Submitted:
07 May 2025
Posted:
07 May 2025
You are already at the latest version
Abstract
Keywords:
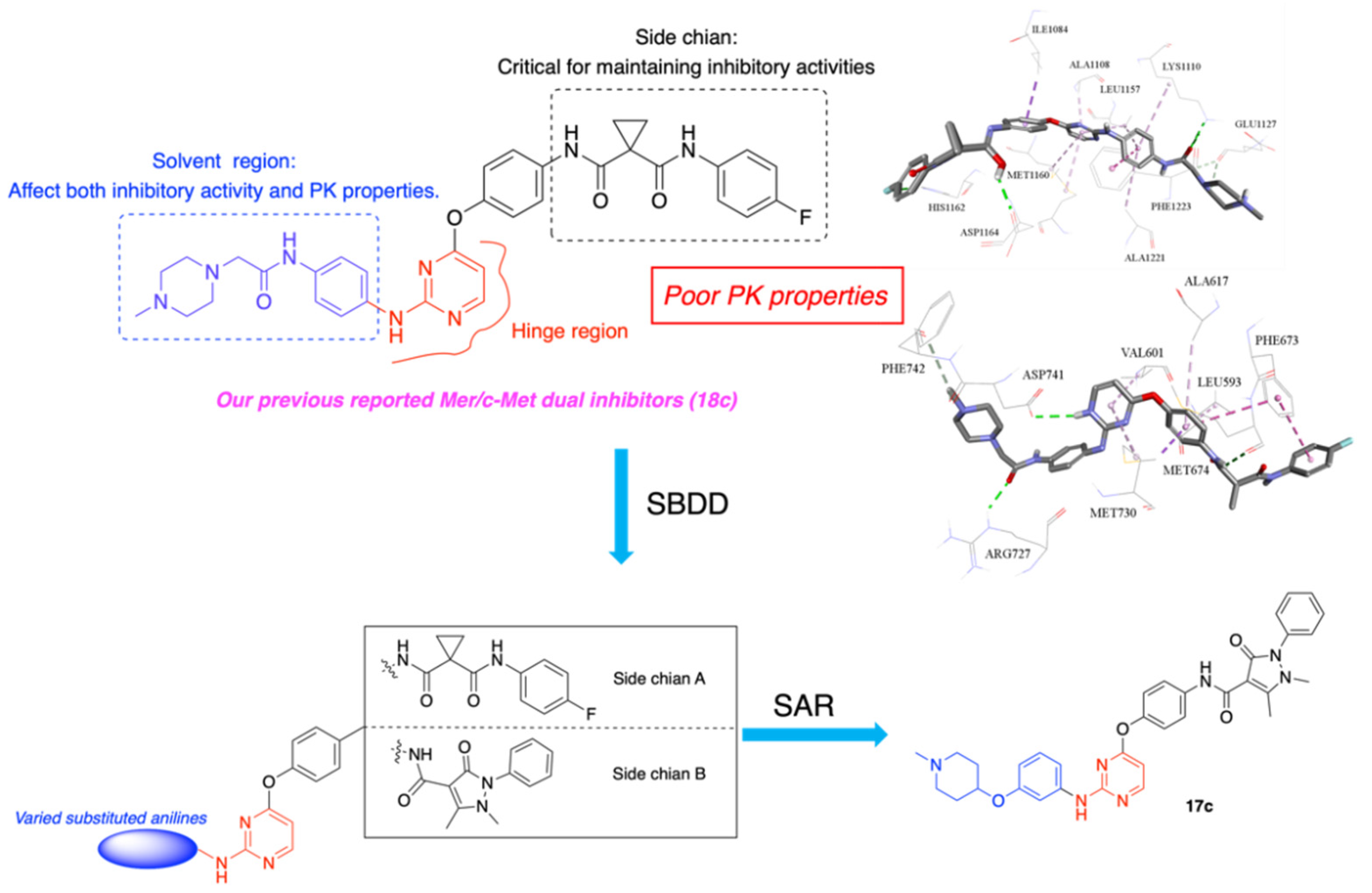
1. Introduction
2. Results and Discussion
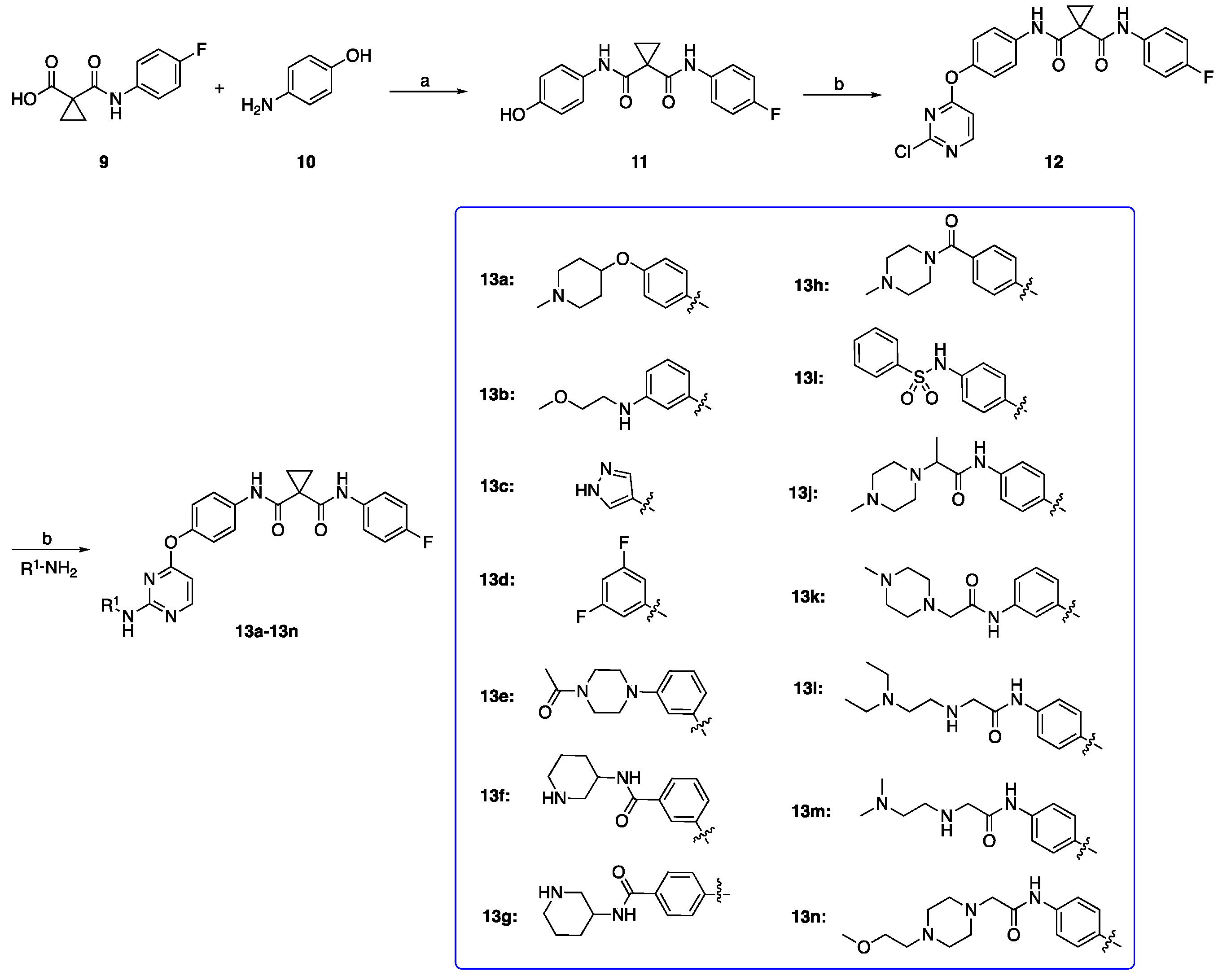
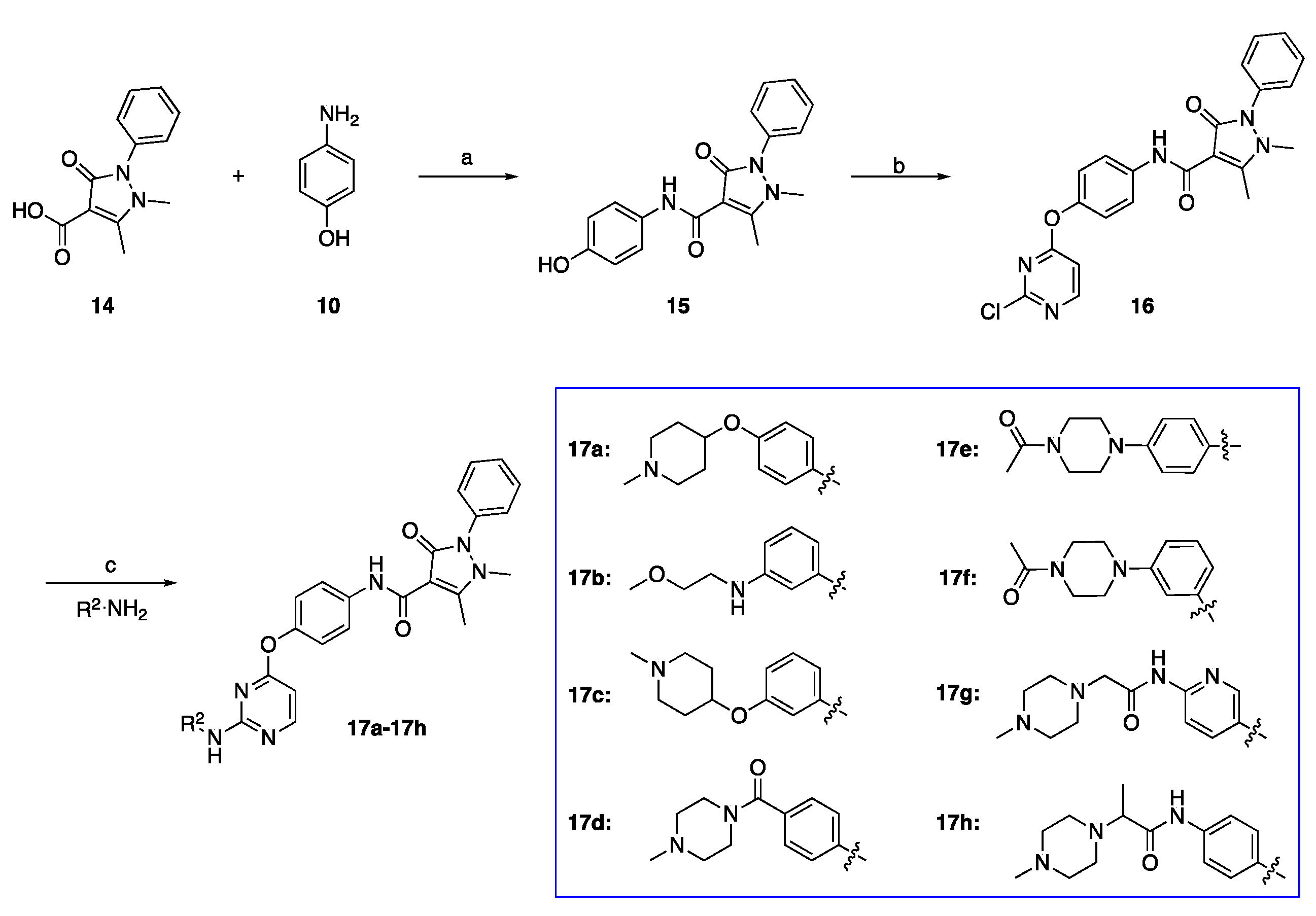
2.1. Chemistry
2.2. Kinase Inhibitory Activities
2.3. In Vitro Liver Microsomal Stability
2.4. Antiproliferation Assay In Vitro
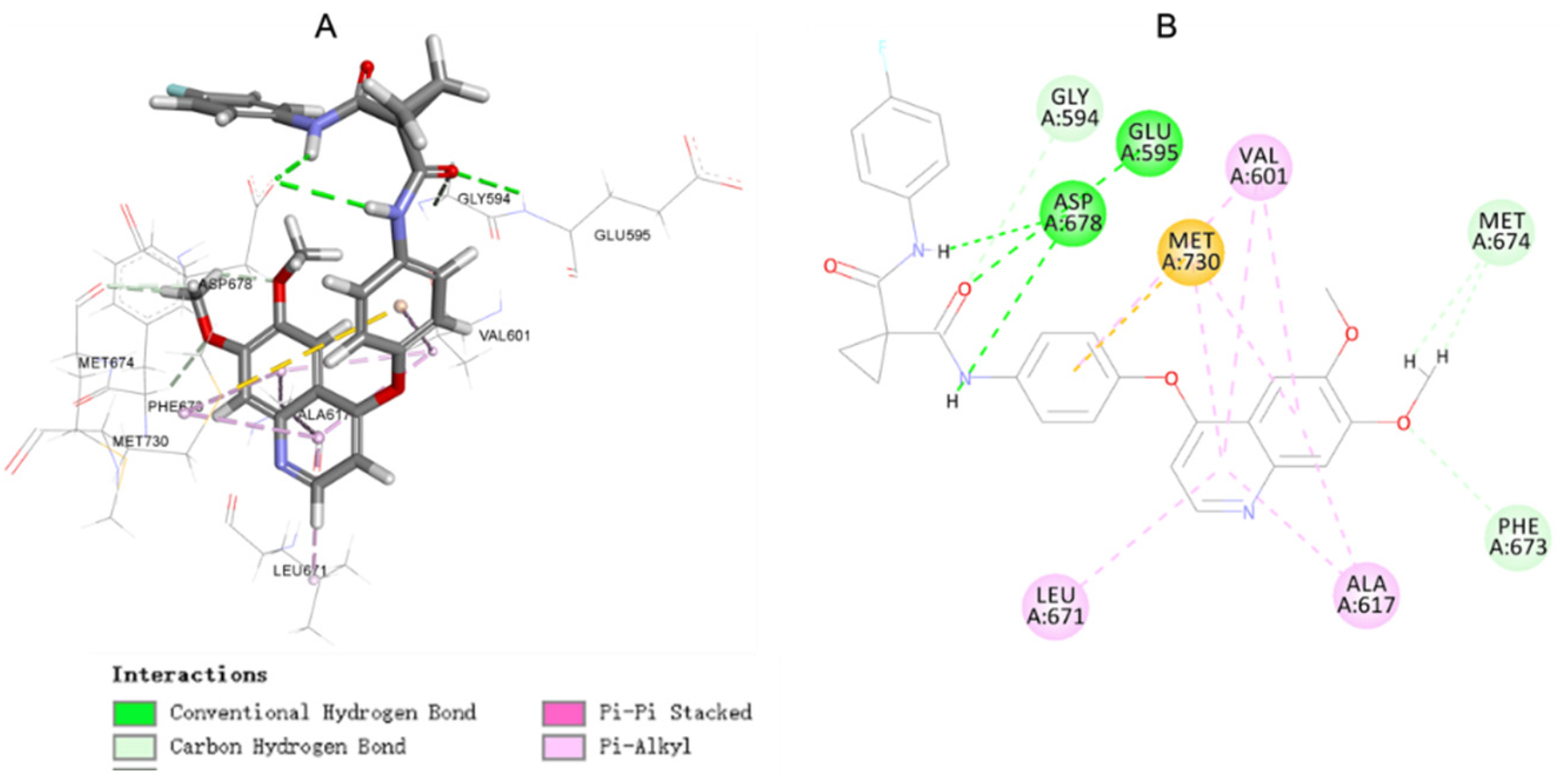
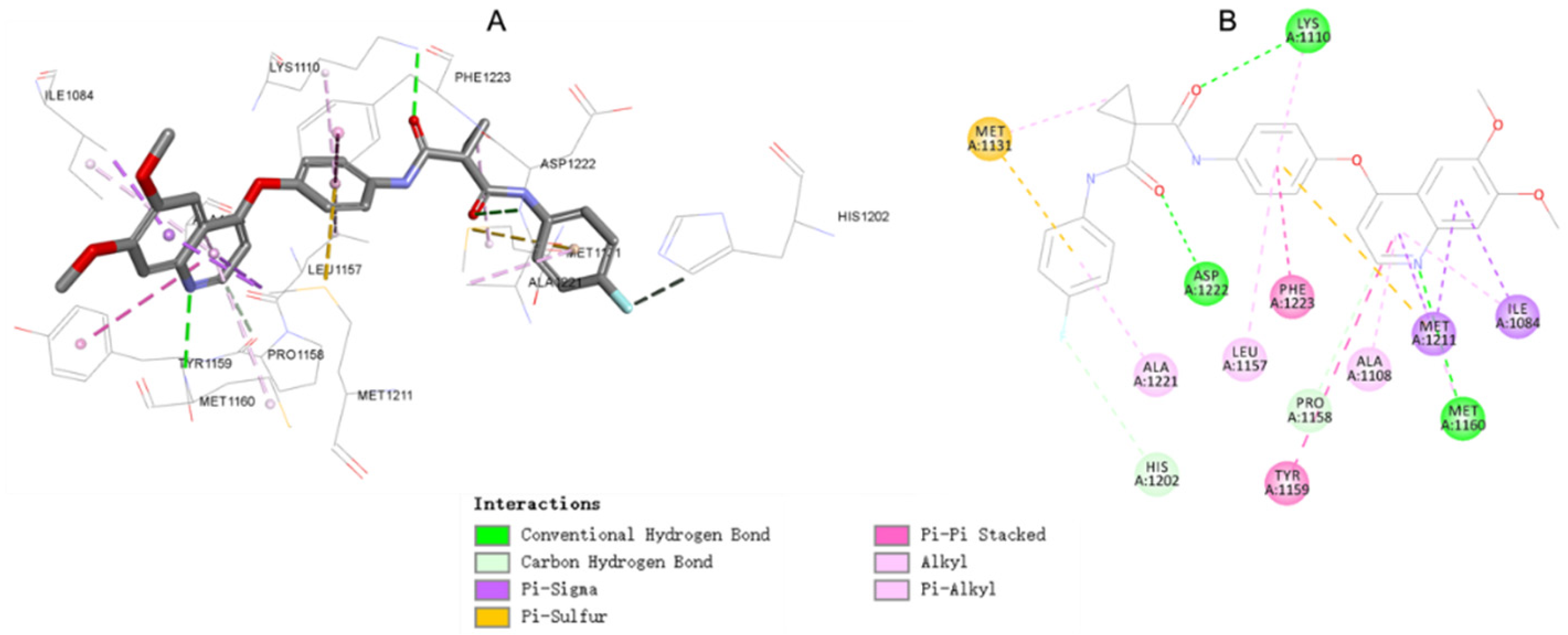
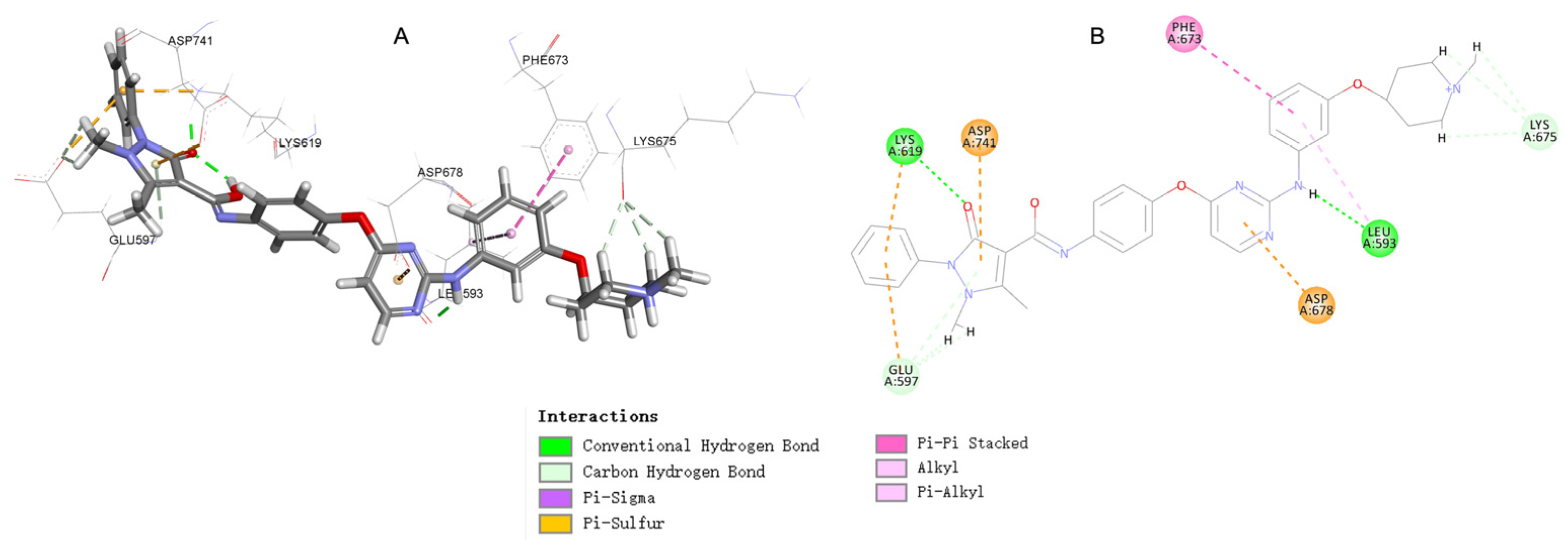
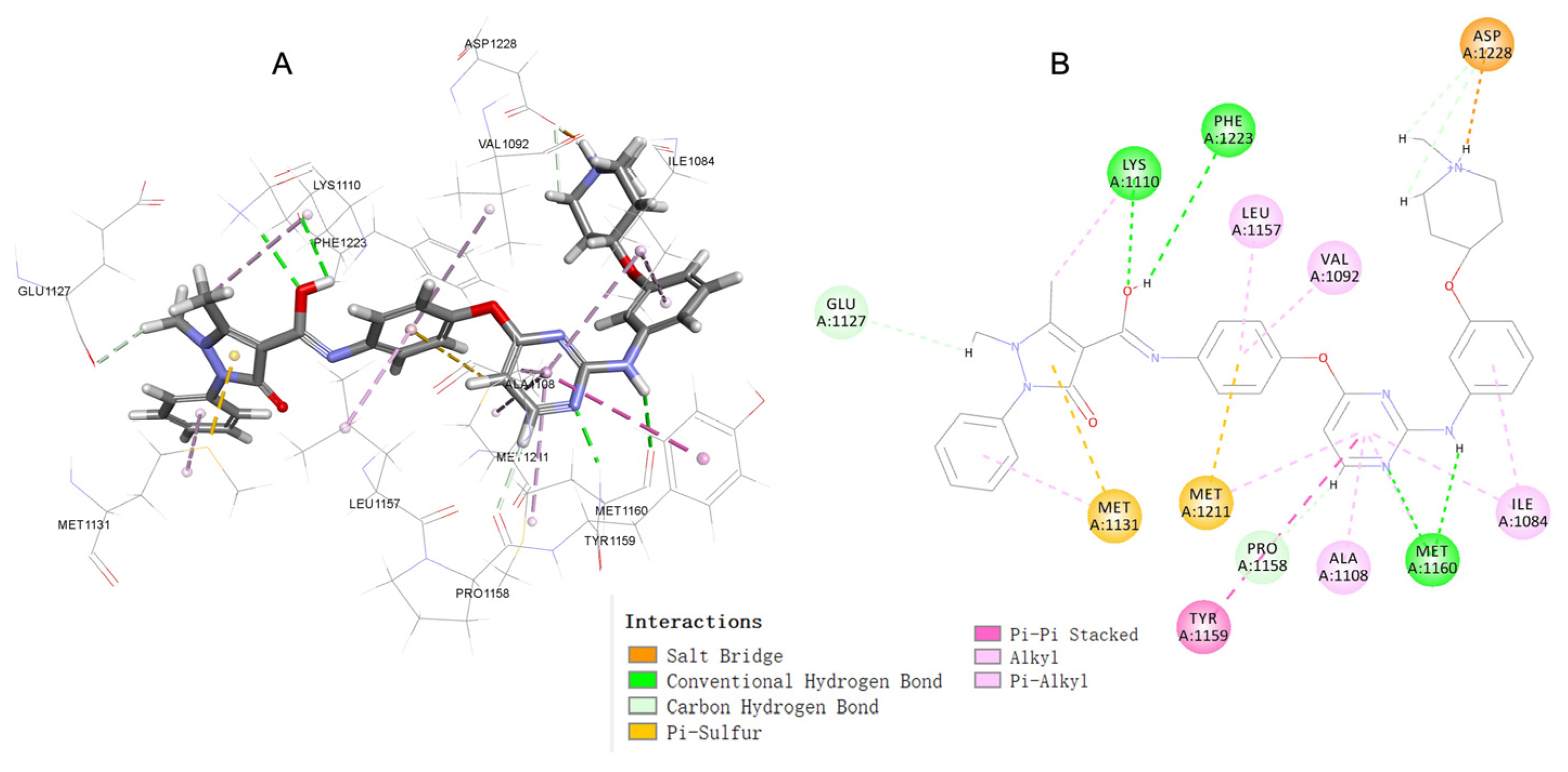
2.5. Molecular Docking Study of Compound 17c
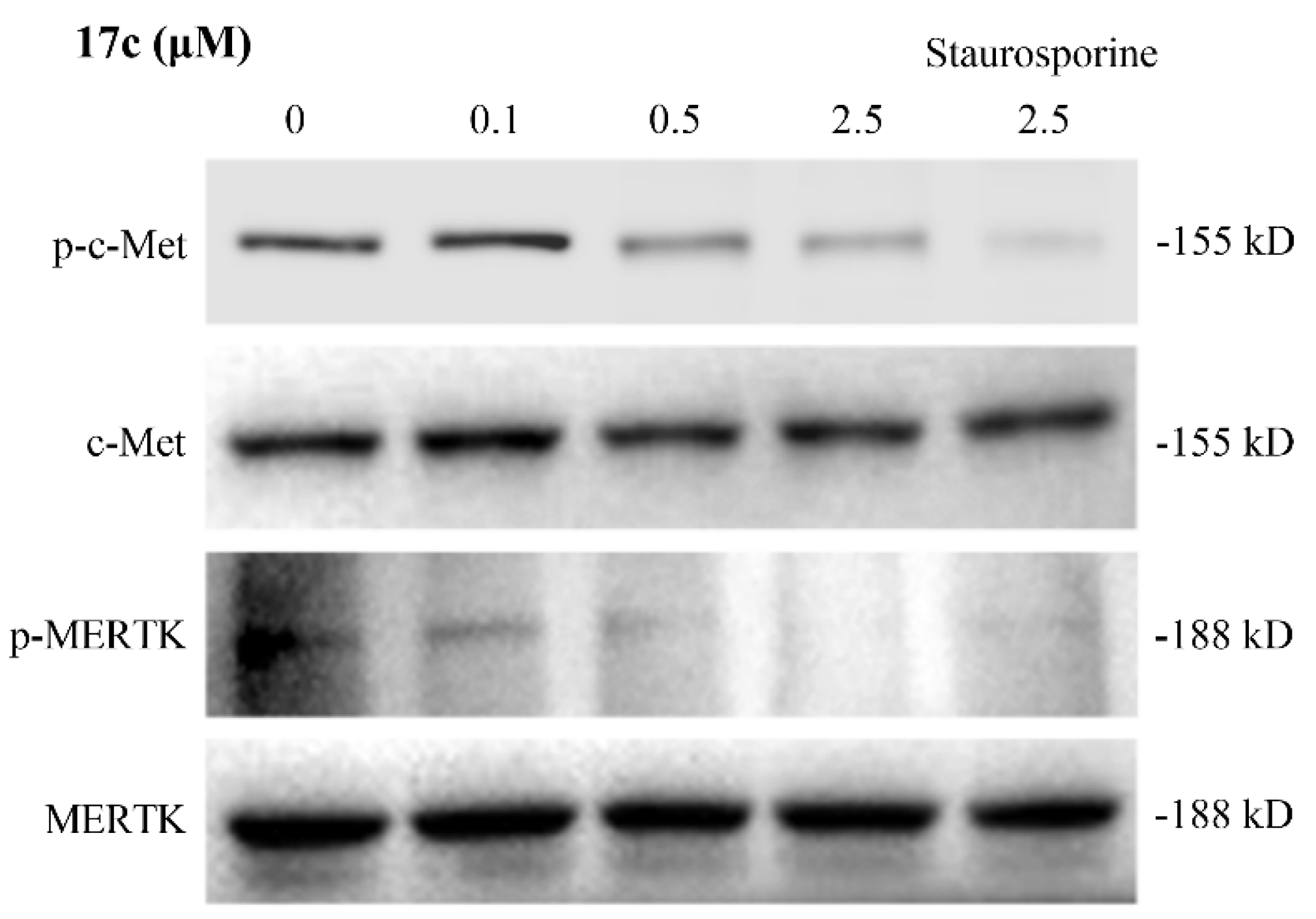
2.6. Western Blot Assay
2.7. The hERG Test
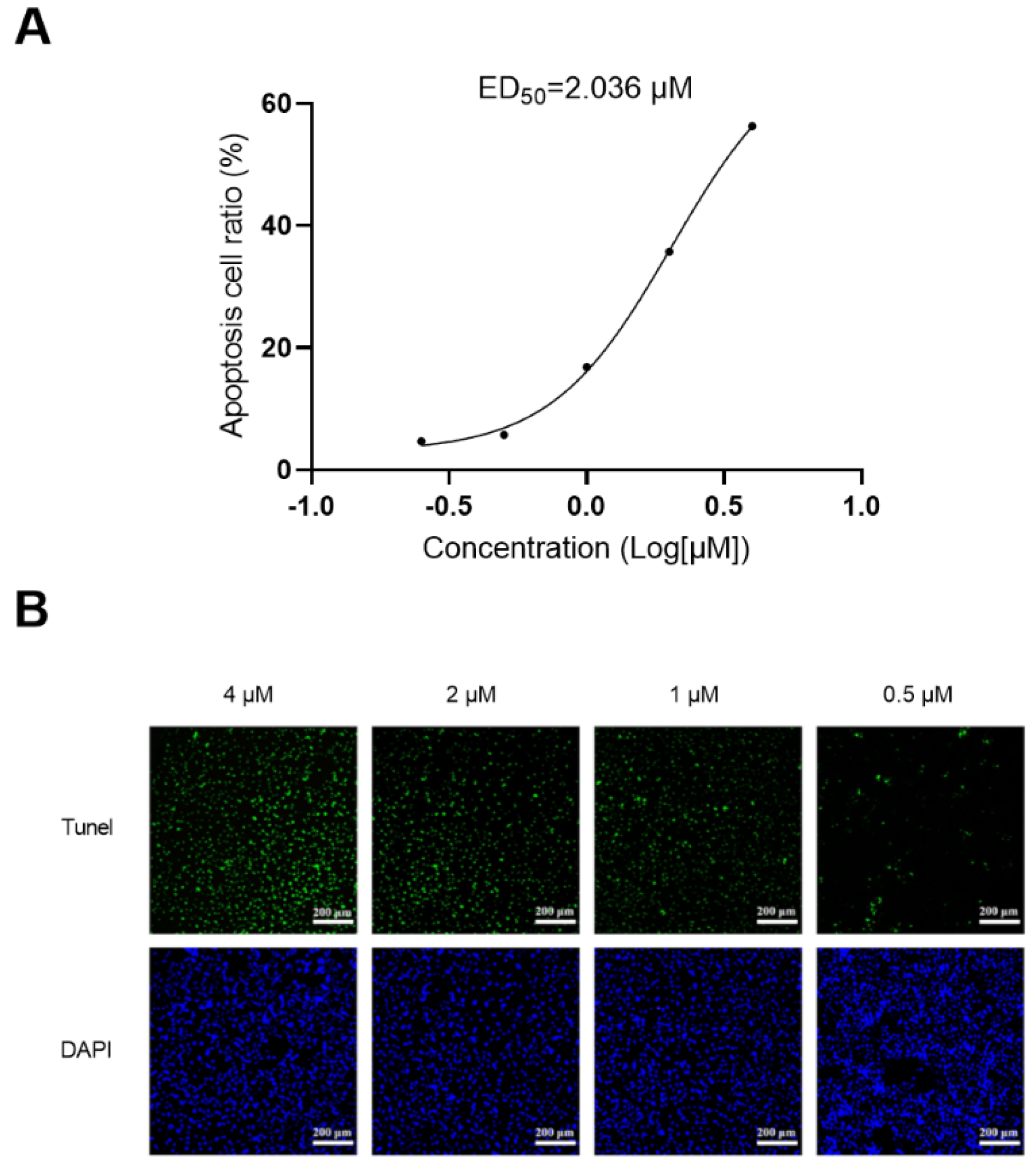
2.8. Apoptosis Assay
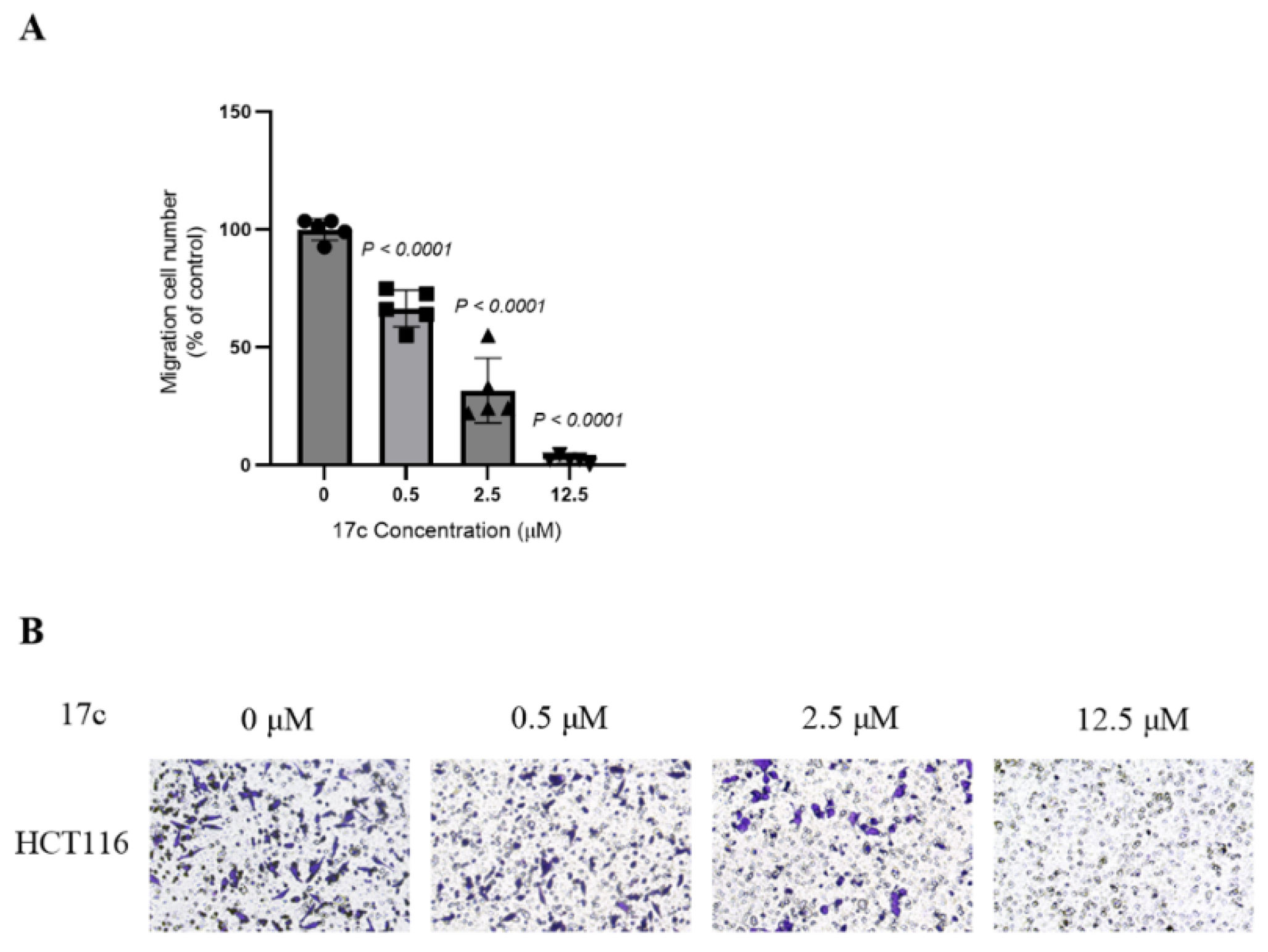
2.9. Transwell Assay
2.10. Plasma Protein Binding Affinity
2.11. Pharmacokinetic Properties
3. Materials and Methods
3.1. Chemical Part
3.1.1. Preparation of N-(4-Fluorophenyl)-N-(4-hydroxyphenyl)cyclopropane-1,1-dicarboxamide (Intermediate 11)
3.1.2. Preparation of N-(4-((2-Chloropyrimidin-4-yl)oxy)phenyl)-N-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide (Intermediate 12)
3.1.3. General Procedure for Preparation of Target Compounds 13a–13n
3.1.4. Preparation of N-(4-Hydroxyphenyl)-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide (Intermediate 15)
3.1.5. Preparation of N-(4-((2-Chloropyrimidin-4-yl)oxy)phenyl)-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide (Intermediate 16)
3.1.6. General Procedure for Preparation of Target Compounds 17a–17h
3.2. Mer and c-Met Inhibitory Activity Assay
3.3. Antiproliferation Assay
3.4. hERG Potassium Currents Assay
3.5. Liver Microsome Stability Assay
3.6. Docking Study
3.7. Western Blot Assay
3.8. Apotosis Study
3.9. Transwell Assay
3.10. Plasma Protein Binding in Plasma
3.11. Pharmacokinetic Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, X.; Frye, S. Chapter Nineteen—Mer Receptor Tyrosine Kinase: Therapeutic Opportunities in Oncology, Virology, and Cardiovascular Indications. Annu. Rep. Med. Chem. 2014, 49, 301–314. [Google Scholar] [CrossRef]
- Graham, D.K.; DeRyckere, D.; Davies, K.D.; Earp, H.S. The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer 2014, 14, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yi, M.; Qin, S.; Wu, K.M. Next generation chimeric antigen receptor T cells: safety strategies to overcome toxicity. Mol. Cancer. 2019, 18, 125. [Google Scholar] [CrossRef] [PubMed]
- Smart, S.; Vasileiadi, E.; Wang, X. The Emerging Role of TYRO3 as a Therapeutic Target in Cancer. Cancers (Basel) 2018, 10, 474. [Google Scholar] [CrossRef]
- Baladi, T.; Abet, V.; Piguel, S. State-of-the-art of small molecule inhibitors of the TAM family: the point of view of the chemist. Eur. J. Med. Chem. 2015, 105, 220–237. [Google Scholar] [CrossRef]
- Davra, V.; Kumar, S.; Geng, K.; Calianese, D.; Mehta, D.; Gadiyar, V.; Kasikara, C.; Lahey, K.C.; Chang, Y.J.; Wichroski, M.; Gao, C.; Lorenzo, M.S.D.; Kotenko, S.V.; Bergsbaken, T.; Mishra, P.; Gause, W.C.; Quigley, M.; Spires, T.E.; Birge, R.B. Axl and Mertk Receptors Cooperate to Promote Breast Cancer Progression by Combined Oncogenic Signaling and Evasion of Host Antitumor Immunity. Cancer Res. 2021, 81, 698–712. [Google Scholar] [CrossRef]
- Park, H.J.; Baen, J.Y.; Lee, Y.J.; Choi, Y.H.; Kang, J.L. The TAM-family receptor Mer mediates production of HGF through the RhoA-dependent pathway in response to apoptotic cells. Mol. Biol. Cell 2012, 23, 3254–3265. [Google Scholar] [CrossRef]
- Nguyen, K.Q.; Tsou, W.I.; Kotenko, S.; Birge, R.B. TAM receptors in apoptotic cell clearance, autoimmunity, and cancer. Autoimmunity 2013, 46, 294–297. [Google Scholar] [CrossRef]
- Sayama, A.; Okado, K.; Yamaguchi, M.; Samata, N.; Imaoka, M.; Kai, K.; Mori, K. The impact of the timing of dosing on the severity of UNC569-induced ultrastructural changes in the mouse retina. Toxicol. Pathol. 2020, 48(5), 669–676. [Google Scholar] [CrossRef]
- Koda, Y.; Itoh, M.; Tohda, S. Effects of MERTK inhibitors UNC569 and UNC1062 on the growth of acute myeloid Leukaemia cells. Anticancer Res. 2018, 38(1), 199–204. [Google Scholar] [CrossRef]
- DeRyckere, D.; Lee-Sherick, A.B.; Huey, M.G.; Hill, A.A.; Tyner, J.W.; Jacobsen, K.M.; Page, L.S.; Kirkpatrick, G.G.; Eryildiz, F.; Montgomery, S.A.; Zhang, W.; Wang, X.D.; Fry, S.V.; Earp, H.S.; Graham, D.K. UNC2025, a MERTK small-molecule inhibitor, is therapeutically effective alone and in combination with methotrexate in leukemia models. Clin. Cancer Res. 2017, 23(6), 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Huelse, J.M.; Kireev, D.; Tan, Z.; Chen, L.; Goyal, S.; Wang, X.; Frye, S.V.; Behera, M.; Schneider, F.; Ramalingam, S.S. MERTK activation drives osimertinib resistance in EGFR-mutant non–small cell lung cancer. J. Clin. Invest. 2022, 132(15). [CrossRef]
- Zhang, W.; DeRyckere, D.; Hunter, D.; Liu, J.; Stashko, M.A.; Minson, K.A.; Cummings, C.T.; Lee, M.; Glaros, T.G.; Newton, D.L.; Sather, S. UNC2025, a potent and orally bioavailable MER/FLT3 dual inhibitor. J. Med. Chem. 2014, 57(16), 7031–7041. [Google Scholar] [CrossRef] [PubMed]
- Lee-Sherick, A.B.; Jacobsen, K.M.; Henry, C.J.; Huey, M.G.; Parker, R.E.; Page, L.S.; Hill, A.A.; Wang, X.; Frye, S.V.; Earp, H.S.; Jordan, C.T.; Deryckere, D.; Hraham, D.K. MERTK inhibition alters the PD-1 axis and promotes anti-leukemia immunity. JCI insight 2018, 3(21). [CrossRef]
- Zhang, W.; Zhang, D.; Stashko, M.A.; DeRyckere, D.; Hunter, D.; Kireev, D.; Miley, M.J.; Cummings, C.; Lee, M.; Norris-Drouin, J.; Stewart, W.M. Pseudo-cyclization through intramolecular hydrogen bond enables discovery of pyridine substituted pyrimidines as new Mer kinase inhibitors. J. Med. Chem. 2013, 56(23), 9683–9692. [Google Scholar] [CrossRef]
- Shi, C.; Li, X.; Wang, X.; Ding, N.; Ping, Y.; Shi, Y.; Mi, L.; Lai, Y.; Song, Y.; Zhu, J. The proto-oncogene Mer tyrosine kinase is a novel therapeutic target in mantle cell lymphoma. J. Hematol. Oncol. 2018, 11, 1–13. [Google Scholar] [CrossRef]
- Hu, L.; Wang, X.; Song, Z.; Chen, F.; Wu, B. Leveraging CAR macrophages targeting c-Met for precision immunotherapy in pancreatic cancer: insights from single-cell multi-omics. Mol. Med. 2024, 30(1), 231. [Google Scholar] [CrossRef]
- Barbosa-Matos, C.; Borges-Pereira, C.; Libório-Ramos, S.; Fernandes, R.; Oliveira, M.; Mendes-Frias, A.; Silvestre, R.; Osório, N.S.; Bastos, H.N.; Santos, R.F.; Guimarães, S. Deregulated immune cell recruitment orchestrated by c-MET impairs pulmonary inflammation and fibrosis. Resp. Res. 2024, 25(1), 1–18. [Google Scholar] [CrossRef]
- Faiella, A.; Riccardi, F.; Cartenì, G.; Chiurazzi, M.; Onofrio, L. The emerging role of c-Met in carcinogenesis and clinical implications as a possible therapeutic target. J. Oncol. 2022, 2022(1), 5179182. [Google Scholar] [CrossRef]
- Lee, M.; Jain, P.; Wang, F.; Ma, P.C.; Borczuk, A.; Halmos, B. MET alterations and their impact on the future of non-small cell lung cancer (NSCLC) targeted therapies. Expert Opin. Ther. Tar. 2021, 25(4), 249–268. [Google Scholar] [CrossRef]
- Gou, Q.; Gou, Q.; Gan, X.; Xie, Y. Novel therapeutic strategies for rare mutations in non-small cell lung cancer. Sci. Rep. 2024, 14(1), 10317. [Google Scholar] [CrossRef]
- Cen, S.Y.; Lin, F.; Li, X.; Hu, Y.; Liu, J.P.; Xue, Z.; Gao, Y.; Sun, Y.P.; Zhu, S.; Dang, Y.; Zhao, Y. Crizotinib and its enantiomer suppress ferroptosis by decreasing PE-O-PUFA content. Cell Death Discov. 2024, 10(1), 360. [Google Scholar] [CrossRef]
- Elghawy, O.; Barsouk, A.; Xu, J.; Chen, S.; Cohen, R.B.; Sun, L. Real word outcomes of cabozantinib therapy in poorly differentiated thyroid carcinoma. Eur. Thyroid J. 2024, 13(6). [CrossRef]
- Patil, J.; Bhattacharya, S.; Saoji, S.D.; Dande, P. Cabozantinib-phospholipid complex for enhanced solubility, bioavailability, and reduced toxicity in liver cancer. Ther. Deliv. 2025, 16(1), 25–41. [Google Scholar] [CrossRef] [PubMed]
- Miao, K.; Zhang, X.; Wang, H.; Si, X.; Zhang, L. Savolitinib versus crizotinib for treating MET positive non-small cell lung cancer. Thorac. Cancer 2023, 14(13), 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Li, J.; Lin, S.; Wu, S.; Chai, K.; Jiang, X.; Qian, J.; Jiang, C. A real-world pharmacovigilance study of FDA adverse event reporting system events for Capmatinib. Sci. Rep. 2024, 14(1), 11388. [Google Scholar] [CrossRef]
- Dhillon, S. Capmatinib: first approval. Drugs, 2020, 80, 1125–1131. [CrossRef]
- Huang, D.W.; Chen, Y.; Yang, J.X.; Zhao, B.Y.; Wang, S.Y.; Chai, T.T.; Cui, J.; Zhou, X. L.; Shang, Z. H. Design, Synthesis, and Biological Evaluation of 2-Substituted Aniline Pyrimidine Derivatives as Potent Dual Mer/c-Met Inhibitors. Molecules, 2024, 29(2), 475. [CrossRef]
- Wei, D.S.; Fan, H.; Zheng, K.; Qin, X.; Yang, L.; Yang, Y.; Duan, Y.; Zhang, Q.; Zeng, C.; Hu, L. Synthesis and anti-tumor activity of [1,4] dioxino [2, 3-f] quinazoline derivatives as dual inhibitors of c-Met and VEGFR-2. Bioorg. Chem. 2019, 88, 102916. [Google Scholar] [CrossRef]
- Gao, G.R.; Li, M.Y.; Lv, Y.C.; Cao, S.F.; Tong, L.J.; Wei, L.X.; Ding, J.; Xie, H.; Duan, W.H. Design, synthesis and biological evaluation of biphenylurea derivatives as VEGFR-2 kinase inhibitors (II). Chinese Chem. Lett. 2016, 27(2), 200–204. [Google Scholar] [CrossRef]
- Park, H.; Jung, H.Y.; Mah, S.; Hong, S. Systematic computational design and identification of low Picomolar inhibitors of Aurora kinase A. J. chem. Inf. Model. 2018, 58(3), 700–709. [Google Scholar] [CrossRef]
- Huang, D.W.; Yang, J.X.; Zhang, Q.W.; Wang, G.; Zhang, Z.X.; Zhang, Y.; Li, J.Q. Structure-guided design and development of novel N-phenylpyrimidin-2-amine derivatives as potential c-Met inhibitors. Eur. J. Med. Chem. 2021, 223, 113648. [Google Scholar] [CrossRef]
- Hwang, S.T.; Um, J.Y.; Chinnathambi, A.; Alharbi, S.A.; Narula, A.S.; Namjoshi, O.A.; Blough, B.E.; Ahn, K.S. Evodiamine mitigates cellular growth and promotes apoptosis by targeting the c-Met pathway in prostate cancer cells. Molecules 2020, 25(6), 1320. [Google Scholar] [CrossRef]
- Li, J.; Tan, G.S.; Cai, Y.B.; Liu, R.H.; Xiong, X.L.; Gu, B.H.; He, W.; Liu, B.; Ren, Q.Y.; Wu, J.P.; Chi, B.; Zhao, Y.H.; Xu, Y.R.; Zou, Z.X.; Kang, F.H.; Xu, K.P. A novel Apigenin derivative suppresses renal cell carcinoma via directly inhibiting wild-type and mutant MET. Biochem. Pharmacol. 2021, 190, 114620. [Google Scholar] [CrossRef]
- Gu, Y.; Chen, Y.; Wei, L.; Wu, S.; Shen, K.; Liu, C.; Dong, Y.; Zhao, Y.; Zhang, Y.; Zhang, C.; Zheng, W. ABHD5 inhibits YAP-induced c-Met overexpression and colon cancer cell stemness via suppressing YAP methylation. Nat. Commun. 2021, 12(1), 6711. [Google Scholar] [CrossRef]
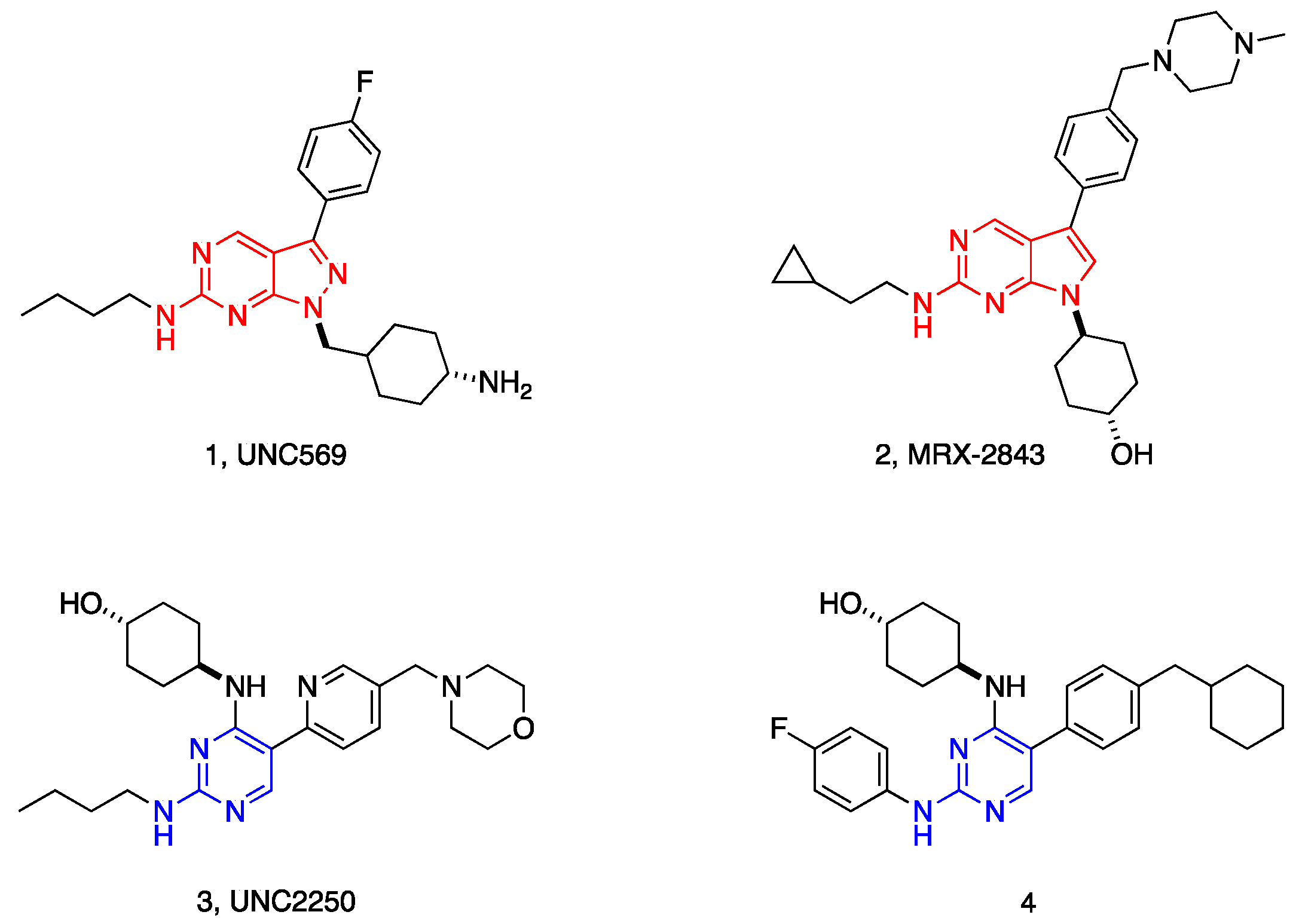
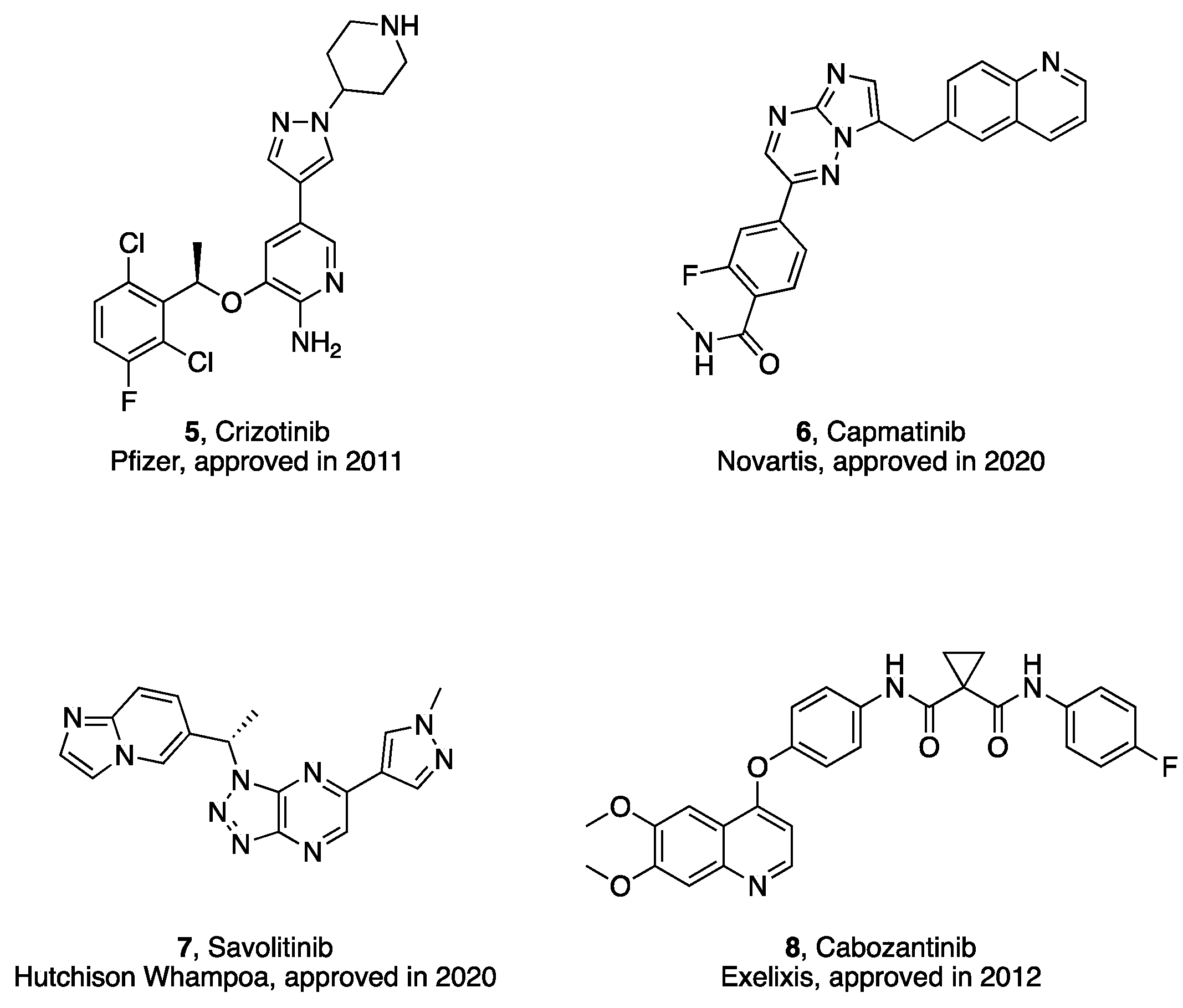
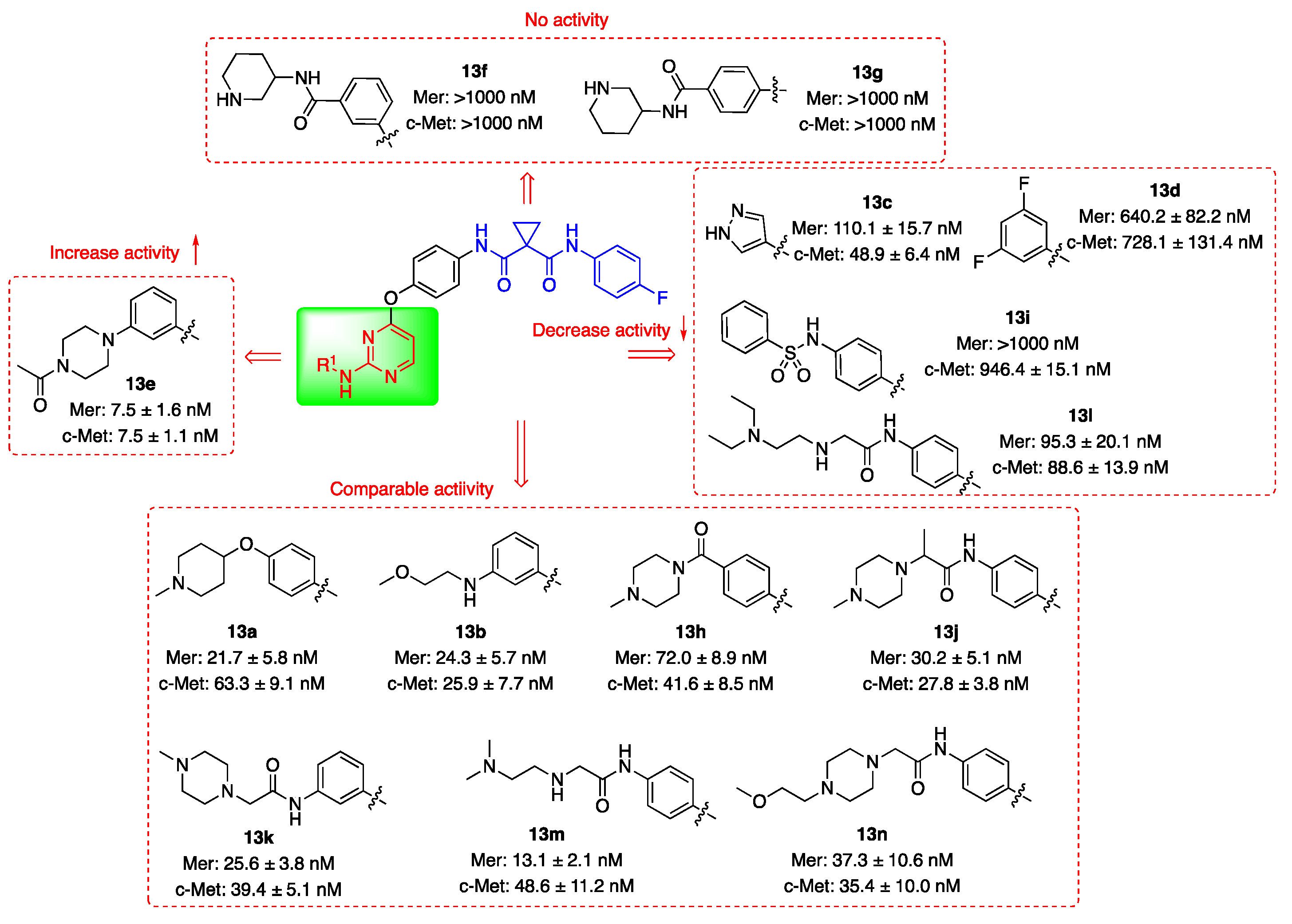
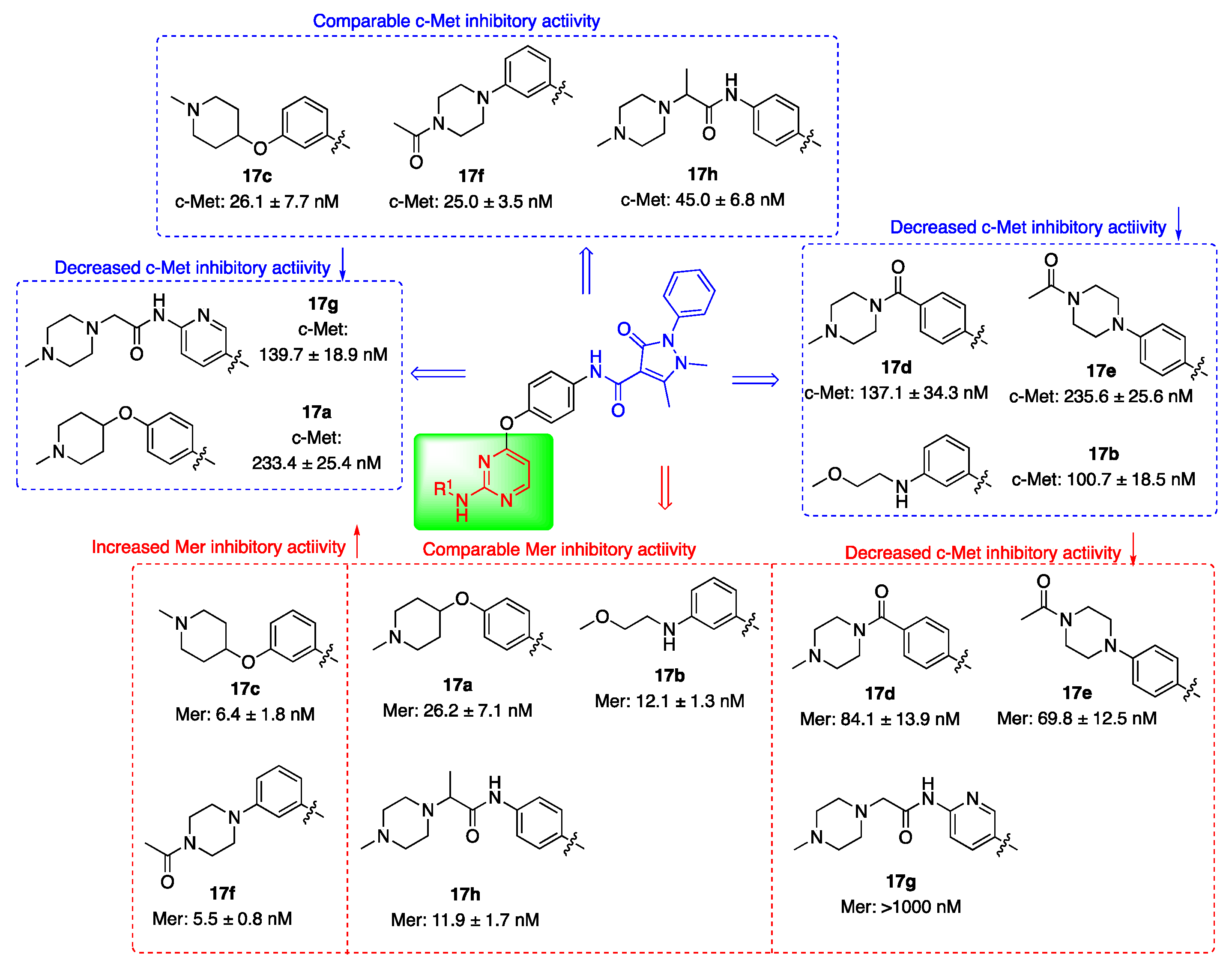
| Compd. | R1 | IC50 (nM)1 | |
| Mer | c-Met | ||
| 13a | 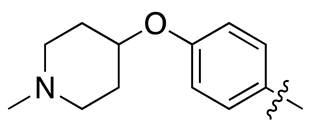 |
21.7 ± 5.8 | 63.3 ± 9.1 |
| 13b | 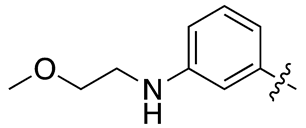 |
24.3 ± 5.7 | 25.9 ± 7.7 |
| 13c | 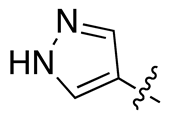 |
110.1 ± 15.7 | 48.9 ± 6.4 |
| 13d | 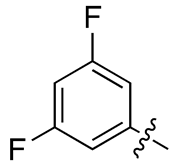 |
640.2 ± 82.2 | 728.1 ± 131.4 |
| 13e | 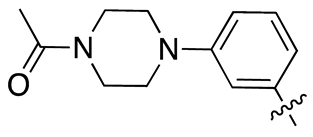 |
7.5 ± 1.6 | 7.5 ± 1.1 |
| 13f | 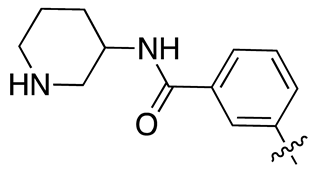 |
>1000 | >1000 |
| 13g | 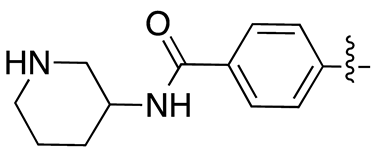 |
>1000 | >1000 |
| 13h | 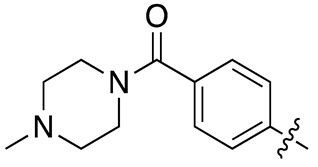 |
72.0 ± 8.9 | 41.6 ± 8.5 |
| 13i | 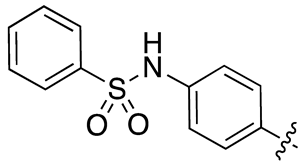 |
>1000 | 946.4 ± 15.1 |
| 13j | 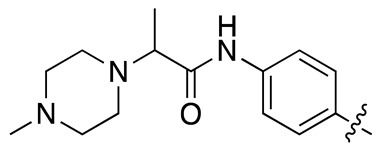 |
30.2 ± 5.1 | 27.8 ± 3.8 |
| 13k | 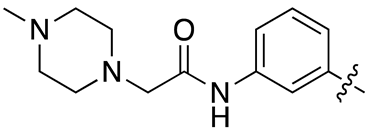 |
25.6 ± 3.8 | 39.4 ± 5.1 |
| 13l | 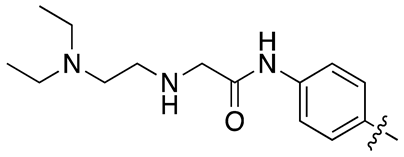 |
95.3 ± 20.1 | 88.6 ± 13.9 |
| 13m | 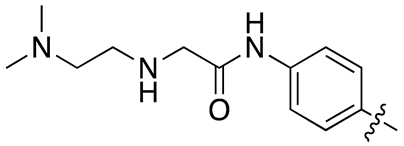 |
13.1 ± 2.1 | 48.6 ± 11.2 |
| 13n | 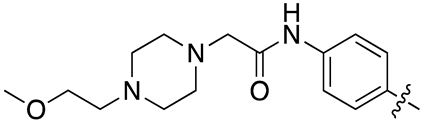 |
37.3 ± 10.6 | 35.4 ± 10.0 |
| 18c | 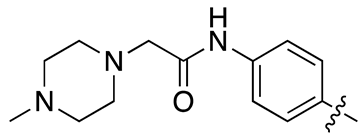 |
18.5 ± 2.3 | 33.6 ± 4.3 |
| Compd. | R2 | IC50 (nM)1 | |
| Mer | c-Met | ||
| 17a | 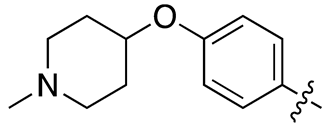 |
26.2 ± 7.1 | 233.4 ± 25.4 |
| 17b | 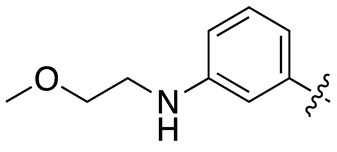 |
12.1 ± 1.3 | 100.7 ± 18.5 |
| 17c | 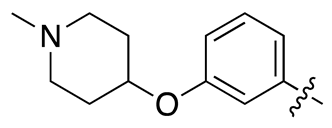 |
6.4 ± 1.8 | 26.1 ± 7.7 |
| 17d | 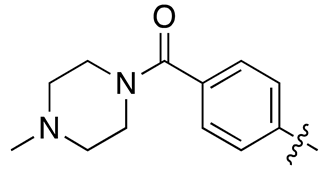 |
84.1 ± 13.9 | 137.1 ± 34.3 |
| 17e | 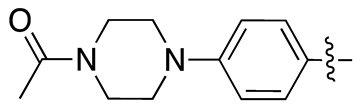 |
69.8 ± 12.5 | 235.6 ± 25.6 |
| 17f | 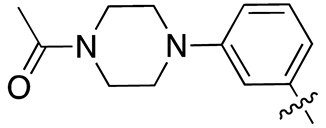 |
5.5 ± 0.8 | 25 ± 3.5 |
| 17g | 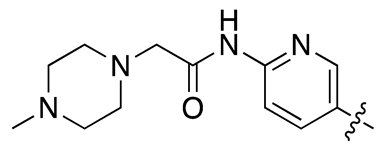 |
>1000 | 139.7 ± 18.9 |
| 17h | 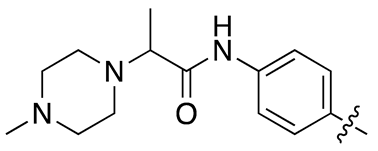 |
11.9 ± 1.7 | 45.0 ± 6.8 |
| 18c | 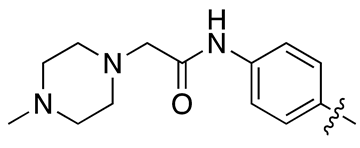 |
18.5 ± 2.3 | 33.6 ± 4.3 |
| Cpd. | HUMAN | |
|---|---|---|
| T1/2 (min) | CL (mL/min/mg) | |
| 13b | 38.3 | 0.0905 |
| 13e | 30.4 | 0.1142 |
| 13j | 11.9 | 0.2912 |
| 13k | 8.6 | 0.4046 |
| 13m | 27.9 | 0.1521 |
| 13n | 38.4 | 0.1243 |
| 17c | 147.0 | 0.0236 |
| 17f | 51.0 | 0.0680 |
| Testosterone | 32.1 | 0.11 |
| Cpds | IC50(μM)1 of 3 cell lines | |||
|---|---|---|---|---|
| HepG2 | MDA-MB0231 | HCT116 | ||
| 17c | 0.46 ± 0.06 | 2.31 ± 0.67 | 3.79 ± 1.09 | |
| Cabozatinib | 7.87 ± 2.11 | 4.62 ± 0.54 | 15.57 ± 4.39 | |
| Cpd. | IC50 (μM) |
| 17c | >40 |
| Cisapride | 0.04 |
| Cpd. |
Con. ( μM) |
Species | Mean Fu | Mean Fb |
Stability (%) |
| 17c | 1 | Human | 0.0158 | 98.4% | 99.3% |
| 17c | 1 | Rat | 0.0178 | 98.2% | 102.2% |
| Warfarin | 1 | Human | 0.00877 | 99.1% | 99.8% |
| Warfarin | 1 | Rat | 0.0102 | 99.0% | 98.2% |
| Adm. | Dose (mg/Kg) |
AUClast (h*ng/mL) |
T1/2 (h) |
Tmax (h) |
Cmax (ng/mL) |
CL_obs (mL/min/kg) | MRTINF_obs(h) | F (%) |
| p.o. | 10 | 4626 | 3.46 | 4.00 | 740 | – | 6.52 | 45.3 |
| i.v. | 2 | 2154 | 3.33 | – | – | 16.4 | 3.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





