Submitted:
23 January 2025
Posted:
24 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Genotyping
2.3. RNA Extraction and cDNA Synthesis
2.4. Quantitative Real-Time PCR
2.5. Analysis of Gene Expression Data
3. Results
3.1. six6 and vgll3 Genotypes
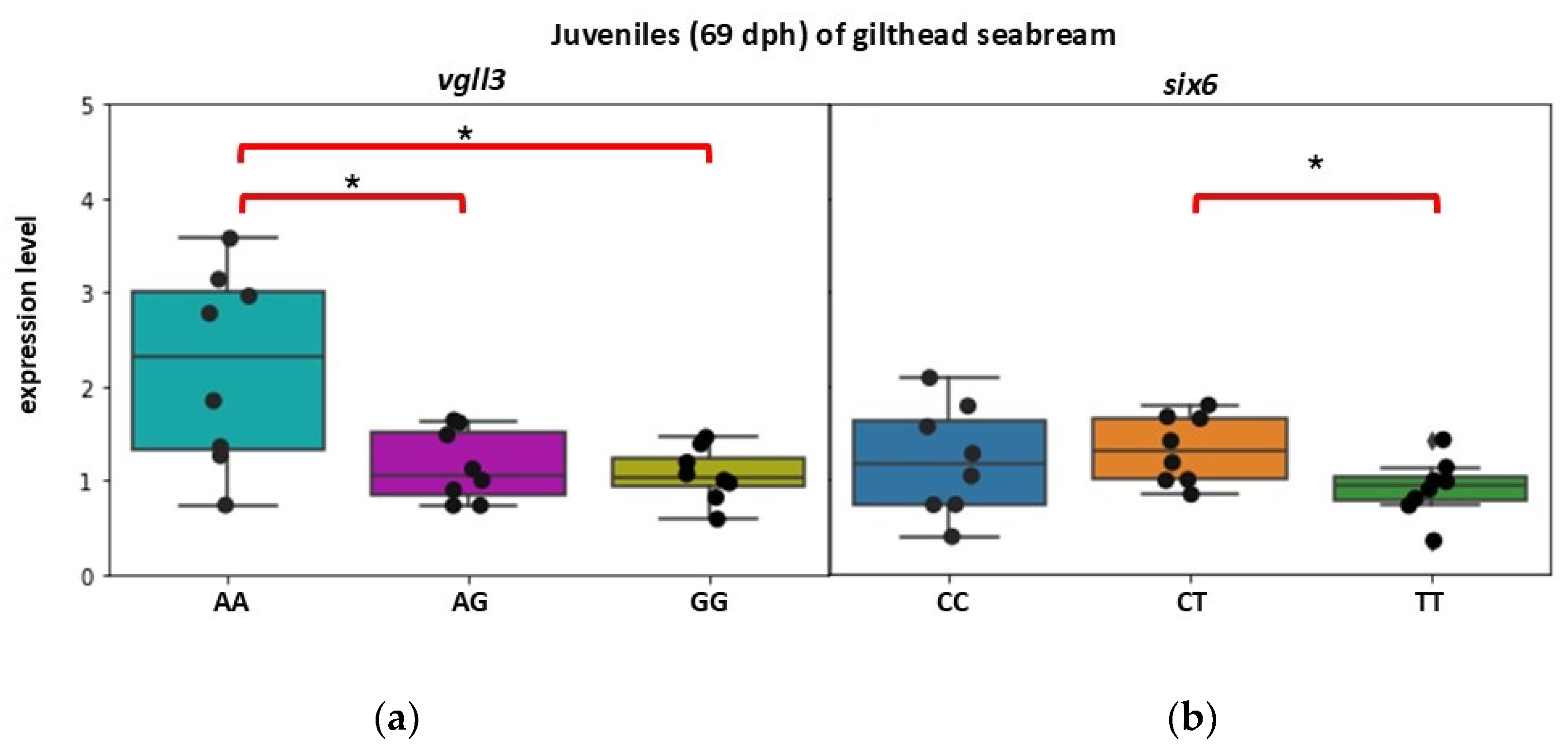
3.2. Expression Patterns of vgll3 and six6 per Genotype in Gilthead Seabream
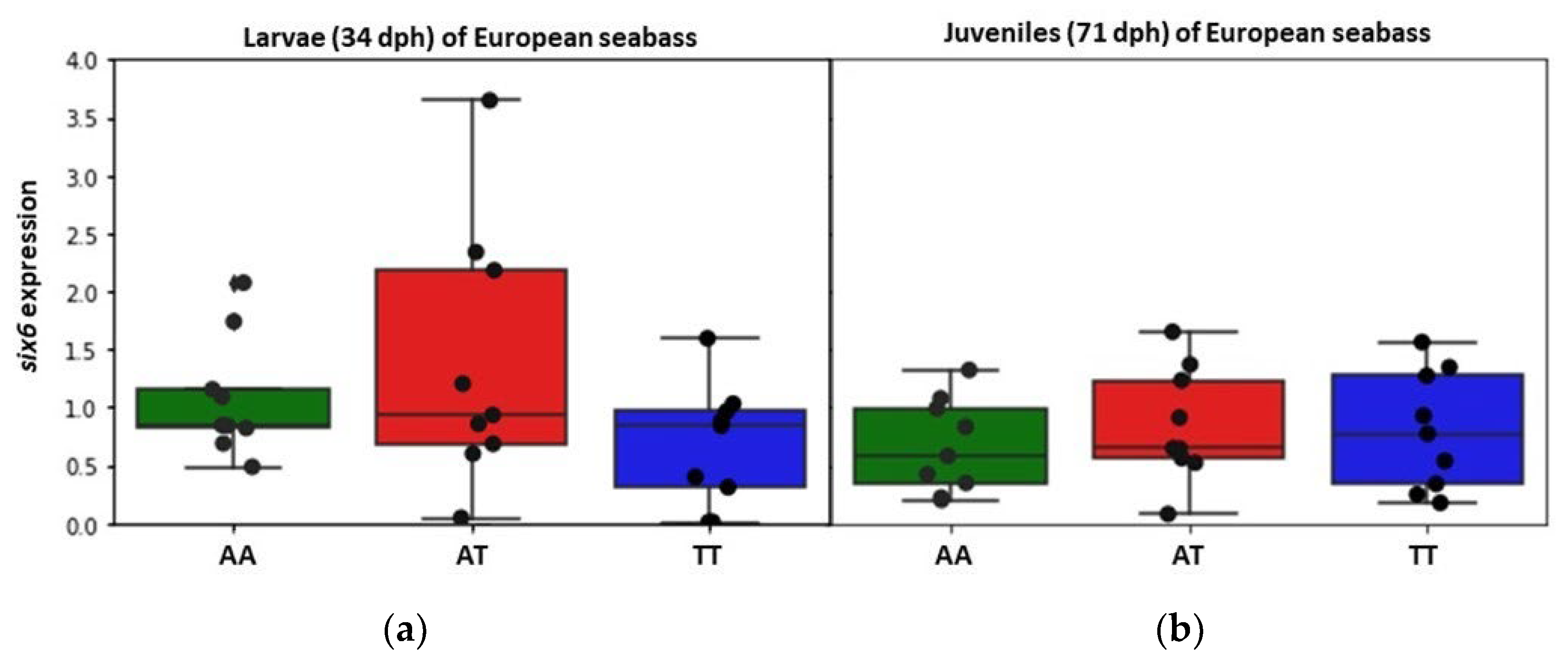
3.3. Expression Patterns of six6 per Genotype and Developmental Stage in European Seabass
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cousminer, D.L.; Berry, D.J.; Timpson, N.J.; Ang, W.; Thiering, E.; Byrne, E.M.; Rob Taal, H.; Huikari, V.; Bradfield, J.P.; Kerkhof, M.; et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet. 2013, 22, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.R.B.; Day, F.; Elks, C.E.; Sulem, P.; Thompson, D.J.; Ferreira, T.; He, C.; Chasman, D.I.; Esko, T.; Thorleifsson, G.; et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 2014, 514, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, A.; Reverter, A.; DeAtley, K.L.; Ashley, R.L.; Colgrave, M.L.; Fortes, M.R.S.; Islas-Trejo, A.; Lehnert, S.; Porto-Neto, L.; Rincón, G.; et al. Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLoS One 2014, 9, e102551. [Google Scholar] [CrossRef]
- Pennonen, J. Characterization of human pubertal timing gene vgll3 in zebrafish development. Master thesis, Degree Programme in Molecular Biosciences, Department of Biosciences, Faculty of Environmental and Biological Sciences, University of Helsinki, Finland, 2017.
- Barson, N.J.; Aykanat, T.; Hindar, K.; Baranski, M.; Bolstad, G.H.; Fiske, P.; Jacq, C.; Jensen, A.J.; Johnston, S.E.; Karlsson, S.; et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 2015, 528, 405–408. [Google Scholar] [CrossRef]
- Ayllon, F.; Kjærner-Semb, E.; Furmanek, T.; Wennevik, V.; Solberg, M.F.; Dahle, G.; Taranger, G.L.; Glover, K.A.; Almén, M.S.; Rubin, C.J.; et al. The vgll3 locus controls age at maturity in wild and domesticated Atlantic salmon (Salmo salar L.) males. PLoS Genet. 2015, 11, 1–15. [Google Scholar] [CrossRef]
- Sinclair-Waters, M.; Ødegård, J.; Korsvoll, S.A.; Moen, T.; Lien, S.; Primmer, C.R.; Barson, N.J. Beyond large-effect loci: large-scale GWAS reveals a mixed large-effect and polygenic architecture for age at maturity of Atlantic salmon. Genet. Sel. Evol. 2020, 52, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.D.; Clemento, A.; Aykanat, T.; Garza, J.C.; Naish, K.A.; Narum, S.; Primmer, C.R. Heterogeneous genetic basis of age at maturity in salmonid fishes. Mol. Ecol. 2021, 30, 1435–1456. [Google Scholar] [CrossRef] [PubMed]
- Kurko, J.; Debes, P. V.; House, A.H.; Aykanat, T.; Erkinaro, J.; Primmer, C.R. Transcription profiles of age-at-maturity-associated genes suggest cell fate commitment regulation as a key factor in the Atlantic salmon maturation process. G3 Genes, Genomes, Genet. 2020, 10, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Kjærner-Semb, E.; Ayllon, F.; Kleppe, L.; Sørhus, E.; Skaftnesmo, K.; Furmanek, T.; Segafredo, F.T.; Thorsen, A.; Fjelldal, P.G.; Hansen, T.; et al. Vgll3 and the Hippo pathway are regulated in Sertoli cells upon entry and during puberty in Atlantic salmon testis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Hori, N.; Okada, K.; Takakura, Y.; Takano, H.; Yamaguchi, N.; Yamaguchi, N. Vestigial-like family member 3 (VGLL3), a cofactor for TEAD transcription factors, promotes cancer cell proliferation by activating the hippo pathway. J. Biol. Chem. 2020, 295, 8798–8807. [Google Scholar] [CrossRef] [PubMed]
- Ahi, E.P.; Sinclair-Waters, M.; Moustakas-Verho, J.; Jansouz, S.; Primmer, C.R. Strong regulatory effects of vgll3 genotype on reproductive axis gene expression in juvenile male Atlantic salmon. Gen. Comp. Endocrinol. 2022, 325, 114055. [Google Scholar] [CrossRef]
- Debes, P. V.; Piavchenko, N.; Ruokolainen, A.; Ovaskainen, O.; Moustakas-Verho, J.E.; Parre, N.; Aykanat, T.; Erkinaro, J.; Primmer, C.R. Polygenic and major-locus contributions to sexual maturation timing in Atlantic salmon. Mol. Ecol. 2021, 30, 4505–4519. [Google Scholar] [CrossRef]
- House, A.H.; Debes, P. V.; Holopainen, M.; Käkelä, R.; Donner, I.; Frapin, M.; Pashay, E.; Kurko, J.; Ruhanen, H.; Primmer, C.R. Seasonal and genetic effects on lipid profiles of juvenile Atlantic salmon. bioRxiv 2023, 1870, 2023.02–22.529528. [Google Scholar] [CrossRef]
- Gjedrem, T.; Baranski, M. Domestication and the application of genetic improvement in aquaculture. In Selective Breeding in Aquaculture: An Introduction; Springer: Dordrecht, The Netherlands, 2009; Volume 10, pp. 5–11. [Google Scholar] [CrossRef]
- Gutierrez, A.P.; Yáñez, J.M.; Davidson, W.S. Evidence of recent signatures of selection during domestication in an Atlantic salmon population. Mar. Genomics 2016, 26, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Houston, R.D. Future directions in breeding for disease resistance in aquaculture species. Rev. Bras. Zootec. 2017, 46, 545–551. [Google Scholar] [CrossRef]
- Moulistanos, A.; Nikolaou, T.; Sismanoglou, S.; Gkagkavouzis, K.; Karaiskou, N.; Antonopoulou, E.; Triantafyllidis, A.; Papakostas, S. Investigating the role of genetic variation in vgll3 and six6 in the domestication of gilthead seabream (Sparus aurata Linnaeus) and European seabass (Dicentrarchus labrax Linnaeus). Ecol. Evol. 2023, 13, 1–17. [Google Scholar] [CrossRef]
- Mobley, K.B.; Aykanat, T.; Czorlich, Y.; House, A.; Kurko, J.; Miettinen, A.; Moustakas-Verho, J.; Salgado, A.; Sinclair-Waters, M.; Verta, J.P.; Primmer, C.R. Maturation in Atlantic salmon (Salmo salar, Salmonidae): A synthesis of ecological, genetic, and molecular processes. Rev Fish Biol Fisheries 2021, 31, 523–571. [Google Scholar] [CrossRef]
- Albert, F.W.; Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015, 16, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Aguet, F.; Brown, A.A.; Castel, S.E.; Davis, J.R.; He, Y.; Jo, B.; Mohammadi, P.; Park, Y.S.; Parsana, P.; Segrè, A. V.; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.J.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using trizol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Bartlett, M.S. Properties of Sufficiency and Statistical Tests. Proc. R. Soc. Lond. A. Math. Phys. Sci. 1937, 160, 268–282. [Google Scholar] [CrossRef]
- Cao, H.; Shockey, J.M. Comparison of TaqMan and SYBR Green QPCR methods for quantitative gene expression in tung tree tissues. J. Agric. Food Chem. 2012, 60, 12296–12303. [Google Scholar] [CrossRef]
- Kesmen, Z.; Gulluce, A.; Sahin, F.; Yetim, H. Identification of meat species by TaqMan-based real-time PCR assay. Meat Sci. 2009, 82, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Fraser, T.W.K.; Hansen, T.J.; Fjelldal, P.G. Environmental and genetic (Vgll3) effects on the prevalence of male maturation phenotypes in domesticated Atlantic salmon. Fishes 2023, 8. [Google Scholar] [CrossRef]
- Fraser, T.W.K.; Tjølsen, A.; Madaro, A.; Hansen, T.J.; Fjelldal, P.G. Vgll3 regulates the timing of puberty in farmed Atlantic salmon, but it does not explain family discordance in male maturation following different smolt production regimes. Aquaculture 2024, 593. [Google Scholar] [CrossRef]
- Moustakas-Verho, J.E.; Kurko, J.; House, A.H.; Erkinaro, J.; Debes, P.; Primmer, C.R. Developmental expression patterns of six6: A gene linked with spawning ecotypes in Atlantic salmon. Gene Expr. Patterns 2020, 38, 119149. [Google Scholar] [CrossRef]
- Chavanne, H.; Janssen, K.; Hofherr, J.; Contini, F.; Haffray, P.; Aquatrace Consortium; Komen, H.; Nielsen, E.E.; Bargelloni, L. A comprehensive survey on selective breeding programs and seed market in the European aquaculture fish industry. Aquac. Int. 2016, 24, 1287–1307. [CrossRef]
- Yue, G.H. Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish. 2014, 15, 376–396. [Google Scholar] [CrossRef]
- Verta, J.-P.; Debes, P.V.; Piavchenko, N.; Ruokolainen, A.; Ovaskainen, O.; Moustakas-Verho, J.E.; Tillanen, S.; Parre, N.; Aykanat, T.; Erkinaro, J.; Primmer, C.R. Cis-regulatory differences in isoform expression associate with life history strategy variation in Atlantic salmon. PLoS Genet. 2020, 16, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ahi, E.P.; Verta, J.P.; Kurko, J.; Ruokolainen, A.; Singh, P.; Debes, P.V.; Erkinaro, J.; Primmer, C.R. Gene co-expression patterns in Atlantic salmon adipose tissue provide a molecular link among seasonal changes, energy balance and age at maturity. Mol. Ecol. 2024, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Pelletier, J. Exploring the impact of single-nucleotide polymorphisms on translation. Front. Genet. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kaitetzidou, E.; Xiang, J.; Antonopoulou, E.; Tsigenopoulos, C.S.; Sarropoulou, E. Dynamics of gene expression patterns during early development of the European seabass (Dicentrarchus labrax). Physiol. Genomics 2015, 47, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Kaitetzidou, E.; Ludwig, A.; Gessner, J.; Sarropoulou, E. Expression patterns of Atlantic sturgeon (Acipenser oxyrinchus) during embryonic development. G3 (Bethesda) 2017, 7, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Melo, D.; Garske, K.M.; Pallares, L.F.; Lea, A.J.; Ayroles, J.F. Characterizing the landscape of gene expression variance in humans. PLoS Genet. 2023, 19, e1010833. [Google Scholar] [CrossRef] [PubMed]
- De Jong, T. V.; Moshkin, Y.M.; Guryev, V. Gene expression variability: The other dimension in transcriptome analysis. Physiol. Genomics 2019, 51, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Papakostas, S.; Vasemägi, A.; Himberg, M.; Primmer, C.R. Proteome variance differences within populations of European whitefish (Coregonus lavaretus) originating from contrasting salinity environments. J. Proteomics 2014, 105, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Piferrer, F.; Miska, E.A.; Anastasiadi, D. Chapter 10 - Epigenetics in fish evolution. In On Epigenetics and Evolution; Guerrero-Bosagna, C.M., Ed.; Translational Epigenetics; Academic Press, 2024; pp. 283–306 ISBN 978-0-443-19051-3.
- Konstantinidis, I.; Sætrom, P.; Mjelle, R.; Nedoluzhko, A. V.; Robledo, D.; Fernandes, J.M.O. Major gene expression changes and epigenetic remodelling in Nile tilapia muscle after just one generation of domestication. Epigenetics 2020, 15, 1052–1067. [Google Scholar] [CrossRef]
- Moulistanos, A.; Papasakellariou, K.; Kavakiotis, I.; Gkagkavouzis, K. Genomic signatures of domestication in European seabass (Dicentrarchus labrax L.) reveal a potential role for epigenetic regulation in adaptation to captivity. Ecol. Evol. [CrossRef]
- Oelschlaeger, P. Molecular mechanisms and the significance of synonymous mutations. Biomolecules 2024, 14. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Ray, S.K.; Banerjee, R. Synonymous codons influencing gene expression in organisms. Res. Reports Biochem. 2016, 6, 57–65. [Google Scholar] [CrossRef]
- Chen, R.; Davydov, E. V.; Sirota, M.; Butte, A.J. Non-synonymous and synonymous coding snps show similar likelihood and effect size of human disease association. PLoS One 2010, 5, e13574. [Google Scholar] [CrossRef]
- Campbell, T.M.; Castro, M.A.A.; de Santiago, I.; Fletcher, M.N.C.; Halim, S.; Prathalingam, R.; Ponder, B.A.J.; Meyer, K.B. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis 2016, 37, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.B.; Brown, S.J.; Cole, M.D. Upregulation of c-myc in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell. Biol. 2010, 30, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Blinova, E.A.; Nikiforov, V.S.; Yanishevskaya, M.A.; Akleyev, A.А. Single nucleotide polymorphism and expression of genes for immune competent cell proliferation and differentiation in radiation-exposed individuals. Vavilovskii Zhurnal Genet. Selektsii 2020, 24, 399–406. [Google Scholar] [CrossRef]
- Verta, J.P.; Jacobs, A. The role of alternative splicing in adaptation and evolution. Trends Ecol. Evol. 2022, 37, 299–308. [Google Scholar] [CrossRef]
| Species | Gene | Genomic coordinates | Annealing temperature | Primer Sequence (5’-3’) | Amplicon size (bp) |
|---|---|---|---|---|---|
| D. labrax | vgll3 | 9203305 – 9203951 | 63°C | TACCCTCCCCGATACCTGG | 646 |
| TGTGTGGACAGTGCAGGAC | |||||
| six6 | 11590632 - 11591488* | 63°C | GGCTACAGGACTTACACCCA | 856 | |
| AAGTACCACAGCAAGATCGC | |||||
| S. aurata | vgll3 | 24911798 - 24912208 * | 63°C | AACGTCTATCACCCTCACCC | 410 |
| ACCAAACTGACGTCTTTGCT | |||||
| six6 | 14406022 - 14406799 * | 63°C | AACCGCAGACAAAGAGACAG | 777 | |
| ACCCCTTATTAAACAACAAGCAC |
| Species | Gene | Annealing temperature | Primers and Probe Sequence (5’-3’) (*probe sequence) |
Amplicon size (bp) |
|---|---|---|---|---|
| D. labrax | six6 | 60°C | GCCTTTCACACCGGCAATTTC | 169 |
| TCCGCACCCTGTATTTGTCCAC | ||||
| TGGAGAACCACAAGTTCACCAAAGAGTCGCAC* | ||||
| rpl13 | 61°C | GACACAAAGTGGTGGTGGTGAG | 130 | |
| AAGTGGTAAGGTCCACGAGAGG | ||||
| AGGCATCAACATCTCCGGCAACTTCTATCGCA* | ||||
| fau | 61°C | TCGGCCCTTAAACAGCCCTTTC | 190 | |
| GCAACAGCACCTGATCCTCAAC | ||||
| CGAGCATCAAGATGCAGCTCTTCTTGCGTGCC* | ||||
| ef1α | 61°C | AGCAGACAACTTCAACGCCC | 156 | |
| TCAAGCTTCTTGCCAGAACGAC | ||||
| CCACACAGCCCACATCGCCTGCAAGTTC* | ||||
| S. aurata | vgll3 | 56°C | TCGATTCAAGCCGGAGATCCAC | 119 |
| GCCAATGTCGCCTTGGAAGTAG | ||||
| TCCGATTCAGAGCTGAAGGACGGCACTCA* | ||||
| six6 | 56°C | CCGCGAACTTTACCACATCCTG | 149 | |
| TCCGCACCCTGTATTTGTCCAC | ||||
| ACCACAAGTTCACCAAAGAGTCGCACACGAAGCTG* | ||||
| rpl13 | 58°C | GTTCAACCAGCCAGCCAGAAAG | 116 | |
| TGGTGGGACATCTGACTTGTGG | ||||
| CGTCAGGCTAAGGCCCGTCGCATTGCTC* | ||||
| fau | 56°C | TGTCCCATGCGGCTAGTTATGC | 108 | |
| AGTGTTCTGGGCACGCAAGAAG | ||||
| AGCTCCCGGCACGTTGCTGTCCTC* | ||||
| ef1α | 56°C | ACTCCACCGAGCCAAACTACAG | 117 | |
| CCGAAATGGGCACAAAGGCAAC | ||||
| AGTGAGCACCTACATCAAGAAGATCGGCTACAATCC* |
| Species | Developmental stage | Gene | Number of individuals for each genotype | ||
| D. labrax | Larvae | six6 | AA (10) | AT (31) | TT (9) |
| Juveniles | sixi6 | AA (9) | AT (23) | TT (17) | |
| S. aurata | Juveniles | six6 | CC (8) | CT (17) | TT (25) |
| Juveniles | vgll3 | AA (8) | AG (30) | GG (12) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





