Submitted:
15 April 2025
Posted:
16 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
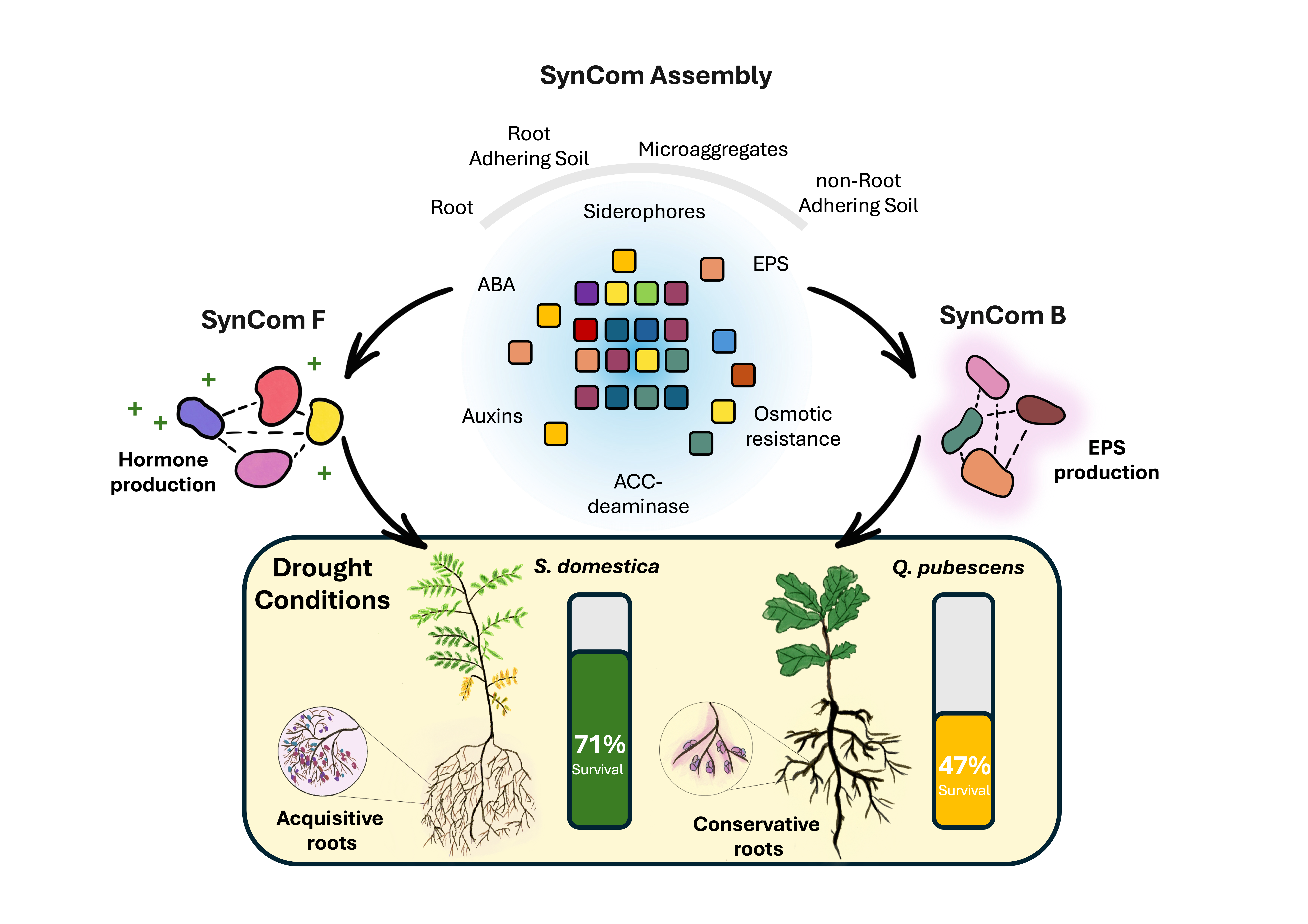
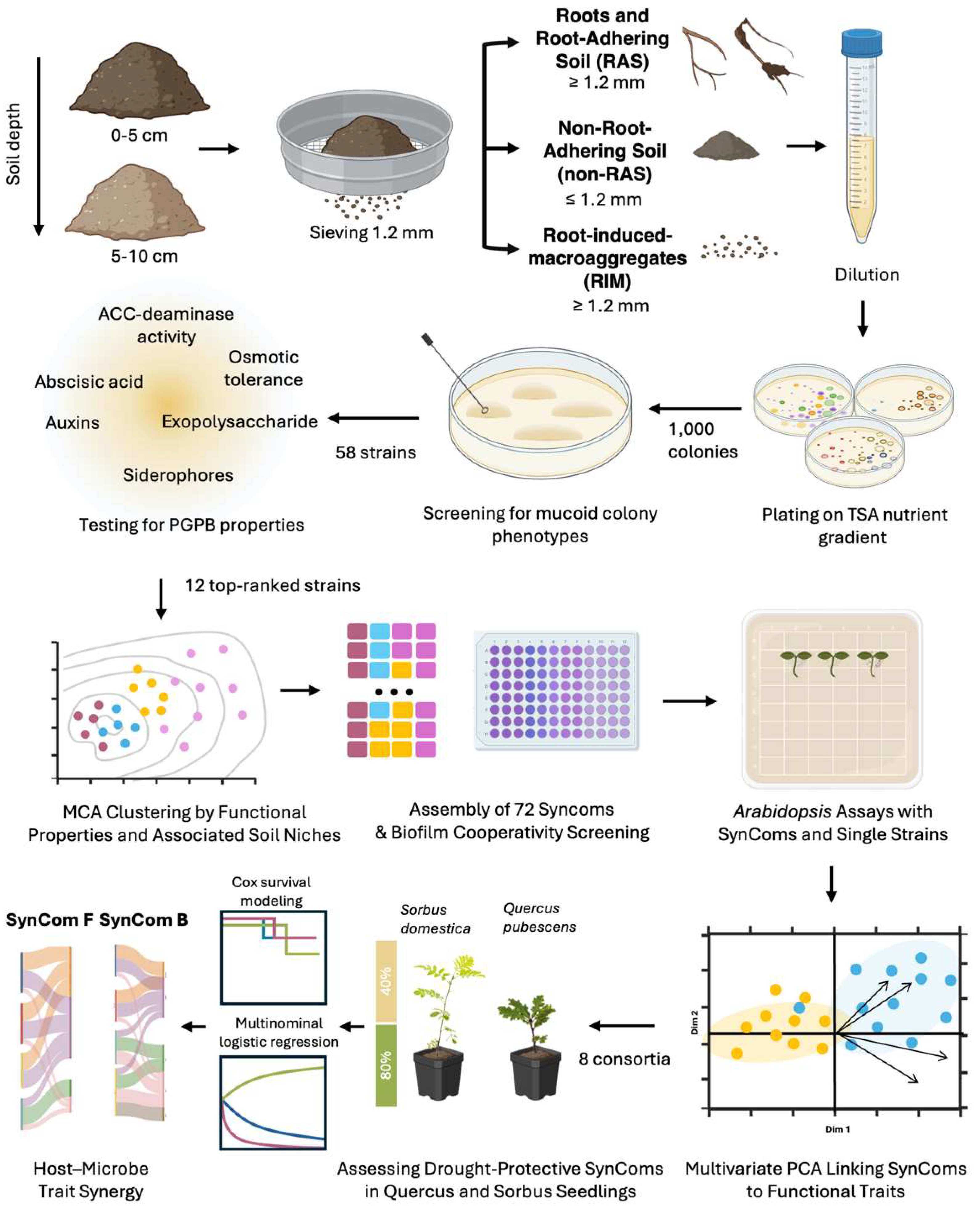
2. Results
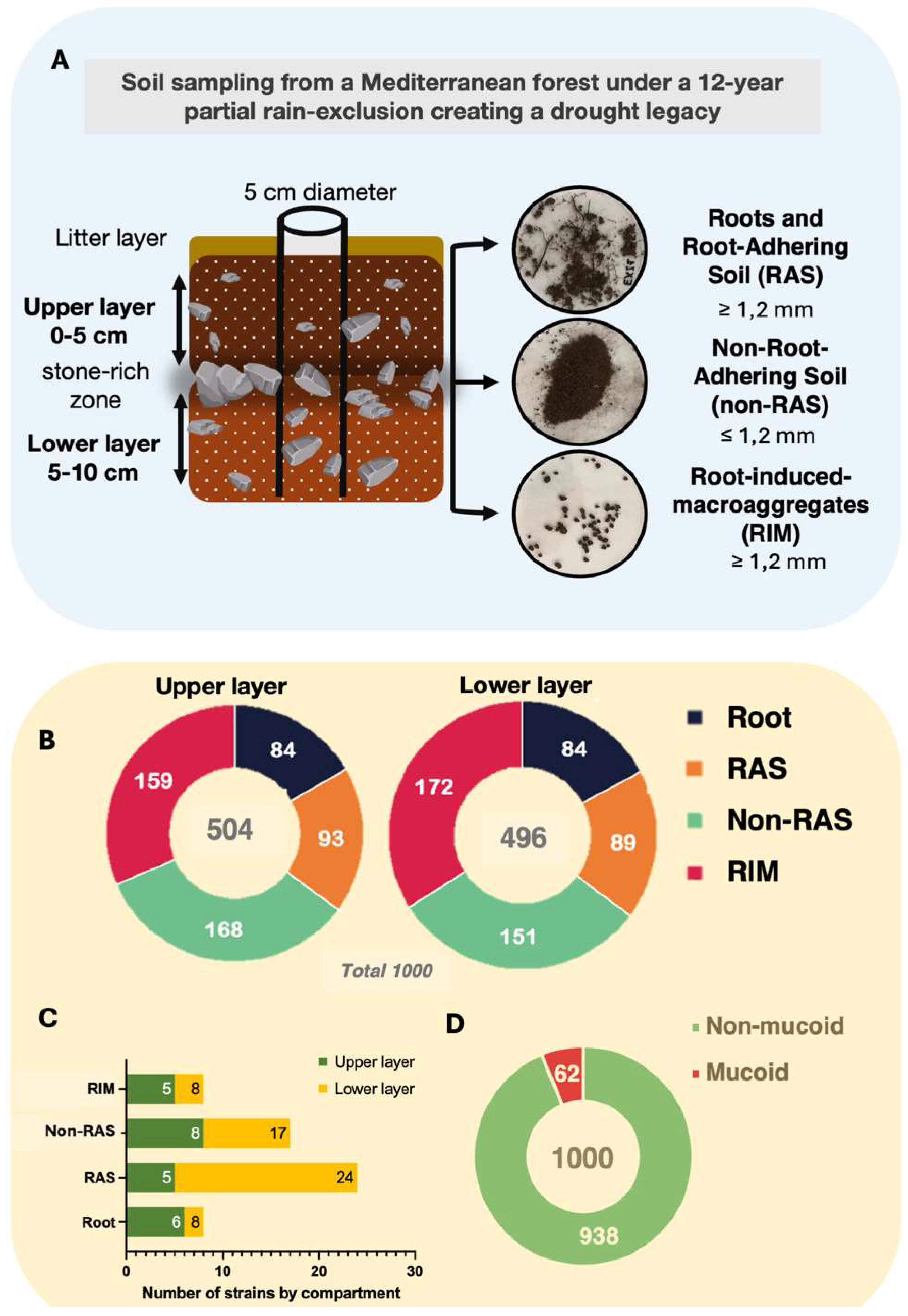
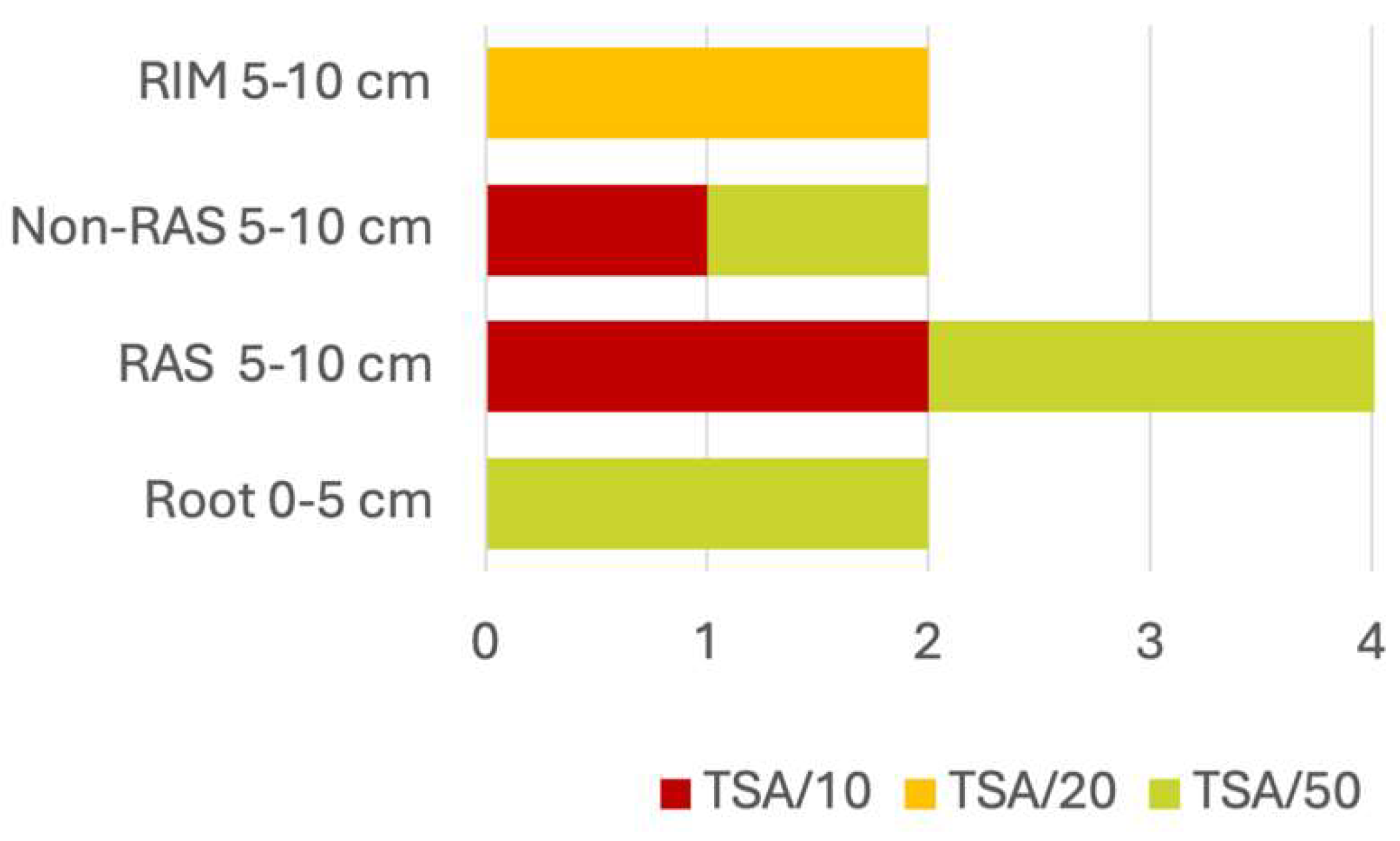
Bacterial Strain Isolation
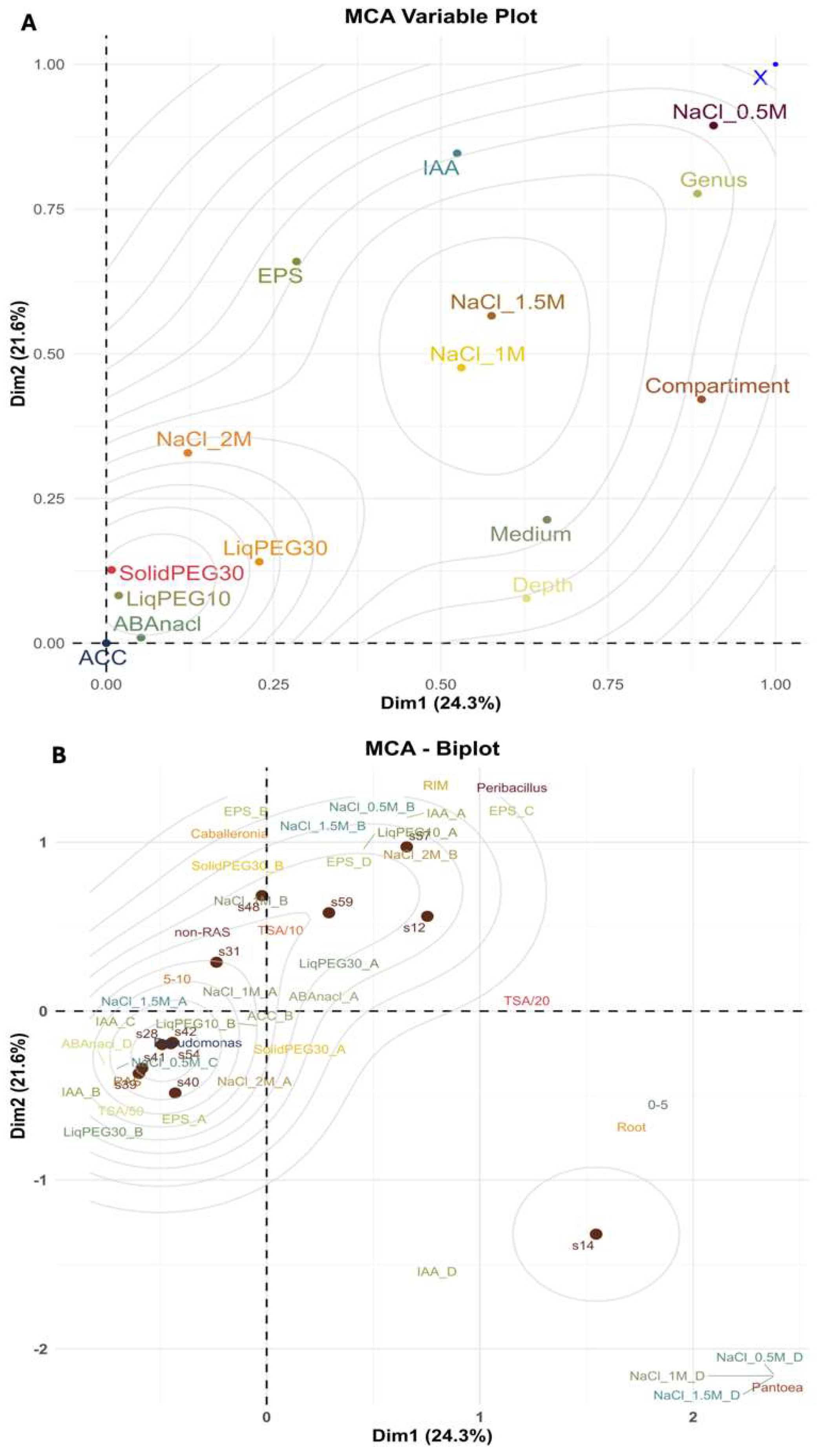
Multivariate Analysis of Strain Clustering Based on Functional Traits, Spatial and Nutrient Gradients
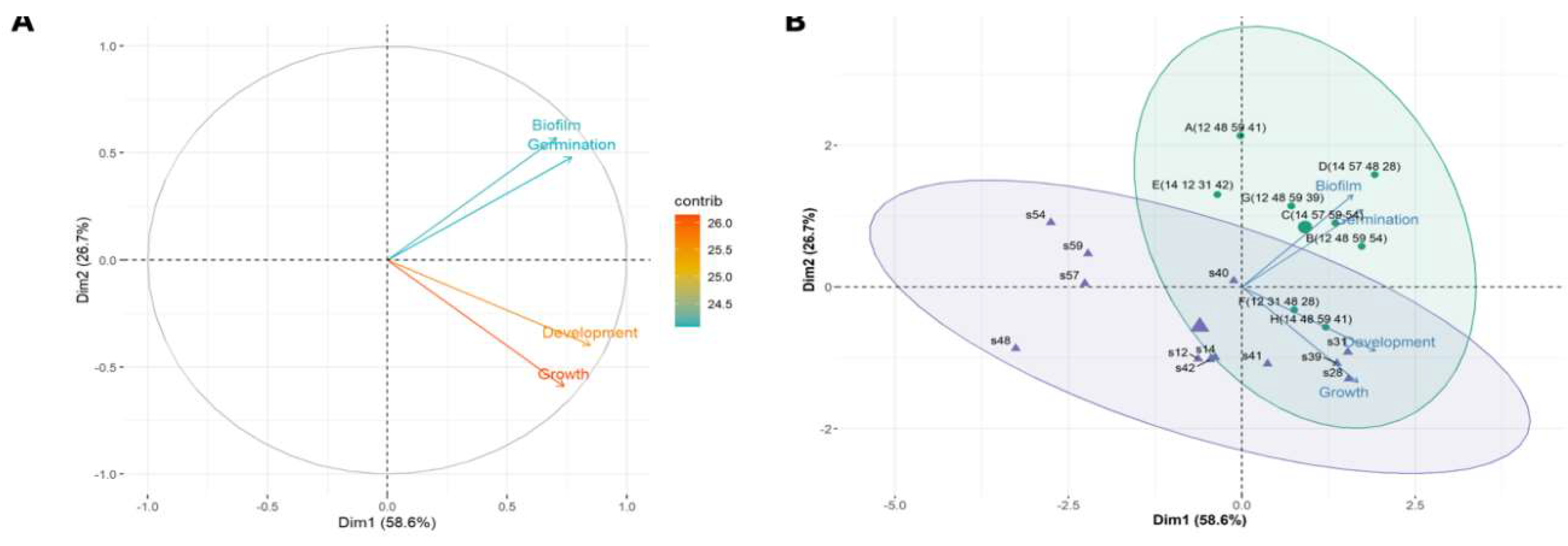
Designing Synthetic Communities (SynComs)
Biofilm Assay
In Vitro Inoculation of Arabidopsis with Single Bacterial Strains or Consortia
Data-Driven Selection of Top-Performing SynCom Candidates
SynComs and Single Inoculants to Mitigate Drought Stress in Tree Seedlings
Prediction of Inoculant Efficacy for Drought Protection of Quercus and Sorbus Seedlings
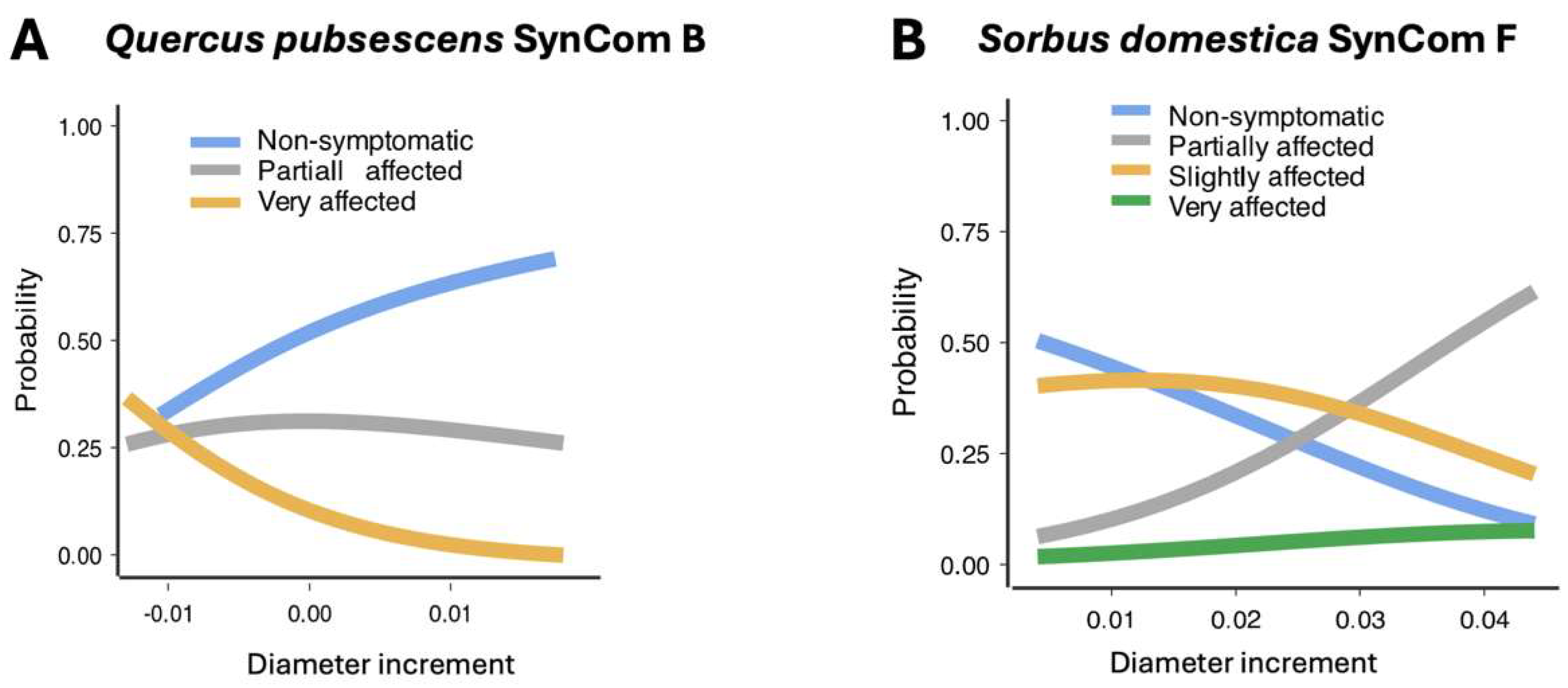
Quercus Pubescens
Sorbus Domestica
3. Discussion
From Soil Niches to Synthetic Communities (SynCom)
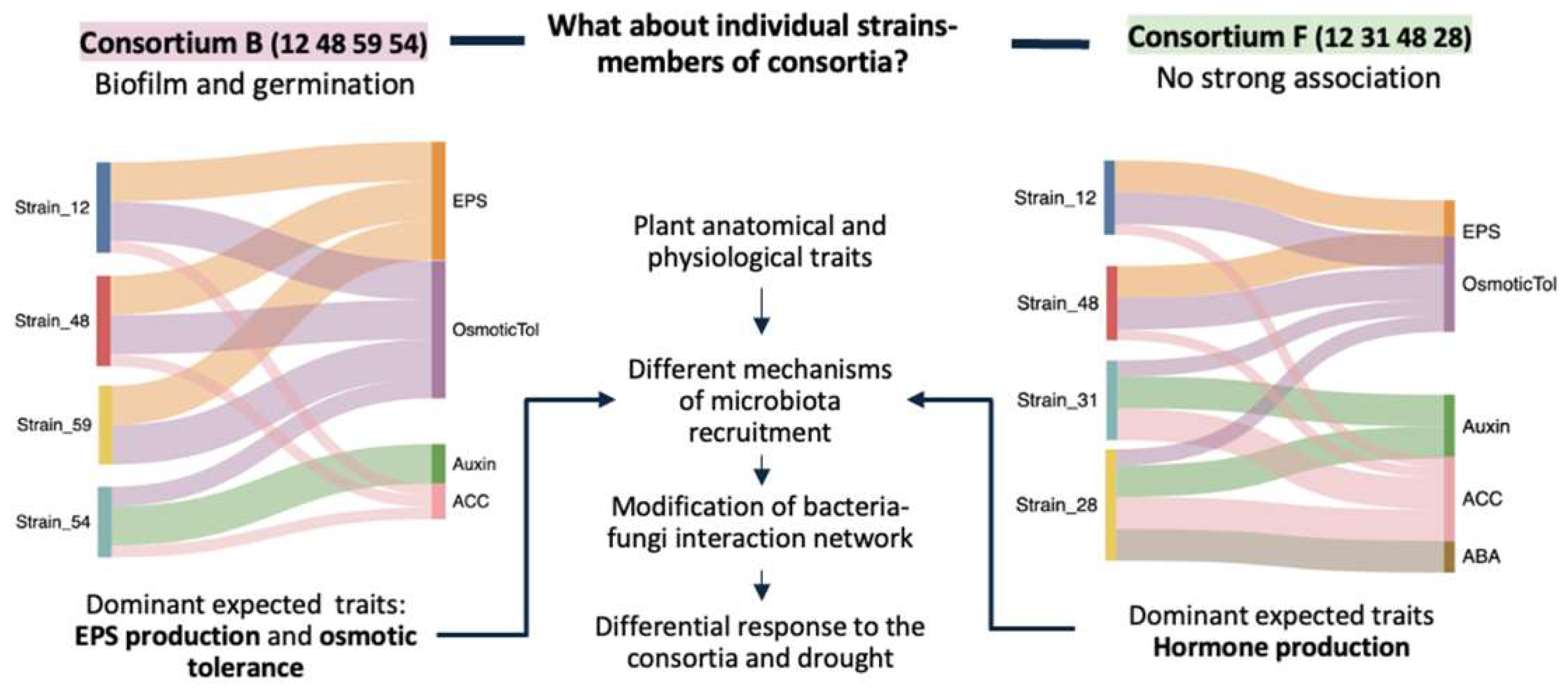
Stronger Together? How Weak Biofilm Formers Drive Collective Performance
Teaming Up Underground: Optimizing SynCom by Matching Microbial Skills to Root Strategies
Beyond Survival: Predictive Insights into SynCom Function and Drought Tolerance in Trees
4. Materials and Methods
Soil Sampling
Strain Isolation and Growth
Strain Screening for Specific Traits
Ability to Form Biofilms
Strain Identification
Test of Single Strains and Consortia on Arabidopsis thaliana
Greenhouse Experiments on Trees
Statistical Analyses
Multivariate Analyses-Driven Selection of the Best Candidate Strains for SynComs Selection
Statistical Analyses on Plant Symptoms and Growth Parameters
Survival Models
Multinomial Logistic Regression
Use of GenAI
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC Climate Change 2022 – Impacts, Adaptation and Vulnerability: Working Group II Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; 1st ed.; Cambridge University Press, 2023; ISBN 978-1-00-932584-4.
- Senf, C.; Buras, A.; Zang, C.S.; Rammig, A.; Seidl, R. Excess Forest Mortality Is Consistently Linked to Drought across Europe. Nat Commun 2020, 11, 6200. [CrossRef]
- Beloiu, M.; Stahlmann, R.; Beierkuhnlein, C. Drought Impacts in Forest Canopy and Deciduous Tree Saplings in Central European Forests. Forest Ecology and Management 2022, 509, 120075. [CrossRef]
- Gazol, A.; Camarero, J.J. Compound Climate Events Increase Tree Drought Mortality across European Forests. Science of The Total Environment 2022, 816, 151604. [CrossRef]
- Bevacqua, E.; Rakovec, O.; Schumacher, D.; Kumar, R.; Thober, S.; Samaniego, L.; Seneviratne, S.; Zscheischler, J. Direct and Lagged Climate Change Effects Strongly Intensified the Widespread 2022 European Drought 2024.
- Dumroese, R.K.; Balachowski, J.A.; Flores, D.; Horning, M.E. Reforestation to Mitigate Changes to Climate: More than Just Planting Seedlings. In In: XV World Forestry Congress: Building a Green, Healthy, and Resilient Future with Forests.; 2-6 May 2022; Coex, Seoul, Republic of Korea. Food and Agriculture Organization of the United Nations; Korean Forest Service 2022, 7.
- Knutzen, F.; Averbeck, P.; Barrasso, C.; Bouwer, L.M.; Gardiner, B.; Grünzweig, J.M.; Hänel, S.; Haustein, K.; Johannessen, M.R.; Kollet, S.; et al. Impacts on and Damage to European Forests from the 2018–2022 Heat and Drought Events. Natural Hazards and Earth System Sciences 2025, 25, 77–117. [CrossRef]
- Puértolas, J.; Villar-Salvador, P.; Andivia, E.; Ahuja, I.; Cocozza, C.; Cvjetković, B.; Devetaković, J.; Diez, J.J.; Fløistad, I.S.; Ganatsas, P.; et al. Die-Hard Seedlings. A Global Meta-Analysis on the Factors Determining the Effectiveness of Drought Hardening on Growth and Survival of Forest Plantations. Forest Ecology and Management 2024, 572, 122300. [CrossRef]
- Ait-El-Mokhtar, M.; Meddich, A.; Baslam, M. Plant-Microbiome Interactions under Drought—Insights from the Molecular Machinist’s Toolbox. Frontiers in Sustainable Food Systems. 2023, 7, 1253735.
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant physiology 2020, 184, 39–52. [CrossRef]
- Zahar Haichar, F.; Santaella, C.; Heulin, T.; Achouak, W. Root Exudates Mediated Interactions Belowground. Soil Biology and Biochemistry 2014, 77, 69–80. [CrossRef]
- Hong, Y.; Zhou, Q.; Hao, Y.; Huang, A.C. Crafting the Plant Root Metabolome for Improved Microbe-assisted Stress Resilience. New Phytologist 2022, 234, 1945–1950. [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for Help with Root Exudates: Adaptive Mechanisms by Which Stressed Plants Assemble Health-Promoting Soil Microbiomes. Current opinion in microbiology 2019, 49, 73–82. [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of Plant Growth Promoting Rhizobacteria (PGPRs) with Multiple Plant Growth Promoting Traits in Stress Agriculture: Action Mechanisms and Future Prospects. Ecotox. Environ. Safe. 2018, 156, 225–246. [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Diaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H.; et al. Phytohormones Trigger Drought Tolerance in Crop Plants: Outlook and Future Perspectives. Front. Plant Sci. 2022, 12, 799318. [CrossRef]
- Campos, E.; Pereira, A.; Aleksieienko, I.; Carmo, G.; Gohari, G.; Santaella, C.; Fraceto, L.; Oliveira, H. Encapsulated Plant Growth Regulators and Associative Microorganisms: Nature-Based Solutions to Mitigate the Effects of Climate Change on Plants. Plant Science 2023, 331, 111688. [CrossRef]
- Tiepo, A.N.; Constantino, L.V.; Madeira, T.B.; Gonçalves, L.S.A.; Pimenta, J.A.; Bianchini, E.; de Oliveira, A.L.M.; Oliveira, H.C.; Stolf-Moreira, R. Plant Growth-Promoting Bacteria Improve Leaf Antioxidant Metabolism of Drought-Stressed Neotropical Trees. Planta 2020, 251, 83. [CrossRef]
- Tiepo, A.N.; Hertel, M.F.; Rocha, S.S.; Calzavara, A.K.; De Oliveira, A.L.M.; Pimenta, J.A.; Oliveira, H.C.; Bianchini, E.; Stolf-Moreira, R. Enhanced Drought Tolerance in Seedlings of Neotropical Tree Species Inoculated with Plant Growth-Promoting Bacteria. Plant Physiology and Biochemistry 2018, 130, 277–288. [CrossRef]
- Khosravi, M.; Heydari, M.; Alikhani, H.A.; Arani, A.M.; Guidi, L.; Bernard, P. Mitigating Negative Impacts of Drought on Oak Seedlings Performances through Plant Growth-Promoting Rhizobacteria. Journal of Environmental Management 2025, 375, 124163. [CrossRef]
- Gillespie, L.M.; Fromin, N.; Milcu, A.; Buatois, B.; Pontoizeau, C.; Hättenschwiler, S. Higher Tree Diversity Increases Soil Microbial Resistance to Drought. Communications biology 2020, 3, 377. [CrossRef]
- Poudel, M.; Mendes, R.; Costa, L.A.; Bueno, C.G.; Meng, Y.; Folimonova, S.Y.; Martins, S.J. The Role of Plant-Associated Bacteria, Fungi, and Viruses in Drought Stress Mitigation. Frontiers in microbiology 2021, 12, 743512. [CrossRef]
- Carballo-Sánchez, M.P.; Alarcón, A.; Pérez-Moreno, J.; Ferrera-Cerrato, R. Agricultural and Forestry Importance of Microorganism-Plant Symbioses: A Microbial Source for Biotechnological Innovations. Reviews in Agricultural Science 2022, 10, 344–355. [CrossRef]
- Santos, R.F.D.; Cruz, S.P.D.; Botelho, G.R.; Flores, A.V. Inoculation of Pinus Taeda Seedlings with Plant Growth-Promoting Rhizobacteria. Floresta e Ambiente 2018, 25, 20160056.
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-Solubilizing Microorganisms: Mechanism and Their Role in Phosphate Solubilization and Uptake. J Soil Sci Plant Nutr 2021, 21, 49–68. [CrossRef]
- Khan, A.L. The Phytomicrobiome: Solving Plant Stress Tolerance under Climate Change. Frontiers in Plant Science 2023, 14, 1219366. [CrossRef]
- Liu, X.; Mei, S.; Salles, J.F. Inoculated Microbial Consortia Perform Better than Single Strains in Living Soil: A Meta-Analysis. Applied Soil Ecology 2023, 190, 105011. [CrossRef]
- Nicotra, D.; Mosca, A.; Dimaria, G.; Massimino, M.E.; Stabile, M.; Bella, E.; Catara, V. Mitigating Water Stress in Plants with Beneficial Bacteria: Effects on Growth and Rhizosphere Bacterial Communities. International Journal of Molecular Sciences 2025, 26, 1467. [CrossRef]
- Jing, J.; Garbeva, P.; Raaijmakers, J.M.; Medema, M.H. Strategies for Tailoring Functional Microbial Synthetic Communities. The ISME journal 2024, wrae049. [CrossRef]
- Mehlferber, E.C.; Arnault, G.; Joshi, B.; Partida-Martinez, L.P.; Patras, K.A.; Simonin, M.; Koskella, B. A Cross-Systems Primer for Synthetic Microbial Communities. Nature microbiology 2024, 9, 2765–2773. [CrossRef]
- Xu, X.; Dinesen, C.; Pioppi, A.; Kovács, Á.T.; Lozano-Andrade, C.N. Composing a Microbial Symphony: Synthetic Communities for Promoting Plant Growth; Trends in Microbiology, 2025;
- Stock, S.C.; Koester, M.; Boy, J.; Godoy, R.; Nájera, F.; Matus, F.; Dippold, M.A. Plant Carbon Investment in Fine Roots and Arbuscular Mycorrhizal Fungi: A Cross-Biome Study on Nutrient Acquisition Strategies. Science of the Total Environment 2021. [CrossRef]
- Bergmann, J.; Weigelt, A.; Plas, F.; Laughlin, D.C.; Kuyper, T.W.; Guerrero-Ramirez, N.; Valverde-Barrantes, O.J.; Bruelheide, H.; Freschet, G.T.; Iversen, C.M.; et al. The Fungal Collaboration Gradient Dominates the Root Economics Space in Plants. Science Advances 2020, 6, 3756. [CrossRef]
- Prada-Salcedo, L.D.; Goldmann, K.; Heintz-Buschart, A.; Reitz, T.; Wambsganss, J.; Bauhus, J.; Buscot, F. Fungal Guilds and Soil Functionality Respond to Tree Community Traits Rather than to Tree Diversity in European Forests. Molecular Ecology 2021, 30, 572–591. [CrossRef]
- Wu, R.; Zeng, X.; McCormack, M.L.; Fernandez, C.W.; Yang, Y.; Guo, H.; Chen, W. Linking Root-Associated Fungal and Bacterial Functions to Root Economics. bioRxiv 2023.
- Kou, X.; Han, W.; Kang, J. Responses of Root System Architecture to Water Stress at Multiple Levels: A Meta-Analysis of Trials under Controlled Conditions. Frontiers in Plant Science 2022, 13, 1085409. [CrossRef]
- Bauke, S.L.; Amelung, W.; Bol, R.; Brandt, L.; Brüggemann, N.; Kandeler, E.; Vereecken, H. Soil Water Status Shapes Nutrient Cycling in Agroecosystems from Micrometer to Landscape Scales. Journal of Plant Nutrition and Soil Science 2022, 185, 773–792. [CrossRef]
- Platt, T.G. Community Outcomes Depend on Cooperative Biofilm Structure. Proceedings of the National Academy of Sciences 2023, 120, 2221624120. [CrossRef]
- Laoué, J.; Havaux, M.; Ksas, B.; Orts, J.P.; Reiter, I.M.; Fernandez, C.; Ormeno, E. A Decade of Rain Exclusion in a Mediterranean Forest Reveals Trade-Offs of Leaf Chemical Defenses and Drought Legacy Effects. Scientific Reports 2024, 14, 24119. [CrossRef]
- Belviso, S.; Reiter, I.M.; Loubet, B.; Gros, V.; Lathière, J.; Montagne, D.; Delmotte, M.; Ramonet, M.; Kalogridis, C.; Lebegue, B.; et al. A Top-down Approach of Surface Carbonyl Sulfide Exchange by a Mediterranean Oak Forest Ecosystem in Southern France. Atmospheric Chemistry and Physics 2016, 16, 14909–14923. [CrossRef]
- Knights, H.E.; Jorrin, B.; Haskett, T.L.; Poole, P.S. Deciphering Bacterial Mechanisms of Root Colonization. Environmental Microbiology Reports 2021, 13, 428–444. [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, Variation, and Assembly of the Root-Associated Microbiomes of Rice. Proceedings of the National Academy of Sciences 2015, 112, 911–920. [CrossRef]
- Li, M.; Song, Z.; Li, Z.; Qiao, R.; Zhang, P.; Ding, C.; Xie, J.; Chen, Y.; Guo, H. Populus Root Exudates Are Associated with Rhizosphere Microbial Communities and Symbiotic Patterns. Frontiers in Microbiology 2022, 13, 1042944. [CrossRef]
- Baumert, V.L.; Vasilyeva, N.A.; Vladimirov, A.A.; Meier, I.C.; Kögel-Knabner, I.; Mueller, C.W. Root Exudates Induce Soil Macroaggregation Facilitated by Fungi in Subsoil. Frontiers in Environmental Science 2018, 6, 140. [CrossRef]
- Deng, Y.; Kong, W.; Zhang, X.; Zhu, Y.; Xie, T.; Chen, M.; Zhu, L.; Sun, J.; Zhang, Z.; Chen, C.; et al. Rhizosphere Microbial Community Enrichment Processes in Healthy and Diseased Plants: Implications of Soil Properties on Biomarkers. Frontiers in Microbiology 2024, 15, 1333076. [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant Growth Promoting Rhizobacteria Isolated from Halophytes and Drought-Tolerant Plants: Genomic Characterisation and Exploration of Phyto-Beneficial Traits. Sci Rep 2020, 10, 14857. [CrossRef]
- Petrillo, C.; Vitale, E.; Ambrosino, P.; Arena, C.; Isticato, R. Plant Growth-Promoting Bacterial Consortia as a Strategy to Alleviate Drought Stress in Spinacia Oleracea. Microorganisms 2022, 10, 1798. [CrossRef]
- Lin, Y.; Zhang, H.; Li, P.; Jin, J.; Li, Z. The Bacterial Consortia Promote Plant Growth and Secondary Metabolite Accumulation in Astragalus Mongholicus under Drought Stress. BMC Plant Biol 2022, 22, 475. [CrossRef]
- Benmrid, B.; Ghoulam, C.; Zeroual, Y.; Kouisni, L.; Bargaz, A. Bioinoculants as a Means of Increasing Crop Tolerance to Drought and Phosphorus Deficiency in Legume-Cereal Intercropping Systems. Commun Biol 2023, 6, 1–15. [CrossRef]
- Ren, D.; Madsen, J.S.; Sørensen, S.J.; Burmølle, M. High Prevalence of Biofilm Synergy among Bacterial Soil Isolates in Cocultures Indicates Bacterial Interspecific Cooperation. The ISME Journal 2015, 9, 81–89. [CrossRef]
- Ren, D.; Madsen, J.S.; Cruz-Perera, C.I.; Bergmark, L.; Sørensen, S.J.; Burmølle, M. High-Throughput Screening of Multispecies Biofilm Formation and Quantitative Pcr-Based Assessment of Individual Species Proportions, Useful for Exploring Interspecific Bacterial Interactions. Microb Ecol 2014, 68, 146–154. [CrossRef]
- Lange, M.A. Climate Change in the Mediterranean: Environmental Impacts and Extreme Events. IEMed Mediterranean Yearbook 2020, 224–229.
- Meisner, A.; Jacquiod, S.; Snoek, B.L.; ten Hooven, F.C.; van der Putten, W.H. Drought Legacy Effects on the Composition of Soil Fungal and Prokaryote Communities. Front. Microbiol. 2018, 9. [CrossRef]
- Vilonen, L.L.; Hoosein, S.; Smith, M.D.; Trivedi, P. Legacy Effects of Intensified Drought on the Soil Microbiome in a Mesic Grassland. Ecosphere 2023, 14, e4545. [CrossRef]
- Tang, Y.; Winterfeldt, S.; Brangarí, A.C.; Hicks, L.C.; Rousk, J. Higher Resistance and Resilience of Bacterial Growth to Drought in Grasslands with Historically Lower Precipitation. Soil Biology and Biochemistry 2023, 177, 108889. [CrossRef]
- Alotaibi, F.; St-Arnaud, M.; Hijri, M. In-Depth Characterization of Plant Growth Promotion Potentials of Selected Alkanes-Degrading Plant Growth-Promoting Bacterial Isolates. Frontiers in Microbiology 2022, 13, 863702. [CrossRef]
- Manetsberger, J.; Caballero Gómez, N.; Soria-Rodríguez, C.; Benomar, N.; Abriouel, H. Simply Versatile: The Use of Peribacillus Simplex in Sustainable Agriculture. Microorganisms 2023, 11, 2540. [CrossRef]
- Ma, Y.; Yin, Y.; Rong, C.; Chen, S.; Liu, Y.; Wang, S.; Xu, F. Pantoea Pleuroti Sp. Nov., Isolated from the Fruiting Bodies of Pleurotus Eryngii. Current Microbiology 2016, 72, 207–212. [CrossRef]
- Sikorski, J.; Pukall, R.; Stackebrandt, E. Carbon Source Utilization Patterns of Bacillus Simplex Ecotypes Do Not Reflect Their Adaptation to Ecologically Divergent Slopes in ‘Evolution Canyon.’ Israel. FEMS microbiology ecology 2008, 66, 38–44.
- Palmer, M.; Steenkamp, E.T.; Coetzee, M.P.; Avontuur, J.R.; Chan, W.Y.; Zyl, E.; Venter, S.N. Mixta Gen. Nov., a New Genus in the Erwiniaceae. International journal of systematic and evolutionary microbiology 2018, 68, 1396–1407.
- Dragone, N.B.; Hoffert, M.; Strickland, M.S.; Fierer, N. Taxonomic and Genomic Attributes of Oligotrophic Soil Bacteria. ISME communications 2024, 4, 081. [CrossRef]
- Verhille, S.; Baida, N.; Dabboussi, F.; Hamze, M.; Izard, D.; Leclerc, H. Pseudomonas Gessardii Sp. Nov. and Pseudomonas Migulae Sp. Nov., Two New Species Isolated from Natural Mineral Waters. International Journal of Systematic and Evolutionary Microbiology 1999, 49, 1559–1572.
- Puri, A.; Padda, K.P.; Chanway, C.P. Evidence of Endophytic Diazotrophic Bacteria in Lodgepole Pine and Hybrid White Spruce Trees Growing in Soils with Different Nutrient Statuses in the West Chilcotin Region of British Columbia, Canada. Forest Ecology and Management 2018, 430, 558–565. [CrossRef]
- Costa, O.Y.A.; Raaijmakers, J.M.; Kuramae, E.E. Microbial Extracellular Polymeric Substances: Ecological Function and Impact on Soil Aggregation. Front. Microbiol. 2018, 9. [CrossRef]
- Waqas, M.; Zelenková, S.; Vogel, C.; Rühmann, B.; Varsadiya, M.; Liebmann, P.; Bárta, J. Stick Together: Isolation and Characterization of Exopolysaccharides Producing Bacteria from Degraded Permafrost Soils. bioRxiv 2024.
- Dobritsa, A.P.; Linardopoulou, E.V.; Samadpour, M. Transfer of 13 Species of the Genus Burkholderia to the Genus Caballeronia and Reclassification of Burkholderia Jirisanensis as Paraburkholderia Jirisanensis Comb. Nov. International Journal of Systematic and Evolutionary Microbiology 2017, 67, 3846–3853. [CrossRef]
- Zolg, W.; Ottow, J.C. Pseudomonas Glathei Sp. Nov., a New Nitrogen-Scavening Rod Isolated from Acid Lateritic Relicts in Germany. J Comp Neurol 1975, 164, 287–299.
- Peeters, C.; Meier-Kolthoff, J.P.; Verheyde, B.; Brandt, E.; Cooper, V.S.; Vandamme, P.; Y., O.; Raaijmakers, J.M.; Kuramae, E.E. Phylogenomic Study of Burkholderia Glathei-like Organisms, Proposal of 13 Novel Burkholderia Species and Emended Descriptions of Burkholderia Sordidicola, Burkholderia Zhejiangensis, and Burkholderia Grimmiae. Frontiers in microbiology 2016, 7, 877 ,.
- Li, R.; Jiang, Y.; Wang, X.; Yang, J.; Gao, Y.; Zi, X.; Hu, N. Psychrotrophic Pseudomonas Mandelii CBS-1 Produces High Levels of Poly-β-Hydroxybutyrate. Springerplus 2013, 2, 1–7. [CrossRef]
- Kimmel, S.A.; Roberts, R.F.; Ziegler, G.R. Optimization of Exopolysaccharide Production by Lactobacillus Delbrueckii Subsp. Bulgaricus RR Grown in a Semidefined Medium. Applied and Environmental Microbiology 1998, 64, 659–664.
- Naseem, H.; Ahsan, M.; Shahid, M.A.; Khan, N. Exopolysaccharides Producing Rhizobacteria and Their Role in Plant Growth and Drought Tolerance. Journal of basic microbiology 2018, 58, 1009–1022. [CrossRef]
- Guo, Y.S.; Furrer, J.M.; Kadilak, A.L.; Hinestroza, H.F.; Gage, D.J.; Cho, Y.K.; Shor, L.M. Bacterial Extracellular Polymeric Substances Amplify Water Content Variability at the Pore Scale. Frontiers in Environmental Science 2018, 6, 93. [CrossRef]
- Bourles, A.; Amir, H.; Gensous, S.; Cussonneau, F.; Medevielle, V. Investigating Some Mechanisms Underlying Stress Metal Adaptations of Two Burkholderia Sensu Lato Species Isolated from New Caledonian Ultramafic Soils. European Journal of Soil Biology 2020, 97, 103166. [CrossRef]
- Paul, S.; Parvez, S.S.; Goswami, A.; Banik, A. Exopolysaccharides from Agriculturally Important Microorganisms: Conferring Soil Nutrient Status and Plant Health. International Journal of Biological Macromolecules 2024, 262, 129954. [CrossRef]
- Vanhaverbeke, C.; Heyraud, A.; Mazeau, K. Conformational Analysis of the Exopolysaccharide from Burkholderia Caribensis Strain MWAP71: Impact on the Interaction with Soils. Biopolymers 2003, 69, 480–497. [CrossRef]
- Lünsdorf, E.; Abraham; Timmis Clay Hutches’: A Novel Interaction between Bacteria and Clay Minerals. Environmental Microbiology 2000, 2, 161–168.
- Santaella, C.; Schue, M.; Berge, O.; Heulin, T.; Achouak, W. The Exopolysaccharide of Rhizobium Sp. YAS34 Is Not Necessary for Biofilm Formation on Arabidopsis Thaliana and Brassica Napus Roots but Contributes to Root Colonization. Environmental Microbiology 2008, 10, 2150–2163. [CrossRef]
- Driouich, A.; Gaudry, A.; Pawlak, B.; Moore, J.P. Root Cap–Derived Cells and Mucilage: A Protective Network at the Root Tip. Protoplasma 2021, 258, 1179–1185. [CrossRef]
- Aznar, A.; Dellagi, A. New Insights into the Role of Siderophores as Triggers of Plant Immunity: What Can We Learn from Animals? Journal of Experimental Botany 2015, 66, 3001–3010. [CrossRef]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Waqas, M.; Kang, S.-M.; Lee, I.-J. Inoculation of Abscisic Acid-Producing Endophytic Bacteria Enhances Salinity Stress Tolerance in Oryza Sativa. Environmental and Experimental Botany 2017, 136, 68–77. [CrossRef]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic Bacteria in Sunflower (Helianthus Annuus L.): Isolation, Characterization, and Production of Jasmonates and Abscisic Acid in Culture Medium. Appl Microbiol Biotechnol 2007, 76, 1145–1152. [CrossRef]
- Boiero, L.; Perrig, D.; Masciarelli, O.; Penna, C.; Cassán, F.; Luna, V. Phytohormone Production by Three Strains of Bradyrhizobium Japonicum and Possible Physiological and Technological Implications. Appl Microbiol Biotechnol 2007, 74, 874–880. [CrossRef]
- Joni, F.R.; Hamid, H.; Yanti, Y. Effect of Plant Growth Promoting Rhizobacteria (PGPR) on Increasing the Activity of Defense Enzymes in Tomato Plants. IJEAB 2020, 5, 1474–1479. [CrossRef]
- Amaya-Gómez, C.V.; Porcel, M.; Mesa-Garriga, L.; Gómez-Álvarez, M.I. A Framework for the Selection of Plant Growth-Promoting Rhizobacteria Based on Bacterial Competence Mechanisms. Appl Environ Microbiol 2020, 86, e00760-20. [CrossRef]
- Fadiji, A.E.; Orozco-Mosqueda, M.D.C.; Santos-Villalobos, S.D.L.; Santoyo, G.; Babalola, O.O. Recent Developments in the Application of Plant Growth-Promoting Drought Adaptive Rhizobacteria for Drought Mitigation. Plants 2022, 11, 3090. [CrossRef]
- Shayanthan, A.; Ordoñez, P.A.C.; Oresnik, I.J. The Role of Synthetic Microbial Communities (SynCom) in Sustainable Agriculture. Frontiers in Agronomy 2022, 4, 896307. [CrossRef]
- Haque, M.M.; Mosharaf, M.K.; Khatun, M.; Haque, M.A.; Biswas, M.S.; Islam, M.S.; Siddiquee, M.A. Biofilm Producing Rhizobacteria with Multiple Plant Growth-Promoting Traits Promote Growth of Tomato under Water-Deficit Stress. Frontiers in Microbiology 2020, 11, 542053. [CrossRef]
- Kumar, A.; Maurya, B.R.; Raghuwanshi, R. Rhizobacteria Genome Sequences: Platform for Defining Functional Traits for Sustainable Agriculture. Journal of Genetic Engineering and Biotechnology 2020, 18, 92.
- Abdelaziz, M.E.; Abdelsattar, M.; Abdeldaym, E.A.; Atia, M.A.M.; Mahmoud, A.W.M.; Saad, M.M.; Hirt, H. Piriformospora Indica Promotes Growth of Maize Plants Under High Salinity Stress: A Study on the Regulatory Mechanisms of Salt Stress Response. Frontiers in Microbiology 2019, 10, 1088.
- Wang, L.; Wang, X.; Wu, H.; Wang, H.; Lu, Z. Interspecies Synergistic Interactions Mediated by Cofactor Exchange Enhance Stress Tolerance by Inducing Biofilm Formation. mSystems 2024, 9, 00884–24.
- Cooperation and Conflict in Microbial Biofilms. Proceedings of the National Academy of Sciences 2007, 104, 876–881. [CrossRef]
- Valiei, A.; Dickson, A.; Aminian-Dehkordi, J.; Mofrad, M.R. Metabolic Interactions Shape Emergent Biofilm Structures in a Conceptual Model of Gut Mucosal Bacterial Communities. npj Biofilms and Microbiomes 2024, 10, 99. [CrossRef]
- Sadiq, F.A.; Reu, K.; Burmølle, M.; Maes, S.; Heyndrickx, M. Synergistic Interactions in Multispecies Biofilm Combinations of Bacterial Isolates Recovered from Diverse Food Processing Industries. Frontiers in microbiology 2023, 14, 1159434. [CrossRef]
- Matthus, E.; Zwetsloot, M.; Delory, B.M.; Hennecke, J.; Andraczek, K.; Henning, T.; Bergmann, J. Revisiting the Root Economics Space—Its Applications, Extensions and Nuances Advance Our Understanding of Fine-Root Functioning; Plant and Soil, 2025;
- Kong, D.; Wang, J.; Wu, H.; Valverde-Barrantes, O.J.; Wang, R.; Zeng, H. Nonlinearity of Root Trait Relationships and the Root Economics Spectrum. Nat. Commun 2019, 10, 1–9. [CrossRef]
- Veiga, R.S.L.; Faccio, A.; Genre, A.; Pieterse, C.M.J.; Bonfante, P.; van der Heijden, M.G.A. Arbuscular Mycorrhizal Fungi Reduce Growth and Infect Roots of the Non-Host Plant Arabidopsis Thaliana. Plant Cell Environ 2013, 36, 1926–1937. [CrossRef]
- Ristova, D.; Rosas, U.; Krouk, G.; Ruffel, S.; Birnbaum, K.D.; Coruzzi, G.M. RootScape: A Landmark-Based System for Rapid Screening of Root Architecture in Arabidopsis. Plant physiology 2013, 161, 1086–1096. [CrossRef]
- Iorio, A.; Lasserre, B.; Scippa, G.S.; Chiatante, D. Root System Architecture of Quercus Pubescens Trees Growing on Different Sloping Conditions. Annals of Botany. 2005, 95, 351–361. [CrossRef]
- Jia, H.; Guan, C.; Zhang, J.; He, C.; Yin, C.; Meng, P. Drought Effects on Tree Growth, Water Use Efficiency, Vulnerability and Canopy Health of Quercus Variabilis-Robinia Pseudoacacia Mixed Plantation 2022.
- Tognetti, R.; Longobucco, A.; Miglietta, F.; Raschi, A. Water Relations, Stomatal Response and Transpiration of Quercus Pubescens Trees during Summer in a Mediterranean Carbon Dioxide Spring. Tree Physiology 1999, 19, 261–270. [CrossRef]
- Pividori, M.; Giannetti, F.; Barbati, A.; Chirici, G. European Forest Types: Tree Species Matrix. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., Ed.; Publications Office of the European Union: Luxembourg, 2016; p. 01 162.
- Šajbidorová, V.; Lichtnerová, H.; Paganová, V. The Impact of Different Water Regime on Chlorophyll Fluorescence of Pyrus Pyraster L. and Sorbus Domestica L. Acta Univ. Agric. Silvic. Mendelianae Brun. 2015, 63, 1575–1579. [CrossRef]
- Paganová, V.; Jureková, Z.; Lichtnerová, H. The Nature and Way of Root Adaptation of Juvenile Woody Plants Sorbus and Pyrus to Drought. Environ Monit Assess 2019, 191, 714. [CrossRef]
- Schmucker, J.; Skovsgaard, J.P.; Uhl, E.; Pretzsch, H. Crown Structure, Growth, and Drought Tolerance of True Service Tree (Sorbus Domestica L.) in Forests and Urban Environments. Urban Forestry & Urban Greening 2024, 91, 128161. [CrossRef]
- Rudrappa, T.; Splaine, R.E.; Biedrzycki, M.L.; Bais, H.P. Cyanogenic pseudomonads influence multitrophic interactions in the rhizosphere. PLoS One 2008, 3, 2073.
- Cheng, J.; Schloerke, B.; Karambelkar, B.; Xie, Y. Leaflet: Create Interactive Web Maps with the JavaScript “Leaflet” Library; 2024;
- Kunz, J.; Räder, A.; Bauhus, J. Effects of Drought and Rewetting on Growth and Gas Exchange of Minor European Broadleaved Tree Species. Forests 2016, 7, 239. [CrossRef]
- Emmenegger, B.; Massoni, J.; Pestalozzi, C.M.; Bortfeld-Miller, M.; Maier, B.A.; Vorholt, J.A. Identifying Microbiota Community Patterns Important for Plant Protection Using Synthetic Communities and Machine Learning. Nature Communications 2023, 14, 7983.
- Polade, S.D.; Pierce, D.W.; Cayan, D.R.; Gershunov, A.; Dettinger, M.D. The Key Role of Dry Days in Changing Regional Climate and Precipitation Regimes. Sci Rep 2014, 4, 4364. [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a Fast and Universal Method for Siderophore Detection. Journal of Microbiological Methods 2007, 70, 127–131. [CrossRef]
- Ustiatik, R.; Nuraini, Y.; Suharjono, S.; Handayanto, E. Siderophore Production of the Hg-Resistant Endophytic Bacteria Isolated from Local Grass in the Hg-Contaminated Soil. J. Ecol. Eng. 2021, 22, 129–138. [CrossRef]
- Gomes, A.F.R.; Sousa, E.; Resende, D.I.S.P. A Practical Toolkit for the Detection, Isolation, Quantification, and Characterization of Siderophores and Metallophores in Microorganisms. ACS Omega 2024, 9, 26863–26877. [CrossRef]
- Gordon, S.A.; Weber, R.P. COLORIMETRIC ESTIMATION OF INDOLEACETIC ACID. Plant Physiol 1951, 26, 192–195. [CrossRef]
- Gang, S.; Sharma, S.; Saraf, M.; Buck, M.; Schumacher, J. Analysis of Indole-3-Acetic Acid (IAA) Production in Klebsiella by LC-MS/MS and the Salkowski Method. BIO-PROTOCOL 2019, 9. [CrossRef]
- Fricke, W.; Akhiyarova, G.; Veselov, D.; Kudoyarova, G. Rapid and Tissue-specific Changes in ABA and in Growth Rate in Response to Salinity in Barley Leaves. Journal of Experimental Botany 2004, 55, 1115–1123. [CrossRef]
- Verslues, P.E. Quantification of Water Stress-Induced Osmotic Adjustment and Proline Accumulation for Arabidopsis Thaliana Molecular Genetic Studies. Methods Mol Biol 2010, 639, 301–315. [CrossRef]
- Siddique, S.; Naveed, M.; Yaseen, M.; Shahbaz, M. Exploring Potential of Seed Endophytic Bacteria for Enhancing Drought Stress Resilience in Maize (Zea Mays L.). Sustainability 2022, 14, 673. [CrossRef]
- Orhan, F.; Efe, D.; Gormez, A. Chapter 7 - Advantages of Using Halotolerant/Halophilic Bacteria in Agriculture. In Unravelling Plant-Microbe Synergy; Chandra, D., Bhatt, P., Eds.; Developments in Applied Microbiology and Biotechnology; Academic Press, 2023; pp. 133–149 ISBN 978-0-323-99896-3.
- Ren, D.; Madsen, J.S.; Sørensen, S.J.; Burmølle, M. High Prevalence of Biofilm Synergy among Bacterial Soil Isolates in Cocultures Indicates Bacterial Interspecific Cooperation. ISME J 2015, 9, 81–89. [CrossRef]
- Yang, N.; Nesme, J.; Røder, H.L.; Li, X.; Zuo, Z.; Petersen, M.; Sørensen, S.J. Emergent Bacterial Community Properties Induce Enhanced Drought Tolerance in Arabidopsis. npj Biofilms and Microbiomes 2021, 7, 82.
- Sprouffske, K.; Wagner, A. Growthcurver: An R Package for Obtaining Interpretable Metrics from Microbial Growth Curves. BMC bioinformatics 2016, 17, 1–4.
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. Journal of Open Source Software 2019, 4, 1686. [CrossRef]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests 2023.
- Lê, S.; Josse, J.; Husson, F. FactoMineR : An R Package for Multivariate Analysis. J. Stat. Soft. 2008, 25. [CrossRef]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Survminer: Drawing Survival Curves Using “Ggplot2” 2024.
- Balcı, S. ClinicoPathJamoviModule: Justice for All 2020.
- The Jamovi Project (2025). Jamovi (Version 2.6).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




