Submitted:
20 April 2025
Posted:
21 April 2025
You are already at the latest version
Abstract
Keywords:
1.0. Introduction
2.0. Methodology
3.0. Strategies for SNP Discovery in Complex Plant Genomes
4.0. SNP Validation and Modern Genotyping Platforms and Chemistries
- Simple SNPs identify allelic differences at corresponding loci within a single subgenome, leading to distinct genotype groupings.
- Hemi-SNPs reveal genetic variations that appear as homozygous in one organism but heterozygous in another.
- Homoeo-SNPs, on the other hand, identify variation at homoeologous or paralogous loci across A and D subgenomes but are often monomorphic in tetraploid species (Mammadov et al., 2012a; Abdelraheem et al., 2017; Akter et al., 2019).
5.0. Comparative Analysis of Four Genotyping Assays and Platforms
5.1. The OpenArray Technology (TaqMan System)
5.2. Illumina’s BeadArray Platform
5.3. High-Resolution Melting (HRM)
5.4. Kompetitive Allele-Specific PCR (KASP)
6.0. Next-Generation Sequencing (NGS)
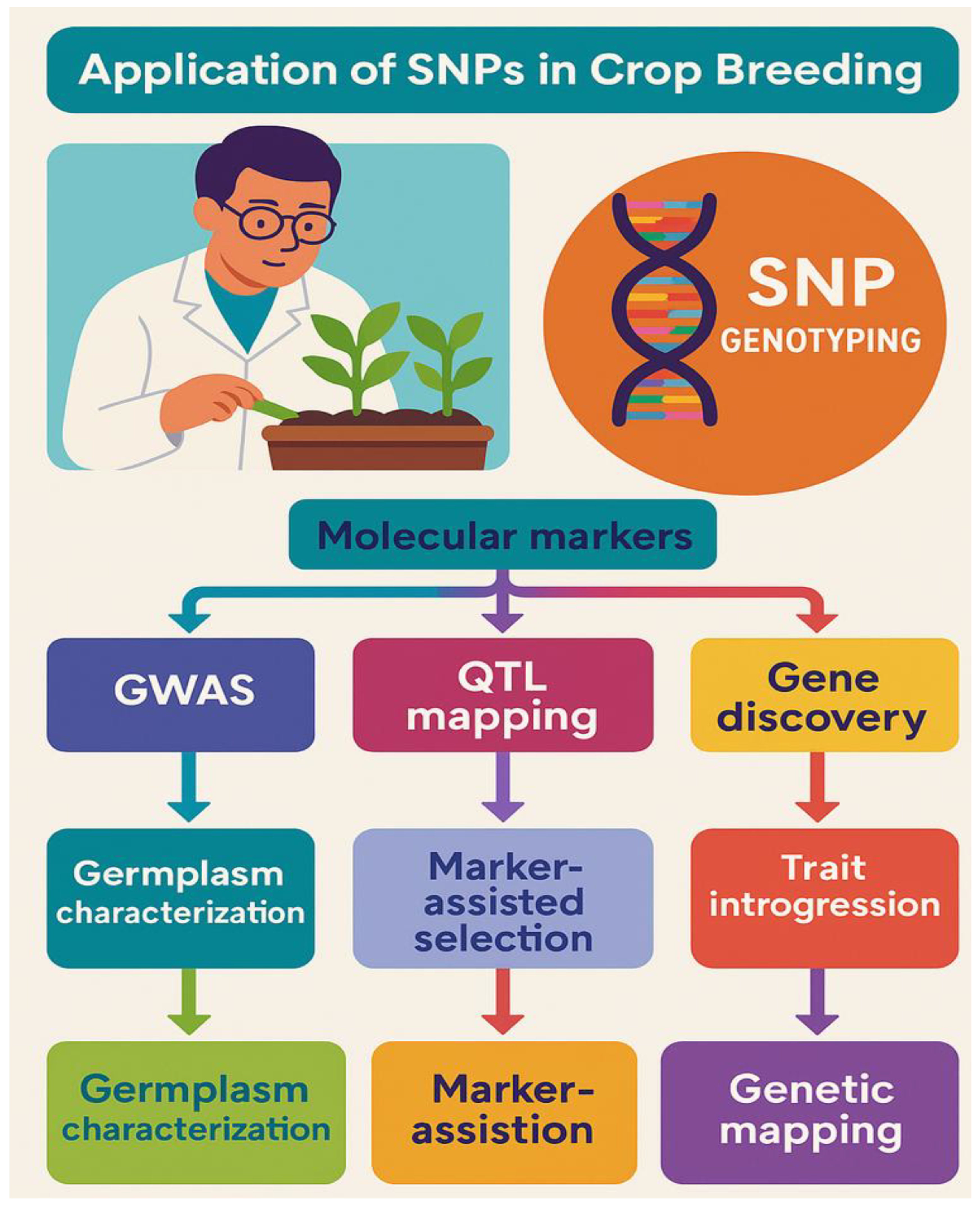
7.0. Application of SNPs in Crop Breeding
8.0. Conclusions
Acknowledgements
Conflict of Interest
References
- Abdelraheem, A.; Fang, D.D.; Zhang, J. “Quantitative trait locus mapping of drought and salt tolerance in an introgressed recombinant inbred line population of upland cotton under the greenhouse and field conditions”. Euphytica 2017, 214, 8. [Google Scholar] [CrossRef]
- Adler, A.J.; Wiley, G.B.; Gaffney, P.M. “Infinium Assay for Large-scale SNP Genotyping Applications”. Journal of Visualized Experiments 2013, 81, e50683. [Google Scholar] [CrossRef]
- Ahn, Y.K.; Manivannan, A.; Karna, S.; Jun, T.H.; Yang, E.Y.; Choi, S.; Kim, J.H.; Kim, D.S.; Lee, E.S. “Whole Genome Resequencing of Capsicum baccatum and Capsicum annuum to Discover Single Nucleotide Polymorphism Related to Powdery Mildew Resistance”. Sci. Rep 2018, 8, 51–88. [Google Scholar] [CrossRef]
- Akter, T; Islam, A.K.M.A.; Rasul, M.G.; Kundu, S.; halequzzaman; Ahmed, J.U. “Evaluation of genetic diversity in short duration cotton (Gossypium hirsutum L.)”. Journal of Cotton Research 2019, 2, 1–10. [Google Scholar] [CrossRef]
- Allen, A.M.; Barker, G.L.A.; Berry, S.T.; Coghill, J.A.; Gwilliam, R.; Kirby, S.; Robinson, P.; et al. “Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.)”. Plant Biotechnology Journal 2011, 9, 1086–1099. [Google Scholar] [CrossRef]
- Alvarez-Fernandez, A.; Bernal, M.J.; Fradejas, I. Alvarez-Fernandez, A., M.J. Bernal, I. Fradejas. 2021. “KASP: a genotyping method to rapid identification of resistance in Plasmodium falciparum”. Malaria Journal. 2021, 20, 1–16. [Google Scholar] [CrossRef]
- Andelkovic, V.; Cvejić, S.; Jocić, S.; Kondic-Spika, A.; Jeromela, A. Marjanović; Mikić, S.; Prodanović, S. “Use of plant genetic resources in crop improvement–example of Serbia”. Genetic Resources and Crop Evolution 2020, 67, 1935–1948. [Google Scholar] [CrossRef]
- Appleby, N.; Edwards, D.; Batley, J. “New technologies for ultra-high throughput genotyping in plants”. Plant Genomics 2009, 513, 19–39. [Google Scholar] [CrossRef]
- Aurélie B., M.C.L. Paslier, M. Dardevet, F. Exbrayat-Vinson, I. Bonnin, A. Cenci, A. Haudry, D. Brunel, and C. Ravel. 2009. “High-throughput single nucleotide polymorphism genotyping in wheat (Triticum spp.)”. Plant Biotechnology Journal (7): 364-374.
- Ayalew, H.; Tsang, P.W.; Chu, C.; Wang, J.; Liu, S.; Chen, C.; Ma, X.F. “Comparison of TaqMan, KASP, and rhAmp SNP genotyping platforms in hexaploid wheat”. PLoS One 2019, 14, e0217222. [Google Scholar] [CrossRef]
- Berdugo-Cely, J.; Valbuena, R.I.; Sánchez-Betancourt, E.; Barrero, L.S.; Yockteng, R. “Genetic diversity and association mapping in the Colombian Central Collection of Solanum tuberosum L. Andigenum group using SNPs markers”. PLoS ONE, 2017, 12, e0173039. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, A.; Wang, S.; Amand, P.S.; Bai, G.; Fang, D.D. “Using next-generation sequencing for multiplexed trait-linked markers in wheat”. PLoS One 2015, 10, e0143890. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, R. “Molecular markers and selection for complex traits in plants: learning from the last 20 years,” Crop Science 48: 1649–1664. 2008. [Google Scholar]
- Brenan, C.; Morrison, T. High throughput, nanoliter quantitative PCR. Drug Discovery Today Technologies 2005, 2, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.E.E.; Flint-Garcia, S.; et al. “The Genetic Architecture of Maize Flowering Time”. Science 2009, 325, 714–718. [Google Scholar] [CrossRef]
- Bui, T.G.T., N.T.L. Hoa, J. Yen, and R. Schafleitner. 2017. “PCR-based assays for validation of single nucleotide polymorphism markers in rice and mungbean”. Hereditas 154: 1-3. [CrossRef]
- Bus, A.; Hecht, J.; Huettel, B.; Reinhardt, R.; Stich, B. “High-throughput polymorphism detection and genotyping in Brassica napus using next-generation RAD sequencing”. BMC Genomics 2012, 13, 281. [Google Scholar] [CrossRef]
- Cheema, J.; Dicks, J. “Computational approaches and software tools for genetic linkage map estimation in plants”. Briefings in Bioinformatics 2009, 10, 595–608. [Google Scholar] [CrossRef]
- Chleinitz, D.; DiStefano, J.K.; Kovacs, P. “Disease gene identification: targeted SNP genotyping using the TaqMan assay”; Humana Press: New York, 2011; pp. 77–87. [Google Scholar]
- Chutimanitsakun, Y., R.W. Nipper, A.C.L. Cuesta-Marcos, A. Corey, T. Filichkina, P.M. Hayes. 2011. “Construction and application for QTL analysis of a Restriction Site Associated DNA (RAD) linkage map in barley”. BMC Genomics 12 (1): 1-4. [CrossRef]
- Clevenger, J., Chavarro, C., Pearl, S. A., Ozias-Akins, P., & Jackson, S. A. 2015. Single nucleotide polymorphism identification in polyploids: A review, example, and recommendations. Molecular Plant, 8:, 831–846. [CrossRef]
- Collard, B.C.; Mackill, D.J. “Marker-assisted selection: an approach for precision plant breeding in the twenty-first century”. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences 2008, 363, 557–72. [Google Scholar] [CrossRef]
- Costa, J. R., Bejcek, B. E., McGee, J. E., et al. 2017. Genome editing using engineered nucleases and their use in genomic screening. In S. Markossian, A. Grossman, M. Arkin, et al. (Eds.), Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences. https://www.ncbi.nlm.nih.gov/books/NBK464635/.
- Dagnall, C.L.; Morton, L.M.; Hicks, B.D.; et al. “Successful use of whole genome amplified DNA from multiple source types for high-density Illumina SNP microarrays”. BMC Genomics 2018, 19, 182. [Google Scholar] [CrossRef]
- Davey, J.W.; Cezard, T.; Fuentes-Utrilla, P.; Eland, C.; Gharbi, K.; Blaxter, M.L. “Special features of RAD sequencing data: implications for genotyping”. Molecular Ecology 2013, 22, 3151–3164. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. “Genome-wide genetic marker discovery and genotyping using next-generation sequencing”. Nature Review Genetics 2011, 12, 499–510. [Google Scholar] [CrossRef]
- De Oliveira, E.J.; Ferreira, C.F.; Santos, V. da Silva; Jesus, O.N.E.; Oliveira, G.A.; Silva, M.S. da. “Potential of SNP markers for the characterization of Brazilian cassava germplasm”. Theroritical and applied genetics 2014, 127, 1423–1440. [Google Scholar] [CrossRef] [PubMed]
- Dean, A. “On a chromosome far, far away: LCRs and gene expression”. Trends in Genetics, 2006, 22, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Ebili, H.O.; Ilyas, M. “Cancer mutation screening: Comparison of high-resolution melt analysis between two platforms”. ecancermedicalscience 2015, 9, 522. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Poland, J.A.; Kawamoto, K.; Buckler, E.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 2011, 6, e19379. [Google Scholar] [CrossRef]
- Emberton, J.; Ma, J.; Yuan, Y.; SanMiguel, P.; Bennetzen, J.L. “Gene enrichment in maize with hypomethylated partial restriction (HMPR) libraries”. Genome Research 2005, 15, 1441–1446. [Google Scholar] [CrossRef]
- Er, T.K.; Chang, J.G. “High-resolution melting: Applications in genetic disorders”. Clinica Chimica Acta 2012, 414, 197–201. [Google Scholar] [CrossRef]
- Esuma, W.; Herselman, L.; Labuschagne, M.T.; Ramu, P.; Lu, F.; Baguma, Y.; Buckler, E.S.; Kawuki, R.S. “Genome-wide association mapping of provitamin A carotenoid content in cassava”. Euphytica. 2016. [Google Scholar] [CrossRef]
- Fan, J. B.; Oliphant, A.; Shen, R.; Kermani, B.G.; Garcia, F.; Gunderson, K.L.; Hansen, M.; et al. “Highly parallel SNP genotyping”. Cold Spring Harbor Symposia on Quantitative Biology 2003, 68, 69–78. [Google Scholar] [CrossRef]
- Ferguson, M.E.; Hearne, S.J.; Close, T.J.; Wanamaker, S.; Moskal, W.A.; Town, C.D.; et al. “Identification, validation, and high-throughput genotyping of transcribed gene SNPs in cassava”. Theoretical and Applied Genetics 2011, 124, 685–695. [Google Scholar] [CrossRef]
- Fernando, H.K.D.H.; Kajenthini, T.J.C.; Rebeira, S.P.; Bamunuarachchige, T.C.; Wickramasinghe, H.A.M. Validation of Molecular Markers for the Analysis of Genetic Diversity of Amylase Content and Gel Consistency among Representative Rice Varieties in Sri Lanka. Tropical Agricultural Research 2015, 26, 317–328. [Google Scholar] [CrossRef]
- Ferri, L., E. Perrin, S. Campana, S. Tabacchioni, G. Taccetti, P. Cocchi, N. Ravenni, C. Dalmastri..(2010). “Application of multiplex single nucleotide primer extension (mSNuPE) to the identification of bacteria: the Burkholderia cepacia complex case”. Journal of Microbiological Methods 80: 251–256. [CrossRef]
- Ganal, M.W.; Altmann, T.; Roder, M.S. “SNP identification in crop plants”. Current Opinion in Plant Biology 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Ganal, M.W.; Durstewitz, G.; Polley, A.; Bérard, A.; Buckler, E.S.; Charcosset, A. “Large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping and genetic mapping to compare with the B73 reference genome”. PLoS One 2011, 6, 28334. [Google Scholar] [CrossRef] [PubMed]
- Gore, M.A.; Chia, J.M.; Elshire, R.J.; Sun, Q.; Ersoz, E.S.; Hurwitz, B.L.; et al. “A First-Generation Haplotype Map of Maize”. Science 2009, 326, 1115–1117. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Holme, J.; nd; Anthony, J. “SNP genotyping: the KASP assay”. Methods Molecular Biology. 2014, 1145, 75–86. [Google Scholar] [CrossRef]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. “Potential etiologic and functional implications of genome-wide association loci for human diseases and traits”. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A; Chamarthi, S.K.; et al. “Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes”. Plant Biotechnology Journal 2012, 10, 716–732. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Kumar, H.A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A.; Chamarthi, K.S; et al. “Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes”. Plant Biotechnology Journal 2012, 10, 716–732. [Google Scholar] [CrossRef]
- Hodges, E.; Xuan, Z.; Balija, V. “Genome-wide in situ exon capture for selective resequencing”. Nature Genetics 2007, 39, 1522–1527. [Google Scholar] [CrossRef]
- Howie, B.N.; Donnelly, P.; Marchini, J. “A flexible and accurate genotype imputation method for the next generation of genome-wide association studies”. PLoS Genetics 2009, 5, e1000529. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. “Natural variations and genome-wide association studies in crop plants”. Annual review of plant biology 2014, 65, 85–551. [Google Scholar] [CrossRef]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C. “Genome-wide association studies of 14 agronomic traits in rice landraces”. Nature Genetics 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Jagtap, A.B.; Vikal, Y.; Johal, G.S. “Genome-Wide Development and Validation of Cost-Effective KASP Marker Assays for Genetic Dissection of Heat Stress Tolerance in Maize”. International Journal of Molecular Sciences 2020, 21, 7386. [Google Scholar] [CrossRef] [PubMed]
- Jatayev, S., A. Kurishbayev, L. Zotova, G. Khasanova, D. Serikbay, A. Zhubatkanov, et al. 2017. “Advantages of Amplifluor-like SNP markers over KASP in plant genotyping”. BMC Plant Biol 17: 254. [CrossRef]
- Jiang, G.L. “Molecular Markers and Marker-Assisted Breeding in Plants”. Plant Breeding from Laboratories to Fields. 2013. [Google Scholar] [CrossRef]
- Jiang, Z.; Wang, H.; Michal, J.J.; Zhou, X.; Liu, B.; Woods, L.C.S.; Fuchs, R.A. “Genome-Wide Sampling Sequencing for SNP Genotyping: Methods, Challenges, and Future Development”. International Journal of Biological Sciences 2016, 12, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Jones, H., J. Kawauchi, P. Braglia. 2007. “RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA”. Nature Structural and Molecular Biology 14: 123–130. [CrossRef]
- Jones, M.A.; Gargano, J.W.; Rhodenizer, D.; Martin, I.; Bhandari, P.; Grotewiel, M. “A forward genetic screen in Drosophila implicates insulin signaling in age-related locomotor impairment”. Experimental Gerontology 2009, 44, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.; Charest, A.; Dowd, J.F.; Blumenstiel, J.P.; Yeh, R.F.; Osman, A.; Housman, D.E.; Landers, J.E. “Genome complexity reduction for SNP genotyping analysis”. Proceedings of the National Academy of Sciences 2002, 99, 2942–2947. [Google Scholar] [CrossRef]
- Kansup, J.; Amawan, S.; Wongtiem, P.; Sawwa, A.; Ngorian, S.; Narkprasert, D.; Hansethasuk, J. “Marker-assisted selection for resistance to cassava mosaic disease in Manihot esculenta Crantz". Thai Agricultural Research Journal 2020, 38, 68–79. [Google Scholar] [CrossRef]
- Karim, K.Y.; Ifie, B.; Dzidzienyo, D.; Danquah, E.Y.; Blay, E.T.; Whyte, J.B.A.; et al. “Genetic characterization of cassava (Manihot esculenta Crantz) genotypes using agro-morphological and single nucleotide polymorphism markers”; 317–330: Physiol Mol Biol Plants 26, 2020. [Google Scholar] [CrossRef]
- Kassa, M.T.; M, You F.; W, Hiebert C.; J, Pozniak C.; R, Fobert P.; G, Sharpe A.; et al. “Highly predictive SNP markers for efficient selection of the wheat leaf rust resistance gene Lr16”. BMC Plant Biology 2017, 17, 45. [Google Scholar] [CrossRef]
- Kim, K.S.; Bellendir, S.; Hudso, K.A.; Hill, C.B.; Hartman, G.L.; Hyten, D.L.; et al. “Fine mapping the soybean aphid resistance gene Rag1 in soybean”. Theoretical and Applied Genetics, 2009, 120, 1063–1071. [Google Scholar] [CrossRef]
- Kim, K.S.; Hill, C.B.; Hartman, G.L.; Hyten, D.L.; Hudson, M.E.; Diers, B.W. “Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538”. Theoretical and Applied Genetics 2010, 121, 599–610. [Google Scholar] [CrossRef]
- Kim, S.I.; Tai, T.H. “Identification of SNPs in closely related Temperate Japonica rice cultivars using restriction enzyme-phased sequencing”. PLoS One 2013, 8, 60176. [Google Scholar] [CrossRef]
- Kim, K.D.; Kang, K.; Kim, C. “Application of Genomic Big Data in Plant Breeding: Past, Present, and Future”. Plants 2020, 9, 1454. [Google Scholar] [CrossRef]
- Kim, K., Choe, D., Cho, S., Palsson, B., and Cho, B.-K. Reduction-to-synthesis: The dominant approach to genome-scale synthetic biology. Trends in Biotechnology 2024, 42, 1048–1063. [CrossRef] [PubMed]
- Komar, A.A. 2009. “[Methods in Molecular Biology] Single Nucleotide Polymorphisms Volume 578 The TaqMan Method for SNP Genotyping”., 10.1007/978-1-60327-411-1(Chapter 19), 293–306. [CrossRef]
- Korsa, F., & Feyissa, T. 2022. Effects of functional single nucleotide polymorphisms on plant phenotypes. Archives of Crop Science, 5: 185–192. [CrossRef]
- Krypuy, M.; Newnham, G.M.; Thomas, D.M.; Conron, M.; Dobrovic, A. “High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer”. BMC Cancer 2006, 6, 295. [Google Scholar] [CrossRef] [PubMed]
- Kumpatla, S.P.; Buyyarapu, R.; Abdurakhmonov, I.Y.; Mammadov, J.A. “Genomics-assisted plant breeding in the 21st century: technological advances and progress in Plant Breeding”. I. Y. Abdurakhmonov, Ed., pp. 131–184. 2012. [Google Scholar]
- Kwok, P.Y. “Single Nucleotide Polymorphisms”. Current Issues in Molecular Biology 2002, 5, 43–60. [Google Scholar]
- Li, H.; Vikram, P.; Singh, R.P.; Kilian, A.; Carling, J.; Song, J.; Burgueno-Ferreira, J.A.; et al. “A high-density GBS map of bread wheat and its application for dissecting complex disease resistance traits”. BMC Genomics 2015, 16, 216. [Google Scholar] [CrossRef]
- Liu, P., Lv, J., Ma, C., Zhang, T., Huang, X., Yang, Z., Zhang, L., Hu, J., Wang, S., and Bao, Z. 2022. Targeted genotyping of a whole-gene repertoire by an ultrahigh-multiplex and flexible HD-Marker approach. Engineering, 13: 186–196. [CrossRef]
- Lopes, U. V.; ires; L, J.; ramacho; P, K.; rattapaglia. Genome-wide SNP genotyping as a simple and practical tool to accelerate the development of inbred lines in outbred tree species: An example in cacao (Theobroma cacao L.). PLOS ONE, 2022, 17, e0270437. [Google Scholar] [CrossRef]
- Mader, E.; Lukas, B.; Novak, J. “A strategy to setup co-dominant microsatellite analysis for high-resolution-melting-curve-analysis (HRM)”. BMC Genetics 2008, 9, 69. [Google Scholar] [CrossRef]
- Mammadov, J.A.; Chen, W.; Mingus, J.; Thompson, S.; Kumpatla, S. “Development of versatile gene-based SNP assays in maize (Zea mays L. )”. Molecular Breeding 2012, 29, 779–790. [Google Scholar] [CrossRef]
- Mammadov, J.A.; Chen, W.; Ren, R. “Development of highly polymorphic SNP markers from the complexity reduced portion of maize (Zea mays L.) genome for use in marker-assisted breeding”. Theoretical and Applied Genetics. 2009, 121, 577–588. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. “SNP Markers and Their Impact on Plant Breeding”. International Journal of Plant Genomics 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Manivannan, A., Choi, S., Jun, T. H., Yang, E. Y., Kim, J. H., Lee, E. S., Lee, H. E., Kim, D. S., and Ahn, Y. K. (2021). Genotyping by sequencing-based discovery of SNP markers and construction of linkage maps from F5 population of pepper with contrasting powdery mildew resistance trait. BioMed Research International 2021: 6673010. [CrossRef]
- Marchini, J.; Howie, B. “Genotype imputation for genome-wide association studies”. Nature Reviews Genetics 2010, 11, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Masouleh, A.K.; Waters, D.L.; Reinke, R.F.; Henry, R.J. “A high1016 throughput assay for the rapid and simultaneous analysis of perfect markers for 1017 important quality and agronomic traits in rice using multiplexed MALDI-TOF 1018 mass spectrometry”. Plant Biotechnology Journal 2009, 7, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Monteros, M.J., B.K. Ha, D.V. Phillips, et al. 2010. “SNP assay to detect the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust”. Theoretical and Applied Genetic 121: 1023–1032. [CrossRef]
- Mora-Márquez, F., Nuño, J. C., Soto, Á., and López de Heredia, U. Missing genotype imputation in non-model species using self-organizing maps. Molecular Ecology Resources, 2025, 25, e13992. [CrossRef] [PubMed]
- Morishige, D.T.; Klein, P.E.; Hilley, J.L.; et al. “Digital genotyping of sorghum – a diverse plant species with a large repeat-rich genome”. BMC Genomics, 2013. [Google Scholar] [CrossRef]
- Myles, S.; Chia, J.M.; Hurwitz, B.; Simon, C.; Zhong, G.Y.; et al. “Rapid genomic characterization of the genus Vitis”. PloS One 2010, 5, e8219. [Google Scholar] [CrossRef]
- Narechania, A.; Gore, M.A.; Buckler, E.S.; et al. “Large-scale discovery of gene-enriched SNPs”. The Plant Genome 2009, 2, 121–133. [Google Scholar]
- Neelam, K; Brown-Guedira, G.; Huang, L. “Development and validation of a breeder-friendly KASPar marker for wheat leaf rust resistance locus Lr21”. Molecular Breeding 2013, 31, 233–237. [Google Scholar] [CrossRef]
- Nelson, J.C.; Wang, S.; Wu, Y.; et al. “Single-nucleotide polymorphism discovery by high-throughput sequencing in sorghum,” BMC Genomics 12: 352. 2011. [Google Scholar]
- Okou, T.; Steinberg, K.M.; Middle, C.; Cutler, D.J.; Albert, T.J.; Zwick, M.E. “Microarray-based genomic selection for high-throughput resequencing”. Nature Methods 2007, 411, 907–909. [Google Scholar] [CrossRef]
- Panahi, B., Mohammadzadeh Jalaly, H., and Hamid, R. 2024. Using next-generation sequencing approach for discovery and characterization of plant molecular markers. Current Plant Biology 40: 100412. [CrossRef]
- Patil, G.; Chaudhary, J.; Vuong, T.D.; Jenkins, B.; Qiu, D.; Kadam, S.; Shannon, G.J.; Nguyen, H.T. “Development of SNP Genotyping Assays for Seed Composition Traits in Soybean”. International Journal of Plant Genomics 2017, 2017, 1–12. [Google Scholar] [CrossRef]
- Poland, J. A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L.; Yin, T. “Development of High-Density Genetic Maps for Barley and Wheat Using a Novel Two-Enzyme Genotyping-by-Sequencing Approach”. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Pootakham, W. 2023. Genotyping by sequencing (GBS) for genome-wide SNP identification in plants. In Plant Genotyping: Methods and Protocols (pp. 1–14). Springer. [CrossRef]
- Powel, W.; Machray, G.C.; Proven, J. “Polymorphism revealed by simple sequence repeats,” Trends in Plant Science 1: 215–222. 1996. [Google Scholar]
- Rabbi, I.; Hamblin, M.; Gedil, M.; et al. “Genetic mapping using genotyping-by-sequencing in the clonally propagated cassava”. Crop Science 2014, 54, 1384–1396. [Google Scholar] [CrossRef]
- Rabbi, I.; Udoh, L.I.; Wolfe, M.; Parkes, E.Y.; Gedil, M.; Dixon, A.; Ramu, P.; Jannink, J.; Kulakow, P. “Genome-Wide Association Mapping of Correlated Traits in Cassava: Dry Matter and Total Carotenoid Content”. The Plant Genome 2017, 10, 0. [Google Scholar] [CrossRef] [PubMed]
- Rabbi, I.Y.; Kayondo, S.I.; Bauchet, G.; et al. “Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological, and quality-related traits in cassava”. Plant Mol Biol, 2020. [Google Scholar] [CrossRef]
- Rafalski, A. “Applications of single nucleotide polymorphisms in crop genetics”. Current Opinion in Plant Biotechnology 2002, 5, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Raj, R. N., Qureshi, N., & Pourkheirandish, M. 2022. Genotyping by sequencing advancements in barley. Frontiers in Plant Science 13: 931423. [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. “Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives”. Molecular Plant. 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Lui, J.; Guo, Q.; et al. “Development and validation of KASP assays for functional genes underpinning key economic traits in wheat”. Theoretical and Applied Genetics 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. “High-resolution DNA melting analysis for simple and efficient molecular diagnostics”. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef]
- Rienzo, D.V.; Bubici, G.; Montemurro, C.; Cillo, F.; Shu-Biao, W. “Rapid identification of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus using high resolution melting and TaqMan SNP Genotyping assays as allelic discrimination techniques”. PLOS ONE 2018, 13, e0196738. [Google Scholar] [CrossRef]
- Ruff, T.M.; Marston, E.J.; Eagle, J.D.; Sthapit, S.R.; Hooker, M.A.; Skinner, D.Z.; et al. “Genotyping by multiplexed sequencing (GMS): A customizable platform for genomic selection”. PLoS ONE 2020, 15, e0229207. [Google Scholar] [CrossRef]
- Satam, B., Maheshwari, S., and Dangi, C. B. Next-generation sequencing technology: Current trends and advancements. Biology 2023, 12, 997. [CrossRef]
- Schaarschmidt, S.; Fischer, A.; Zuther, E.; Hincha, D.K. “Evaluation of Seven Different RNA-Seq Alignment Tools Based on Experimental Data from the Model Plant Arabidopsis thaliana”. Int. J. Mol. Sci 2020, 21, 1720. [Google Scholar] [CrossRef]
- Scheben, A.; Batley, J.; Edwards, D. “Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application”. Plant Biotechnology Journal 2016, 15, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Schilbert, H.M.; Rempel, A.; Pucker, B. “Comparison of Read Mapping and Variant Calling Tools for the Analysis of Plant NGS Data”. Plants 2020, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Schleinitz, D, J.K. DiStefano, and P. Kovacs. 2011. “Disease gene identification: targeted SNP genotyping using the TaqMan assay”. New York: Humana Press; p. 77–87.
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. “Single nucleotide polymorphism genotyping using Kompetitive Allele-Specific PCR (KASP): an overview of the technology and its application in crop improvement”. Molecular breeding 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Semagn, K.; Beyene, Y.; Babu, R.; Nair, S.; Gowda, M.; Das, B. “Quantitative Trait Loci Mapping and Molecular Breeding for Developing Stress Resilient Maize for Sub-Saharan Africa”. Crop Science 2015, 55, 1449. [Google Scholar] [CrossRef]
- Senthilvel, S.; Ghosh, A.; Shaik, M.; et al. “Development and validation of an SNP genotyping array and construction of a high-density linkage map in castor”. Sci Rep. 2019, 9, 3003. [Google Scholar] [CrossRef]
- Shahabzadeh, Z., Darvishzadeh, R., Mohammadi, R., Jafari, M., & Alipour, H. High-throughput single nucleotide polymorphism genotyping reveals population structure and genetic diversity of tall fescue (Festuca arundinacea) populations. Crop and Pasture Science 2022, 73, 1070–1084. [CrossRef]
- Shai, Z.; Song, W.; Xing, J.; et al. “Molecular mapping of quantitative trait loci for three kernel-related traits in maize using a doubled haploid population”. Molecular Breeding 2017, 37, 108. [Google Scholar] [CrossRef]
- Shen, G.Q.; Abdullah, K.G.; Wang, Q.K. “The TaqMan method of SNP genotyping. In: Komar A. (eds) Single Nucleotide Polymorphisms”. Methods in Molecular Biology™ (Methods and Protocols), Humana Press, Totowa, NJ 2009, 578, 293–306. [Google Scholar] [CrossRef]
- Singh, R.R.; Bains, A.; Patel, K.P.; Rahimi, H.; Barkoh, B.A.; Paladugu, A.; et al. “Detection of high-frequency and novel DNMT3A mutations in acute myeloid leukemia by high-resolution melting curve analysis”. J Mol Diagn. 2012, 14, 336–45. [Google Scholar] [CrossRef]
- Slomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. “High-Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies”. International Journal of Molecular Sciences 2017, 18, 2316. [Google Scholar] [CrossRef]
- Song, C.; Castellanos-Rizaldos, E.; Bejar, R.; Ebert, B.L.; Makrigiorgos, G.M. “DMSO Increases Mutation Scanning Detection Sensitivity of High-Resolution Melting in Clinical Samples”. Clin Chem. 2015, 61, 1354–62. [Google Scholar] [CrossRef] [PubMed]
- Steele, K.A.; Quinton-Tulloch, M.J.; Amgai, R.B.; Dhakal, R.; Khatiwada, S.P.; Vyas, D.; et al. “Accelerating public sector rice breeding with high-density KASP markers derived from whole-genome sequencing of indica rice”. Molecular Breeding 2018, 38, 38. [Google Scholar] [CrossRef] [PubMed]
- Steemers, F.J.; Gunderson, K.L. “Whole-genome genotyping technologies on the BeadArray platform”. Biotechnology Journal 2007, 2, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Stolle, E.; Moritz, R.F. “RESTseq-efficient benchtop population genomics with restriction Fragment sequencing”. PLoS One 2013, 8, 63960. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Crouch, J.H.; Xu, Y. “The efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement”. Molecular Breeding 2010, 26, 493–511. [Google Scholar] [CrossRef]
- Suo, W.; Shi, X.; Xu, S.; Li, X.; Lin, Y. “Towards low cost, multiplex clinical genotyping: 4-fluorescent Kompetitive Allele-Specific PCR and its application on pharmacogenetics”. PLoS ONE 2020, 15, e0230445. [Google Scholar] [CrossRef]
- Thomson, M.J. “High-throughput SNP genotyping to accelerate crop improvement”. Plant Breeding and Biotechnology 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Thottathil, G.P.; Jayasekaran, K.; Othman, A.S. “Sequencing crop genomes: a gateway to improve tropical agriculture”. Tropical life science research 2016, 27, 93–114. [Google Scholar]
- Tian, H., Y. Yang, H. Yi, L. Xu, H. He, Y. Fan, and J. Zhao. 2020. “New resources for genetic studies in maize (Zea mays L.): a genome-wide Maize6H-60K SNP array and its application”. The Plant Journal. [CrossRef]
- Tucker, E.J.; Huynh, B.L. “Genotyping by high-resolution melting analysis”. Methods Mol. Biol. 2014, 1145, 59–66. [Google Scholar] [CrossRef]
- Udoh, L.I.; Melaku, G.; Parkes, Y.E.; Kulakow, P.; Adesoye, A.; Nwuba, C.; Rabbi, Y.I. “Candidate gene sequencing and validation of SNP markers linked to carotenoid content in cassava (Manihot esculenta Crantz)”. Molecular breeding. 2017, 37, 123. [Google Scholar] [CrossRef]
- Unterseer, S.; Bauer, E.; Haberer, G.; Seidel, M.; Knaak, C.; Ouzunova, M.; Meitinger, T. “A powerful tool for genome analysis in maize, development, and evaluation of the high-density 600k SNP genotyping array”. BMC Genomics 2014, 15, 823. [Google Scholar] [CrossRef] [PubMed]
- Van, O.N.J.; Hogers, R.C. J.; Janssen, A.; Yalcin, F.; Snoeijers, S.; Verstege, E. “Complexity reduction of polymorphic sequences (CRoPS): a novel approach for large-scale polymorphism discovery in complex genomes”. PLoS ONE 2007, 2, 1172. [Google Scholar] [CrossRef]
- Van, P.R.M.; Maccaferri, M.; Tang, J.; T.Truong; Janssen, A.; van-Orsouw, N.J.; Salvi, S.; et al. “Sequence-based SNP genotyping in durum wheat”. Plant Biotechnology Journal 2013, 11, 809–817. [Google Scholar] [CrossRef]
- Varshney, R.K.; Singh, V.K.; Hickey, J.M.; Xun, X.; Marshall, D.F.; Wang, J.; Ribaut, J.M. “Analytical and Decision Support Tools for Genomics-Assisted Breeding”. Trends in Plant Science 2016, 21, 354–363. [Google Scholar] [CrossRef]
- Varshney, R.K.; Hiremath, P.J.; Kashiwagi, P. Lekha. J.; Balaji, J.; Deokar, A.A.; Vadez, V.; Xiao, Y. “A comprehensive resource of drought- and salinity- responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.)”. BMC Genomics 2009, 10, 523. [Google Scholar] [CrossRef]
- Velazco, J.G., M. Malosetti, C.H. Hunt. “Combining pedigree and genomic information to improve prediction quality: an example in sorghum”. Theoritical and Applied Genetics 2019, 132, 2055–2067. [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. “Microsatellite markers: what they mean and why they are so useful”. Genetics and Molecular Biology 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Vossen, R.H.; Aten, E.; Roos, A.; Dunnen, J.T. “High-resolution melting analysis (HRMA): More than just sequence variant screening”. Hum. Mutat. 2009, 30, 860–866. [Google Scholar] [CrossRef]
- Wang, D.G.; Fan, J.B.; Siao, C.J.; Berno, A.; Young, P.; Sapolsky, R.; Ghandour, G.; Perkins, N.; et al. “Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome”. Science 1998, 280, 1077–1082. [Google Scholar] [CrossRef]
- Wang, N., Yuan, Y., Wang, H., Yu, D., Liu, Y., Zhang, A., Gowda, M., Nair, S. K., Hao, Z., Lu, Y., San Vicente, F., Prasanna, B. M., Li, X., & Zhang, X. Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Scientific Reports 2020, 10, 16308. [CrossRef]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. “DNA polymorphisms amplified by arbitrary primers are useful as genetic markers”. Nucleic Acids Research 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.J., I.Y. Rabbi, C. Egesi, M. Hamblin, R. Kawuki, P. Kulakow, and J.L. Janniink. 2016. “Genome-wide association and prediction reveals genetic architecture of cassava mosaic disease resistance and prospects for rapid genetic improvement”. The plant genome 9(2). [CrossRef]
- Wosula, E.N., W. Chen, M. Amour, Z. Fei, and J.P. Legg. 2020. “KASP Genotyping as a Molecular Tool for Diagnosis of Cassava-Colonizing”. Bemisia tabaci. Insects 11: 305. [CrossRef]
- Wright, S. I., I.V. Bi, S.C. Schroeder, M. Yamasaki, J.F. Doebley, M.D McMullen, and B.S. Gaut..2005. “Evolution: the effects of artificial selection on the maize genome”. Science 308: 1310–1314. [CrossRef]
- Xu, Y., X. Liu, J. Fu, H. Wang, J. Wang, C. Huang, B.M. Prasanna, M.S. Olsen, G. Wang, and A. Zhang. 2019. “Enhancing genetic gain through genomic selection: from livestock to plants”. Plant Communications. 100005–. [CrossRef]
- Xu, C., Y. Ren, Y. Jian, Z. Guo, Y. Zhang, C. Xie, J. Fu, H. Wang, G. Wang, Y. Xu, and P. Li. 2017. “Development of a maize 55K SNP array with improved genome coverage for molecular breeding”. Molecular Breeding, 37 (3): 20. [CrossRef]
- Yan, J., X. Yang, T. Shah, H. Sanchez-Villeda, J. Li, M. Warburton, Y. Zhou, J.H. Crouch, and Y. Xu. 2009. “High-throughput SNP genotyping with the GoldenGate assay in maize”. Mol. Breed. 15: 441– 451.
- Yang, S., W. Yu, X. Wei, Z. Wang, Y. Zhao, X. Zhao, B. Tian, Y. Yuan, and X. Zhang.2020. “An extended KASP-SNP resource for molecular breeding in Chinese cabbage (Brassica rapa L. ssp. pekinensis)”. PLoS ONE 15: e0240042. [CrossRef]
- You, Q., Yang, X., Peng, Z., Xu, L., and Wang, J. 2018. Development and applications of a high-throughput genotyping tool for polyploid crops: Single nucleotide polymorphism (SNP) array. Frontiers in Plant Science, 9, Article 104. [CrossRef]
- Yu, H.; Xie, W.; Wang, J.; Xing, Y.; Xu, C.; Li, X.; Xiao, J.; Zhang, O. “Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers”. PLoS ONE 2011, 6, 10–13. [Google Scholar] [CrossRef]
- Yu, J.; Holland, J.B.; McMullen, M.D.; Buckler, E.S. “Genetic design and statistical power of nested association mapping in maize”. Genetics 2008, 178, 539. [Google Scholar] [CrossRef]
- Yuan, Y., SanMiguel, P.J. and Bennetzen, J.L. “High-Cot sequence analysis of the maize genome”. The Plant Journal 2003, 34, 249–255. [CrossRef]
- Zhang Z., X. Guo; Liu, B.; Tang, L.; Chen, F. “Genetic diversity and genetic relationship of Jatropha curcas between China and Southeast Asia revealed by amplified fragment length polymorphisms”. African Journal of Biotechnology 2011, 10, 2825–2832. [Google Scholar] [CrossRef]
- Zhang, S., Ye J., Lu K., Zou M., Chen X. and Zhou X. Genome-wide association studies of 11 Agronomical Traits in Cassava (Manihot esculenta Crantz). Frontier in plant science 2018, 9, 503. [CrossRef]
- Zhang, J., Yang, J., Zhang, L., Chen, L., Luo, J., Zhao, H., and Zhao, J. (2020). A new SNP genotyping technology Target SNP-seq and its application in genetic analysis of cucumber varieties. Scientific Reports 10: 5623. [CrossRef]
- Zhao Y., M. Gowda, W. Liu, T. Würschum, H.P. Maurer, F.H. Longin, N. Ranc, and J.C. Reif.2012. “Accuracy of genomic selection in European maize elite breeding populations”. Theoretical and Applied Genetics 124: 769–776. [CrossRef]
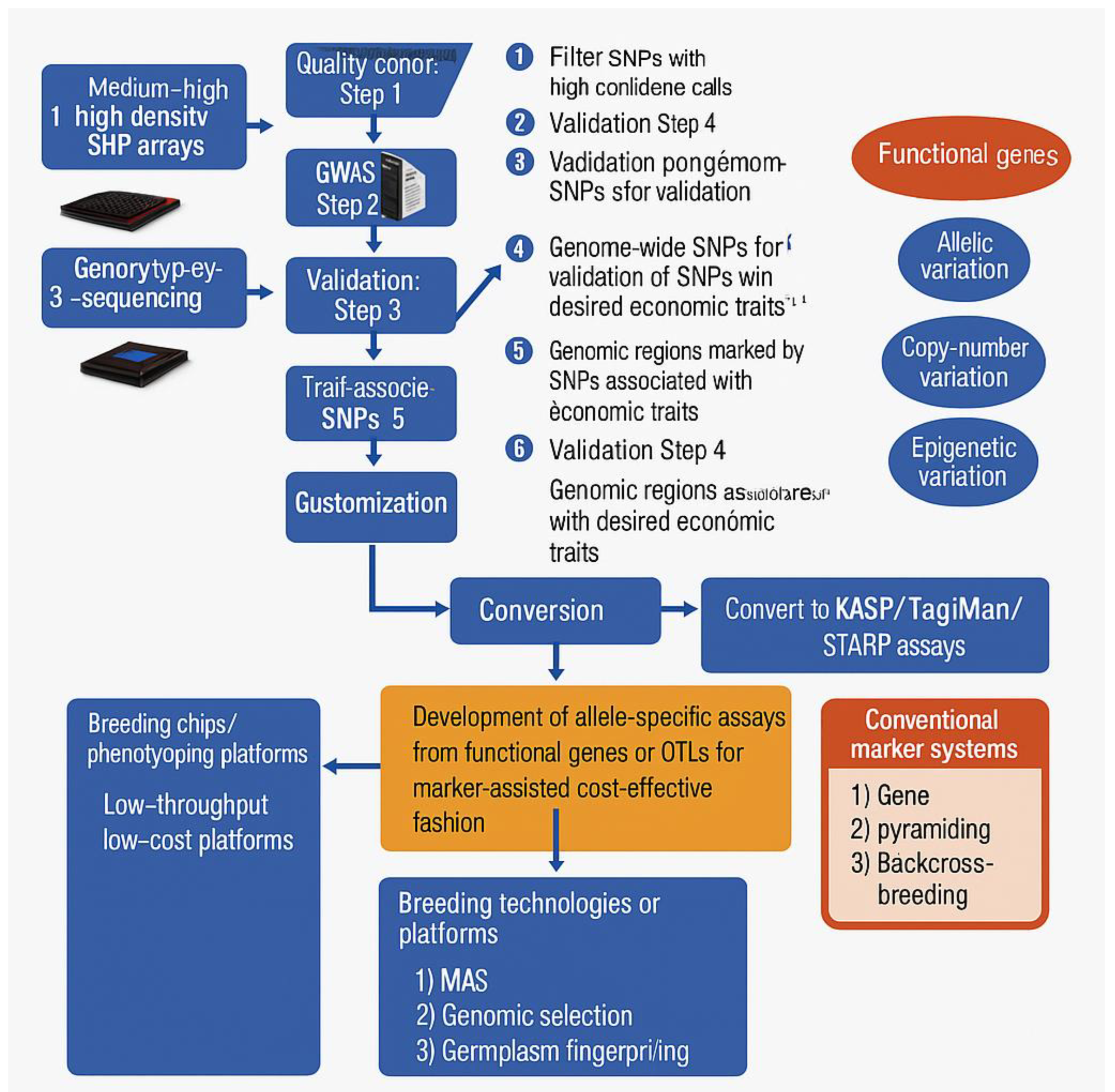
| Platform | Technology | Provider | Cost Per Sample | Cost Per Data Point | Analysis of Complexity | Prior Genomic Knowledge | Throughput | Flexibility | Application |
|---|---|---|---|---|---|---|---|---|---|
| Array-based | GoldenGate | Illumina | High | Moderate | Moderate | Yes | 172 × 1.5K | No | Tier 1 |
| Infinium XT | Illumina | Moderate | Low | Moderate | Yes | 96 × 50K | No | Tier 1 | |
| Infinium HD/HTS | Illumina | High | Low | Moderate | Yes | 24 × 90K/ 24 × 700K | No | Tier 1 | |
| Axiom | Affymetrix | Moderate to high | low | Moderate | Yes | 96 × 1000K Or 384 ×55K | No | Tier 1 | |
| NGS based | GBS | Non-commercial | Moderate | Low | Difficult | No | depend on sample multiples | Low | Tier 1 |
| RAD-seq | Non-commercial | Moderate | Low | Difficult | No | -do- | Low | Tier 1 | |
| SLAF-seq | Biomarker Tech | High | Low | Difficult | No | -do- | Low | Tier 1 | |
| Exome capture | Agilent/Nim-bleGen | High | Low | Difficult | Yes | -do- | Low to moderate | Tier 1 | |
| DArT-seq | DiveraityArray | Moderate | Low | Commercial support available | No | 96 × 50-100K | Low | ||
| rAmseq | Non-commercial | Very low | Low | Difficult | Yes | Multiplex | Low | Tier 1 | |
| Targeted GBS/low density array | fluidgm | fluidgm | Moderate | moderate | moderate | yes | 96 × 96/ 24 × 192/ 48 × 48 | moderate | Tier 2 |
| Sequenom MassARRAY | Agena Bioscience | Moderate | Moderate | Moderate | yes | 96 × 48 | Low | Tier 2 | |
| Eureka | Affymetrix | Moderate | Moderate | Moderate | Yes | At least 5K × 3K | Low | Tier 2 | |
| AmpliSeq | Thermo Fisher | Moderate | Moderate | Moderate | Yes | customizable | Moderate | Tier 2 | |
| Single Markers | KASP | LGC Group | Depend on reaction volume and assay number | High | Easy | Yes | Single-plex (up to ~150K data points/day) | Scalable | Tier 2 |
| TaqMan | Roche Molecular System | -do- | High | Easy | Yes | -do- | -do- | Tier 2 | |
| STARP | Non-commercial | -do- | Moderate | Easy | yes | -do- | -do- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





