Research and Future Perspectives of GH and IGFs Family Genes in Somatic Growth and Reproduction of Teleost Fish. Aquaculture Reports 2022, 26, 101289. [Google Scholar] [CrossRef]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The Myogenic Regulatory Factors, Determinants of Muscle Development, Cell Identity and Regeneration. Seminars in Cell & Developmental Biology 2017, 72, 10–18. [Google Scholar] [CrossRef]
- Liu, T.; Deng, Y.; Zhang, Z.; Cao, B.; Li, J.; Sun, C.; Hu, Z.; Zhang, J.; Li, J.; Wang, Y. Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia. IJMS 2020, 21, 7036. [Google Scholar] [CrossRef]
- Blanco, A.M.; Bertucci, J.I.; Sánchez-Bretaño, A.; Delgado, M.J.; Valenciano, A.I.; Unniappan, S. Ghrelin Modulates Gene and Protein Expression of Digestive Enzymes in the Intestine and Hepatopancreas of Goldfish (Carassius Auratus) via the GHS-R1a: Possible Roles of PLC/PKC and AC/PKA Intracellular Signaling Pathways. Molecular and Cellular Endocrinology 2017, 442, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Sun, X. Genetic and Genomic Analyses for Economically Important Traits and Their Applications in Molecular Breeding of Cultured Fish. Sci. China Life Sci. 2015, 58, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Rhode, C.; Jackson, T.K.; Le Cordeur, N.S.; Jenkins, S.F.; Sampson, J.E.; Vervalle, J. Performance, Heritability, and Candidate Genes for Growth in Dusky Kob (Argyrosomus Japonicus): Implications for Genetic Improvement during Early Phase Domestication. Aquaculture 2023, 577, 739971. [Google Scholar] [CrossRef]
- Wei, J.; Chen, Y.; Wang, W. A High-Density Genetic Linkage Map and QTL Mapping for Sex and Growth-Related Traits of Large-Scale Loach (Paramisgurnus Dabryanus). Front. Genet. 2019, 10, 1023. [Google Scholar] [CrossRef]
- Palaiokostas, C.; Bekaert, M.; Khan, M.G.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Penman, D.J. A Novel Sex-Determining QTL in Nile Tilapia (Oreochromis Niloticus). BMC Genomics 2015, 16, 171. [Google Scholar] [CrossRef]
- Li, H.L.; Gu, X.H.; Li, B.J.; Chen, C.H.; Lin, H.R.; Xia, J.H. Genome-Wide QTL Analysis Identified Significant Associations Between Hypoxia Tolerance and Mutations in the GPR132 and ABCG4 Genes in Nile Tilapia. Mar Biotechnol 2017, 19, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, S.; Jiang, C.; Geng, X.; Zhou, T.; Li, N.; Bao, L.; Li, Y.; Yao, J.; Yang, Y.; et al. Multiple Across-Strain and within-Strain QTLs Suggest Highly Complex Genetic Architecture for Hypoxia Tolerance in Channel Catfish. Mol Genet Genomics 2017, 292, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Li, B.J.; Zhu, Z.X.; Gu, X.H.; Lin, H.R.; Xia, J.H. QTL Mapping for Red Blotches in Malaysia Red Tilapia (Oreochromis Spp.). Mar Biotechnol 2019, 21, 384–395. [Google Scholar] [CrossRef] [PubMed]
- O’Quin, C.T.; Drilea, A.C.; Conte, M.A.; Kocher, T.D. Mapping of Pigmentation QTL on an Anchored Genome Assembly of the Cichlid Fish, Metriaclima Zebra. BMC Genomics 2013, 14, 287. [Google Scholar] [CrossRef]
- Lv, W.; Zheng, X.; Kuang, Y.; Cao, D.; Yan, Y.; Sun, X. QTL Variations for Growth-Related Traits in Eight Distinct Families of Common Carp (Cyprinus Carpio). BMC Genet 2016, 17, 65. [Google Scholar] [CrossRef]
- Zhang, X.; Li, P.; Hu, G.; Li, C.; Lu, C.; Chao, D. QTL Analysis of Body Weight, Body Length, Body Depth and Body Thickness in Mirror Carp Cyprinus Carpio. Chinese Journal of Fisheries 2023, 36, 1–9. [Google Scholar] [CrossRef]
- Rockman, M.V. THE QTN PROGRAM AND THE ALLELES THAT MATTER FOR EVOLUTION: ALL THAT’S GOLD DOES NOT GLITTER. Evolution 2012, 66, 1–17. [Google Scholar] [CrossRef]
- Cai, C.; Yang, P.; Shi, Y.; Wang, X.; Chen, G.; Zhang, Q.; Cheng, G.; Kong, W.; Xu, Z. Transcriptomic and Metabolomic Analysis Revealed Potential Mechanisms of Growth and Disease Resistance Dimorphism in Male and Female Common Carp (Cyprinus Carpio). Fish & Shellfish Immunology 2025, 158, 110150. [Google Scholar] [CrossRef]
- Chen, F.; Ouyang, X.; Liao, Z.; Huang, T.; Tong, G.; Tan, H.; Zhou, M.; Lu, X.; Wei, X.; Yang, X.; et al. Comprehensive Transcriptomic, Proteomic, and Intestinal Microbiota Analyses of Largemouth Bass (Micropterus Salmoides) Intestines Reveal New Insights into Immune Responses to Aeromonas Hydrophila Infection. Fish & Shellfish Immunology 2025, 156, 110057. [Google Scholar] [CrossRef]
- Salem, M.; Vallejo, R.L.; Leeds, T.D.; Palti, Y.; Liu, S.; Sabbagh, A.; Rexroad, C.E.; Yao, J. RNA-Seq Identifies SNP Markers for Growth Traits in Rainbow Trout. PLoS ONE 2012, 7, e36264. [Google Scholar] [CrossRef]
- Ding, W.; Cao, L.; Cao, Z.; Bing, X. Characterization of the Growth-Related Transcriptome in the Liver and Brain of Mandarin Fish (Siniperca Chuatsi) through RNA-Seq Analysis. Journal of Applied Animal Research 2024, 52, 2440045. [Google Scholar] [CrossRef]
- Mendez, K.N.; Zuloaga, R.; Valenzuela, C.A.; Bastias-Molina, M.; Meneses, C.; Vizoso, P.; Valdés, J.A.; Molina, A. RNA-Seq Analysis of Compensatory Growth in the Skeletal Muscle of Fine Flounder (Paralichthys Adspersus). Aquaculture 2018, 490, 270–280. [Google Scholar] [CrossRef]
- Wu, L.; Yang, Y.; Wang, X.; Weng, Z.; Hua, S.; Li, D.; Xia, J.; Liu, X.; Meng, Z. Genome-Wide QTL Mapping and RNA-Seq Reveal the Genetic Variation Influencing Growth Traits in Giant Grouper (Epinephelus Lanceolatus). Aquaculture 2023, 563, 738944. [Google Scholar] [CrossRef]
- Ma, B.; Liu, Y.; Zhang, X.; Chen, T.; Zhang, L.; Hu, C.; Yu, S.; Chen, G.; Liu, L.; Zhu, J.; et al. Genome-Wide QTL Mapping and RNA-Seq Reveal Genetic Mechanisms behind Discrepant Growth Traits in Pacific Whiteleg Shrimp, Litopenaeus Vannamei. Aquaculture 2025, 599, 742084. [Google Scholar] [CrossRef]
- Tian, C.-X.; Lin, X.-H.; Zhou, D.-Y.; Chen, Y.; Shen, Y.-J.; Ye, M.-H.; Duan, C.-Y.; Zhang, Y.-L.; Yang, B.-L.; Deng, S.-P.; et al. A Chromosome-Level Genome Assembly of Hong Kong Catfish (Clarias Fuscus) Uncovers a Sex-Determining Region. BMC Genomics 2023, 24, 291. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lin, X.; Zhu, Y.; Zhou, D.; Zhang, Y.; Huang, Y.; Chen, H.; Li, G.; Tian, C. A Time-Course Transcriptome Analysis of Gonads from HongKong Catfish (Clarias Fuscus) Reveals Genes and Pathways Associated with Gonadal Development. Aquaculture Reports 2024, 37, 102247. [Google Scholar] [CrossRef]
- Anderson, M.J.; Fast, A.W. Temperature and Feed Rate Effects on Chinese Catfish, Clarias Fuscus (Lacepède), Growth. Aquaculture Res 1991, 22, 435–442. [Google Scholar] [CrossRef]
- Lin, X.; Tan, J.; Shen, Y.; Yang, B.; Zhang, Y.; Liao, Y.; Wang, P.; Zhou, D.; Li, G.; Tian, C. A High-Density Genetic Linkage Map and QTL Mapping for Sex in Clarias Fuscus. Aquaculture 2022, 561, 738723. [Google Scholar] [CrossRef]
- Van, O. MapQTL6, Software for the Mapping of Quantitative Trait Loci in Experimental Population of Diploid Species. Wageningen, Netherlands, Kyazma B.V. 2009. [Google Scholar]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome Analysis by Strand-Specific Sequencing of Complementary DNA. Nucleic Acids Research 2009, 37, e123–e123. [Google Scholar] [CrossRef]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-Cell RNA-Seq Profiling of Human Preimplantation Embryos and Embryonic Stem Cells. Nat Struct Mol Biol 2013, 20, 1131–1139. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat Biotechnol 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Valente, L.M.P.; Moutou, K.A.; Conceição, L.E.C.; Engrola, S.; Fernandes, J.M.O.; Johnston, I.A. What Determines Growth Potential and Juvenile Quality of Farmed Fish Species? Reviews in Aquaculture 2013, 5. [Google Scholar] [CrossRef]
- Laghari, M.Y.; Lashari, P.; Zhang, Y.; Sun, X. Identification of Quantitative Trait Loci (QTLs) in Aquaculture Species. Reviews in Fisheries Science & Aquaculture 2014, 22, 221–238. [Google Scholar] [CrossRef]
- Mackay, T.F.C. The Genetic Architecture of Quantitative Traits. Annu. Rev. Genet. 2001, 35, 303–339. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fu, B.; Pang, M.; Feng, X.; Yu, X.; Tong, J. A High-Density Genetic Linkage Map and QTL Fine Mapping for Body Weight in Crucian Carp (Carassius Auratus) Using 2b-RAD Sequencing. G3 Genes|Genomes|Genetics 2017, 7, 2473–2487. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, H.; Pan, Z.; Chang, G.; Wang, H.; Wu, N.; Ding, H.; Yu, X. Construction of a High-Density Genetic Linkage Map and QTL Mapping for Growth Traits in Pseudobagrus Ussuriensis. Aquaculture 2019, 511, 734213. [Google Scholar] [CrossRef]
- Jackson, T.K.; Rhode, C. A High-Density Genetic Linkage Map and QTL Identification for Growth Traits in Dusky Kob (Argyrosomus Japonicus). Aquaculture 2024, 586, 740786. [Google Scholar] [CrossRef]
- Guo, J.; Li, C.; Teng, T.; Shen, F.; Chen, Y.; Wang, Y.; Pan, C.; Ling, Q. Construction of the First High-Density Genetic Linkage Map of Pikeperch (Sander Lucioperca) Using Specific Length Amplified Fragment (SLAF) Sequencing and QTL Analysis of Growth-Related Traits. Aquaculture 2018, 497, 299–305. [Google Scholar] [CrossRef]
- Lu, X.; Chen, H.-M.; Qian, X.-Q.; Gui, J.-F. Transcriptome Analysis of Grass Carp (Ctenopharyngodon Idella) between Fast- and Slow-Growing Fish. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 2020, 35, 100688. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, Z.; Solberg, M.F.; Chen, Z.; Wei, M.; Zhu, F.; Jia, C.; Meng, Q.; Zhang, Z. Comparative Transcriptome Analysis of Mixed Tissues of Black Porgy (Acanthopagrus Schlegelii) with Differing Growth Rates. Aquaculture Research 2021, 52, 5800–5813. [Google Scholar] [CrossRef]
- Yang, J.; Lu, B.; Yu, Z.; Zhang, L.; Chen, Y.; Chen, Z.; Han, C.; Shu, H. Multiple Tissues Transcriptome of Zig-Zag Eel (Mastacembelus Armatus) with Different Growth Rates. Animals 2024, 14, 248. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-Citrate Lyase Links Cellular Metabolism to Histone Acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef]
- Kleiber, M. BODY SIZE AND METABOLIC RATE. Physiological Reviews 1947, 27, 511–541. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R. Metabolism and Functions of Lipids and Fatty Acids in Teleost Fish. Reviews in Fisheries Science 2003, 11, 107–184. [Google Scholar] [CrossRef]
- Varanasi, U.; Chu, R.; Chu, S.; Espinosa, R.; LeBeau, M.M.; Reddy, J.K. Isolation of the Human Peroxisomal Acyl-CoA Oxidase Gene: Organization, Promoter Analysis, and Chromosomal Localization. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 3107–3111. [Google Scholar] [CrossRef]
- Patel, M.S.; Roche, T.E. Molecular Biology and Biochemistry of Pyruvate Dehydrogenase Complexes. FASEB J. 1990, 4, 3224–3233. [Google Scholar] [CrossRef]
- Linster, C.L.; Noël, G.; Stroobant, V.; Vertommen, D.; Vincent, M.-F.; Bommer, G.T.; Veiga-da-Cunha, M.; Van Schaftingen, E. Ethylmalonyl-CoA Decarboxylase, a New Enzyme Involved in Metabolite Proofreading. Journal of Biological Chemistry 2011, 286, 42992–43003. [Google Scholar] [CrossRef]
- Panov, A.V.; Mayorov, V.I.; Dikalov, S.I. Role of Fatty Acids β-Oxidation in the Metabolic Interactions Between Organs. IJMS 2024, 25, 12740. [Google Scholar] [CrossRef]
- Trefely, S.; Lovell, C.D.; Snyder, N.W.; Wellen, K.E. Compartmentalised Acyl-CoA Metabolism and Roles in Chromatin Regulation. Molecular Metabolism 2020, 38, 100941. [Google Scholar] [CrossRef]
- Modrell, M.S.; Baker, C.V.H. Evolution of Electrosensory Ampullary Organs: Conservation of Eya4 Expression during Lateral Line Development in Jawed Vertebrates. Evolution and Development 2012, 14, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sewell, W.F.; Kim, S.D.; Shin, J.T.; MacRae, C.A.; Zon, L.I.; Seidman, J.G.; Seidman, C.E. Eya4 Regulation of Na+/K+-ATPase Is Required for Sensory System Development in Zebrafish. Development 2008, 135, 3425–3434. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.B.; Rogers, S.; Roychoudhury, K.; Tan, Y.S.; Atkinson, C.J.; Sobinoff, A.P.; Tomlinson, C.G.; Hsu, A.; Lu, R.; Dray, E.; et al. The Eyes Absent Family Members EYA4 and EYA1 Promote PLK1 Activation and Successful Mitosis through Tyrosine Dephosphorylation. Nat Commun 2024, 15, 1385. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razek, O.; Marzouk, A.; MacKinnon, M.; Guy, E.T.; Pohar, S.A.; Zhushma, E.; Liu, J.; Sia, I.; Gokey, J.J.; Tay, H.G.; et al. Calcium Signaling Mediates Proliferation of the Precursor Cells That Give Rise to the Ciliated Left-Right Organizer in the Zebrafish Embryo. Front. Mol. Biosci. 2023, 10, 1292076. [Google Scholar] [CrossRef]
- Nakamura, T.; Hamada, H. Left-Right Patterning: Conserved and Divergent Mechanisms. Development 2012, 139, 3257–3262. [Google Scholar] [CrossRef]
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors with Pleiotropic Functions in Cell Physiology and Development. Physiological Reviews 2017, 97, 1529–1554. [Google Scholar] [CrossRef]
- Wu, N.; Li, C.-J.; Gui, J.-F. Molecular Characterization and Functional Commonality of Nucleophosmin/Nucleoplasmin in Two Cyprinid Fish. Biochem Genet 2009, 47, 749–762. [Google Scholar] [CrossRef]
- Stooke-Vaughan, G.A.; Obholzer, N.D.; Baxendale, S.; Megason, S.G.; Whitfield, T.T. Otolith Tethering in the Zebrafish Otic Vesicle Requires Otogelin and α-Tectorin. Development 2015, 142, 1137–1145. [Google Scholar] [CrossRef]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a Negative Regulator of Muscle Growth, Functions by Inhibiting Myoblast Proliferation. Journal of Biological Chemistry 2000, 275, 40235–40243. [Google Scholar] [CrossRef]
- Coogan, M.; Alston, V.; Su, B.; Khalil, K.; Elaswad, A.; Khan, M.; Simora, R.M.C.; Johnson, A.; Xing, D.; Li, S.; et al. CRISPR/Cas-9 Induced Knockout of Myostatin Gene Improves Growth and Disease Resistance in Channel Catfish (Ictalurus Punctatus). Aquaculture 2022, 557, 738290. [Google Scholar] [CrossRef]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y.; et al. Targeted Disruption of Sp7 and Myostatin with CRISPR-Cas9 Results in Severe Bone Defects and More Muscular Cells in Common Carp. Sci Rep 2016, 6, 22953. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, R.A.; Scott, H.; Walters, K. Immunocytochemical Localization of Epidermal Growth Factor Receptor in Early Embryos of the Japanese Medaka Fish (Oryzias Latipes). Histochem. J. 2001, 33, 37–42. [Google Scholar] [CrossRef]
- Xie, L.; Tang, Q.; Yang, L.; Chen, L. Insulin-like Growth Factor I Promotes Oocyte Maturation through Increasing the Expression and Phosphorylation of Epidermal Growth Factor Receptor in the Zebrafish Ovary. Molecular and Cellular Endocrinology 2016, 419, 198–207. [Google Scholar] [CrossRef]
- Zhang, Q.; He, X.; Yao, S.; Lin, T.; Zhang, L.; Chen, D.; Chen, C.; Yang, Q.; Li, F.; Zhu, Y.-M.; et al. Ablation of Mto1 in Zebrafish Exhibited Hypertrophic Cardiomyopathy Manifested by Mitochondrion RNA Maturation Deficiency. Nucleic Acids Research 2021, 49, 4689–4704. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lu, D. The Ikaros Family of Zinc-Finger Proteins. Acta Pharmaceutica Sinica B 2016, 6, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Weatherbee, S.D.; Niswander, L.A.; Anderson, K.V. A Mouse Model for Meckel Syndrome Reveals Mks1 Is Required for Ciliogenesis and Hedgehog Signaling. Human Molecular Genetics 2009, 18, 4565–4575. [Google Scholar] [CrossRef]
- Kozłowski, J.; Konarzewski, M.; Czarnoleski, M. Coevolution of Body Size and Metabolic Rate in Vertebrates: A Life-history Perspective. Biological Reviews 2020, 95, 1393–1417. [Google Scholar] [CrossRef]
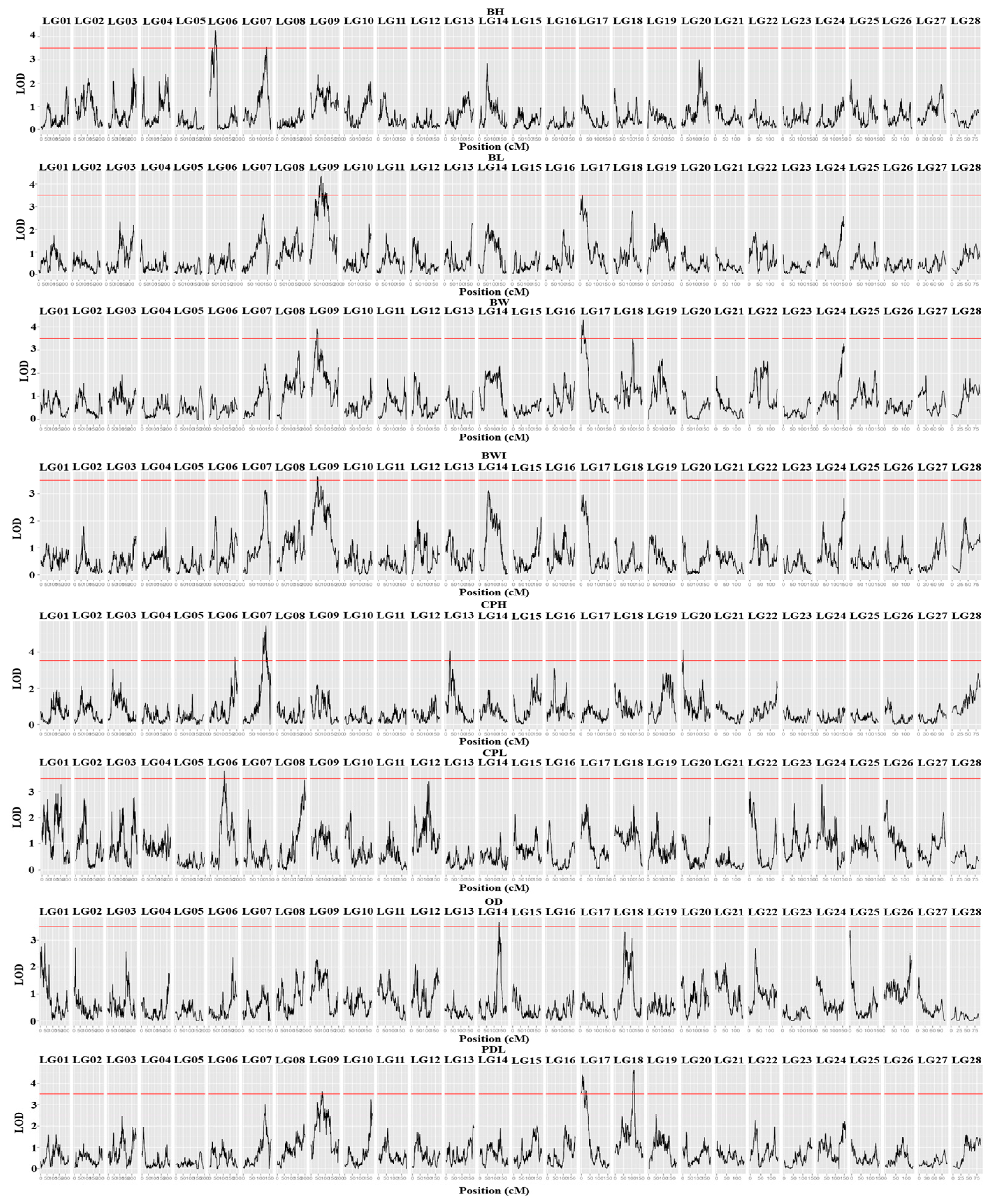
| Trait | QTL | Linkage group | Marking interval (cM) | peak marker (cM) | LOD | PVE (%) | LOD Peek (cM) | Corresponding markers for LOD peaks | Candidate gene |
|---|---|---|---|---|---|---|---|---|---|
| BW | qBW-1 | LG09 | 41.931-45.835 | np27 | 3.910 | 8.70 | 44.543 | LG9 6545823 | --- |
| qBW-2 | LG17 | 13.52-17.098 | np1240 | 4.280 | 9.50 | 14.832 | LG17 28458728 | slc4a1 | |
| BL | qBL-3 | LG09 | 82.998-85.093 | lm55 | 4.350 | 9.80 | 83.615 | LG9 13097329 | --- |
| qBL-4 | LG17 | 11.779-14.832 | lm1144 | 3.510 | 8.00 | 13.520 | LG17 28458720 | slc4a1 | |
| CPH | qCPH-5 | LG06 | 184.207-186.967 | lm3244 | 3.730 | 8.30 | 186.516 | LG6 4577776 | pgap1 |
| qCPH-6 | LG07 | 128.571-129.622 | lm3434 | 5.440 | 11.90 | 128.590 | LG7 8175232 | --- | |
| qCPH-7 | LG13 | 24.138-31.262 | np671 | 4.060 | 9.00 | 26.118 | LG13 30699773 | ltk | |
| qCPH-8 | LG20 | 5.037-6.976 | lm1639 | 4.110 | 9.10 | 5.619 | LG20 1586316 | --- | |
| CPL | qCPL-9 | LG06 | 102.507-103.569 | np3359 | 3.790 | 9.50 | 102.644 | LG6 22085749 | pdk1 |
| OD | qOD-10 | LG14 | 132.742-134.846 | hk257 | 3.580 | 8.00 | 134.365 | LG14 25875874 | qrich1 |
| PDL | qPDL-11 | LG09 | 77.872-80.447 | lm52 | 3.610 | 8.10 | 79.160 | LG9 12492904 | pp2a |
| qPDL-12 | LG17 | 2.533-6.695 | np1243 | 4.400 | 9.80 | 5.627 | LG17 29429338 | mto1 | |
| qPDL-13 | LG18 | 112.055-115.500 | lm1230 | 4.580 | 10.20 | 112.554 | LG18 25096181 | --- | |
| BWI | qBWI-14 | LG09 | 44.543-46.478 | np28 | 3.630 | 8.10 | 45.835 | LG9 6720412 | --- |
| BH | qBH-15 | LG06 | 21.046-22.095 | np3410 | 3.500 | 8.00 | 21.431 | LG6 36044360 | cps |
| qBH-16 | LG06 | 41.649-44.768 | np3396 | 4.220 | 9.60 | 41.702 | LG6 34731920 | --- | |
| qBH-17 | LG07 | 134.41-134.773 | lm3428 | 3.550 | 8.10 | 134.410 | LG7 7360877 | --- |
| Sample | Raw-reads | Clean-reads | Clean-reads rate (%) | Total-map | Total-map rate (%) | Q20 | Q30 |
|---|---|---|---|---|---|---|---|
| S1 | 49640720 | 47460188 | 95.61 | 41564096 | 87.58 | 98.16 | 95.10 |
| S2 | 46547234 | 44737262 | 96.11 | 35866634 | 80.17 | 98.12 | 95.23 |
| S3 | 42787644 | 40724206 | 95.18 | 33787631 | 82.97 | 98.01 | 94.76 |
| B1 | 51865132 | 50973770 | 98.38 | 46109511 | 90.46 | 98.61 | 95.99 |
| B2 | 50052990 | 49099298 | 98.09 | 43891232 | 89.39 | 98.76 | 96.40 |
| B3 | 52370048 | 51467180 | 98.28 | 47125928 | 91.57 | 98.71 | 96.22 |
| Gene ID | Gene name | Gene annotation | log2 (FC) |
|---|---|---|---|
| cfu_9G0006470 | trim32 | E3 ubiquitin-protein ligase TRIM32 | 9.534 |
| cfu_15G0008560 | bloc-1 | Biogenesis of lysosome-related organelles complex 1 | 9.487 |
| cfu_16G0005300 | wwtr1 | WW domain-containing transcription regulator protein 1 | 8.941 |
| cfu_26G0005640 | scfd2 | Sec1 family domain-containing protein 2 | 8.833 |
| cfu_1G0009900 | nipsnap3a | Protein NipSnap homolog 3A | 8.720 |
| cfu_27G0004410 | camlg | Guided entry of tail-anchored proteins factor | 8.636 |
| cfu_10G0006000 | slc16a4 | Monocarboxylate transporter 5 | 8.621 |
| cfu_26G0004610 | atraid | All-trans retinoic acid-induced differentiation factor | 8.559 |
| cfu_27G0003260 | efemp2 | EGF-containing fibulin-like extracellular matrix protein 2 | 8.472 |
| cfu_5G0009050 | aadac | Arylacetamide deacetylase | 8.306 |
| cfu_3G0006820 | rims4 | Regulating synaptic membrane exocytosis protein 4 | -6.820 |
| cfu_9G0008290 | shc3 | SHC-transforming protein 3 | -6.933 |
| cfu_6G0004030 | lpa1 | High-affinity lysophosphatidic acid receptor | -6.988 |
| cfu_15G0007480 | nos2 | Nitric oxide synthase | -7.002 |
| cfu_3G0009150 | tcf23 | Transcription factor 23 | -7.015 |
| cfu_14G0005780 | hrh1 | Histamine H1 receptor | -7.032 |
| cfu_4G0003010 | apoc-1 | Apolipoprotein C-I | -7.104 |
| cfu_14G0006580 | chia | Acidic mammalian chitinase | -7.329 |
| cfu_11G0001850 | fgfbp2 | Fibroblast growth factor-binding protein 2 | -7.474 |
| cfu_22G0000210 | itln | Intelectin | -7.841 |
| Gene ID | Gene name | Gene annotation | log2 (FC) |
|---|---|---|---|
| cfu_6G0008310 | calcrla | calcitonin gene-related peptide type 1 receptorisoform X1 | 2.397 |
| cfu_6G0008340 | wdr75 | WD repeat-containing protein 75 | 1.554 |
| cfu_6G0008350 | asnsd1 | asparagine synthetase domain-containing protein 1 | -1.380 |
| cfu_6G0008380 | osgepl1 | probable tRNA N6-adenosine threonylcarbamoyltransferase | -1.102 |
| cfu_6G0008410 | pms1 | PMS1 protein homolog 1 | -1.197 |
| cfu_6G0008420 | mstnb | myostatin | -2.353 |
| cfu_6G0008440 | pofut2 | GDP-fucose protein O-fucosyltransferase 2 | 6.582 |
| cfu_6G0008510 | ikzf2 | IKAROS Family Zinc Finger 2 | -2.744 |
| cfu_6G0008520 | egfr | Epidermal Growth Factor Receptor | -2.210 |
| cfu_6G0008530 | cps1 | carbamoyl-phosphate synthase | -1.581 |
| cfu_9G0003920 | c9orf78 | Telomere length and silencing protein 1 homolog | -1.118 |
| cfu_9G0003990 | tecta | alpha-tectorin | 2.098 |
| cfu_9G0004000 | tbcel | Tubulin-specific chaperone cofactor E-like protein | 1.174 |
| cfu_9G0004050 | f11r | Junctional adhesion molecule A | 4.879 |
| cfu_9G0003810 | glipr2 | Golgi-associated plant pathogenesis-related protein 1 | -2.428 |
| cfu_9G0002330 | mks1 | Meckel syndrome type 1 protein | -1.804 |
| cfu_14G0007230 | qrich1 | glutamine-rich protein 1 | 1.404 |
| cfu_17G0007930 | atxn7l3 | ataxin-7protein 3 | -1.171 |
| cfu_17G0008220 | mto1 | protein MTO1 homolog | -2.294 |
| cfu_17G0008240 | tctn3 | tectonic-3 | -1.298 |
| cfu_17G0008260 | sult1c1 | sulfotransferase family cytosolic 1B member 1 | -1.707 |
| cfu_17G0008270 | atp2a1 | sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | 1.895 |
| cfu_17G0008530 | ccser2 | serine-rich coiled-coil domain-containing protein 2 | 2.420 |
| cfu_18G0007570 | lca5 | lebercilin | 7.172 |
| cfu_18G0007610 | eya4 | eyes absent homolog 4 isoform X2 | 1.161 |
| cfu_20G0000670 | tlx3 | T-cell leukemia homeobox protein 3 | -6.172 |
| cfu_20G0000680 | npm1 | nucleophosmin | 1.503 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





