Submitted:
14 April 2025
Posted:
15 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Genome-Wide Identification and Analysis of GA-Oxidase Genes in Heracleum sosnowskyi
2.2. Phylogenetic Analysis of the HsGAox Genes
2.3. Conserved Motif and Gene Structure Analysis of the HsGAox Genes
2.4. Analysis of HsGAoxs Expression in Response to GA3 Within Heracleum sosnowskyi Developing Ovaries
2.5. Changes in Endogenous GA levels in Heracleum sosnowskyi Ovaries After Treatment with Exogenous GA3
2.6. GA3 Impact on the Phenotype of Heracleum sosnowskyi Ovaries
3. Discussion
4. Materials and Methods
4.1. Research Object and Growth Conditions
4.2. Application of Plants with GA3
4.3. Sample Harvesting
4.4. Bioinformatic Analysis of Heracleum sosnowskyi GAoxs Subfamilies
4.5. Multiple Sequence Alignment and Phylogenetic Analysis
4.6. Extraction of Endogenous GAs
4.7. Endogenous GA Analysis by Gas Chromatography-Mass Spectrometry
4.8. Identification of GA Biosynthesis Gene Fragments
4.9. Real-Time Quantitative PCR
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sponsel, V.M. Signal Achievements in Gibberellin Research: The Second Half-century. In Annual Plant Reviews, Volume 49: The Gibberellins; Wiley Online Library, 2016; pp. 1–36. [Google Scholar]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol 2020, 61, 1832–1849. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Thomas, S.G. Gibberellin Biosynthesis and Its Regulation. Biochemical Journal 2012, 444, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P. Gibberellins. In Encyclopedia of Applied Plant Sciences; Elsevier Inc., 2016; Vol. 1, pp. 411–420. ISBN 9780123948083. [Google Scholar]
- Hedden; Thomas. In The Gibberellins; 2016; Vol. 1, ISBN 9788578110796.
- Lange, T.; Hedden, P.; Graebe, J.E. Gibberellin Biosynthesis in Cell-Free Extracts from Developing Cucurbita maxima Embryos and the Identification of New Endogenous Gibberellins. Planta 1993, 189, 350–358. [Google Scholar] [CrossRef]
- MacMillan, J. Occurrence of Gibberellins in Vascular Plants, Fungi, and Bacteria. J Plant Growth Regul 2001, 20, 387–442. [Google Scholar] [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a Key Gene Responsible for Bioactive Gibberellin Biosynthesis, Is Regulated during Embryogenesis by Leafy Cotyledon2 and FUSCA3 in Arabidopsis. Plant Physiol 2004, 136, 3660–3669. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.A.; MacMillan, J.; Gong, F.; Phillips, A.L.; Hedden, P. Gibberellin 3-Oxidases in Developing Embryos of the Southern Wild Cucumber, Marah macrocarpus. Phytochemistry 2010, 71, 2010–2018. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Kourmpetli, S.; Ward, D.A.; Thomas, S.G.; Gong, F.; Powers, S.J.; Carrera, E.; Taylor, B.; Gonzalez, F.N. de C.; Tudzynski, B.; et al. Characterization of the Fungal Gibberellin Desaturase as a 2-Oxoglutarate-Dependent Dioxygenase and Its Utilization for Enhancing Plant Growth. Plant Physiol 2012, 160, 837–845. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin Metabolism and Its Regulation. Annu Rev Plant Biol 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Aach, H.; Bode, H.; Robinson, D.G.; Graebe, J.E. Ent-Kaurene Synthase Is Located in Proplastids of Meristematic Shoot Tissues. Planta 1997, 211–219. [Google Scholar] [CrossRef]
- Sun, T.P.; Gubler, F. Molecular Mechanism of Gibberellin Signalling in Plants. Annu Rev Plant Biol 2004, 55, 197–223. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Sullivan, J.A.; Mould, R.M.; Gray, J.C.; James Peacock, W.; Dennis, E.S. A Plastid Envelope Location of Arabidopsis Ent-Kaurene Oxidase Links the Plastid and Endoplasmic Reticulum Steps of the Gibberellin Biosynthesis Pathway. Plant Journal 2001, 28, 201–208. [Google Scholar] [CrossRef]
- Nelson, D.R.; Schuler, M.A.; Paquette, S.M.; Werck-Reichhart, D.; Bak, S. Comparative Genomics of Rice and Arabidopsis. Analysis of 727 Cytochrome P450 Genes and Pseudogenes from a Monocot and a Dicot. Plant Physiol 2004, 135, 756–772. [Google Scholar] [CrossRef]
- Pimenta Lange, M.J.; Lange, T. Gibberellin Biosynthesis and the Regulation of Plant Development. Plant Biol 2006, 8, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Sponsel, V.M.; Hedden, P. Gibberellin Biosynthesis and Inactivation. In Plant Hormones; Springer, 2010; pp. 63–94. [Google Scholar]
- Pimenta Lange, M.J.; Liebrandt, A.; Arnold, L.; Chmielewska, S.M.; Felsberger, A.; Freier, E.; Heuer, M.; Zur, D.; Lange, T. Functional Characterization of Gibberellin Oxidases from Cucumber, Cucumis sativus L. Phytochemistry 2013, 90, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and Diversity of the 2-Oxoglutarate-Dependent Dioxygenase Superfamily in Plants. Plant Journal 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Pearce, S.; Huttly, A.K.; Prosser, I.M.; Li, Y.D.; Vaughan, S.P.; Gallova, B.; Patil, A.; Coghill, J.A.; Dubcovsky, J.; Hedden, P.; et al. Heterologous Expression and Transcript Analysis of Gibberellin Biosynthetic Genes of Grasses Reveals Novel Functionality in the GA3ox Family. BMC Plant Biol 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Rebers, M.; Kaneta, T.; Kawaide, H.; Yamaguchi, S.; Yang, Y.Y.; Imai, R.; Sekimoto, H.; Kamiya, Y. Regulation of Gibberellin Biosynthesis Genes during Flower and Early Fruit Development of Tomato. Plant Journal 1999, 17, 241–250. [Google Scholar] [CrossRef]
- Lange, T.; Kappler, J.; Fischer, A.; Frisse, A.; Padeffke, T.; Schmidtke, S.; Lange, M.J.P. Gibberellin Biosynthesis in Developing Pumpkin Seedlings. Plant Physiol 2005, 139, 213–223. [Google Scholar] [CrossRef]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An Overview of Gibberellin Metabolism Enzyme Genes and Their Related Mutants in Rice. Plant Physiol 2004, 134, 1642–1653. [Google Scholar] [CrossRef]
- Lange, T. Purification and Partial Amino-Acid Sequence of Gibberellin 20-Oxidase from Cucurbita Maxima L. Endosperm. 1994; Vol. 195. [Google Scholar]
- Chiang, H.-H.; Hwang, L.; Goodman, H.M. Lsolation of the Arabidopsis GA4 Locus; American Society of Plant Physiologists, 1995; Vol. 7. [Google Scholar]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular Cloning and Functional Expression of Gibberellin 2-Oxidases, Multifunctional Enzymes Involved in Gibberellin Deactivation. Plant Biol 1999, 96, 4698–4703. [Google Scholar] [CrossRef]
- Kang, H.-G.; Jun, S.-H.; Kim, J.; Kawaide, H.; Kamiya, Y.; An, G. Cloning and Molecular Analyses of a Gibberellin 20-Oxidase Gene Expressed Specifically in Developing Seeds of Watermelon. Plant Physiol 1999, 121, 373–382. [Google Scholar] [CrossRef]
- García-Hurtado, N.; Carrera, E.; Ruiz-Rivero, O.; López-Gresa, M.P.; Hedden, P.; Gong, F.; García-Martínez, J.L. The Characterization of Transgenic Tomato Overexpressing Gibberellin 20-Oxidase Reveals Induction of Parthenocarpic Fruit Growth, Higher Yield, and Alteration of the Gibberellin Biosynthetic Pathway. J Exp Bot 2012, 63, 5803–5813. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Bello, L.; Moritz, T.; López-Díaz, I. Silencing C19-GA 2-Oxidases Induces Parthenocarpic Development and Inhibits Lateral Branching in Tomato Plants. J Exp Bot 2015, 66, 5897–5910. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, X.; Zhang, L.; Lin, S.; Liu, D.; Wang, Q.; Cai, S.; El-Tanbouly, R.; Gan, L.; Wu, H.; et al. Identification and Characterization of Tomato Gibberellin 2-Oxidases (GA2oxs) and Effects of Fruit-Specific SlGA2ox1 Overexpression on Fruit and Seed Growth and Development. Hortic Res 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shan, X.; Jiang, Z.; Zhao, L.; Jin, F. Genome-Wide Identification and Expression Analysis of the GA2ox Gene Family in Maize (Zea mays L.) under Various Abiotic Stress Conditions. Plant Physiology and Biochemistry 2021, 166, 621–633. [Google Scholar] [CrossRef]
- Toyomasu, T.; Kawaide, H.; Sekimoto, H.; von Numers, C.; Phillips, A.L.; Hedden, P.; Kamiya Toyomasu, Y. ; Numers, von Frontier Research Pro-Gram, The Institute of Physical and Chemical Research. 1997.
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.D.; Chen, L.-J.; Yu, S.-M. A Novel Class of Gibberellin 2-Oxidases Control Semidwarfism, Tillering, and Root Development in Rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef]
- Cheng, J.; Ma, J.; Zheng, X.; Lv, H.; Zhang, M.; Tan, B.; Ye, X.; Wang, W.; Zhang, L.; Li, Z. Functional Analysis of the Gibberellin 2-Oxidase Gene Family in Peach. Front Plant Sci 2021, 12, 619158. [Google Scholar] [CrossRef]
- Giacomelli, L.; Rota-Stabelli, O.; Masuero, D.; Acheampong, A.K.; Moretto, M.; Caputi, L.; Vrhovsek, U.; Moser, C. Gibberellin Metabolism in Vitis Vinifera L. During Bloom and Fruit-Set: Functional Characterization and Evolution of Grapevine Gibberellin Oxidases. J Exp Bot 2013, 64, 4403–4419. [Google Scholar] [CrossRef]
- Sabir, I.A.; Manzoor, M.A.; Shah, I.H.; Abbas, F.; Liu, X.; Fiaz, S.; Shah, A.N.; Jiu, S.; Wang, J.; Abdullah, M.; et al. Evolutionary and Integrative Analysis of Gibberellin-Dioxygenase Gene Family and Their Expression Profile in Three Rosaceae Genomes (F. Vesca, P. Mume, and P. Avium) Under Phytohormone Stress. Front Plant Sci 2022, 13. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J.R. Breadfruit (Artocarpus Altilis) Gibberellin 2-Oxidase Genes in Stem Elongation and Abiotic Stress Response. Plant Physiology and Biochemistry 2016, 98, 81–88. [Google Scholar] [CrossRef]
- Fos, M.; Nuez, F.; Garcı́a-Martı́nez, J.L. The Gene Pat-2, Which Induces Natural Parthenocarpy, Alters the Gibberellin Content in Unpollinated Tomato Ovaries. Plant Physiol 2000, 122, 471–480. [Google Scholar] [CrossRef]
- Wang, H.; Wu, T.; Liu, J.; Cong, L.; Zhu, Y.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; Xu, L. PbGA20ox2 Regulates Fruit Set and Induces Parthenocarpy by Enhancing GA4 Content. Front Plant Sci 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Gederaas, L.; Loennechen Moen, T.; Skjelseth, S.; Larsen, L.K. Alien Species in Norway - with the Norwegian Black List 2012; Norwegian Biodiversity Infomation Centre (NBIC): Trondheim, 2012; ISBN 9788292838372. [Google Scholar]
- Gudžinskas, Z.; Kazlauskas, M.; Pilate, D.; Balalaikins, M.; Pilats, M.; Šaulys, A.; Šaulienė, I.; Šukienė, L. Invasive Organisms the Border Region of Lithuania and Latvia; BMK Press: Vilnius, 2014. [Google Scholar]
- Tkachenko, K.G. Peculiarities and Seed Productivity in Some Heracleum Species Grown in Leningrad Area. Rastitelnye Resursy 1989, 1, 52–61. [Google Scholar]
- Gudžinskas, Z.; Žalneravičius, E. Seedling Dynamics and Population Structure of Invasive Heracleum Sosnowskyi (Apiaceae) in Lithuania. Ann Bot Fenn 2018. [Google Scholar] [CrossRef]
- Zangerl, A.R.; Berenbaum, M.R.; Nitao, J.K. Parthenocarpic Fruits in Wild Parsnip: Decoy Defence against a Specialist Herbivore. Evol Ecol 1991, 5, 136–145. [Google Scholar] [CrossRef]
- Koryznienė, D.; Jurkonienė, S.; Žalnierius, T.; Gavelienė, V.; Jankovska-Bortkevič, E.; Bareikienė, N.; Būda, V. Heracleum Sosnowskyi Seed Development under the Effect of Exogenous Application of GA3. PeerJ 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Žalnierius, T.; Šveikauskas, V.; Aphalo, P.J.; Gavelienė, V.; Būda, V.; Jurkonienė, S. Gibberellic Acid (GA3) Applied to Flowering Heracleum Sosnowskyi Decreases Seed Viability Even If Seed Development Is Not Inhibited. Plants 2022, 11, 314. [Google Scholar] [CrossRef]
- Schelkunov, M.I.; Shtratnikova, V.Y.; Klepikova, A. V; Makarenko, M.S.; Omelchenko, D.O.; Novikova, L.A.; Obukhova, E.N.; Bogdanov, V.P.; Penin, A.A.; Logacheva, M.D. The Genome of the Toxic Invasive Species Heracleum Sosnowskyi Carries an Increased Number of Genes despite Absence of Recent Whole-genome Duplications. The Plant Journal 2024, 117, 449–463. [Google Scholar] [CrossRef]
- Rieu, I.; Eriksson, S.; Powers, S.J.; Gong, F.; Griffiths, J.; Woolley, L.; Benlloch, R.; Nilsson, O.; Thomas, S.G.; Hedden, P. Genetic Analysis Reveals That C19-GA 2-Oxidation Is a Major Gibberellin Inactivation Pathway in Arabidopsis. Plant Cell 2008, 20, 2420–2436. [Google Scholar] [CrossRef]
- Dorcey, E.; Urbez, C.; Blázquez, M.A.; Carbonell, J.; Perez-Amador, M.A. Fertilization-Dependent Auxin Response in Ovules Triggers Fruit Development through the Modulation of Gibberellin Metabolism in Arabidopsis. Plant Journal 2009, 58, 318–332. [Google Scholar] [CrossRef]
- Vivian-Smith, A.; Koltunow, A.M. Genetic Analysis of Growth-Regulator-Induced Parthenocarpy in Arabidopsis. Plant Physiol 1999, 121, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Serrani, J.C.; Fos, M.; Atarés, A.; García-Martínez, J.L. Effect of Gibberellin and Auxin on Parthenocarpic Fruit Growth Induction in the Cv Micro-Tom of Tomato. J Plant Growth Regul 2007, 26, 211–221. [Google Scholar] [CrossRef]
- Tiwari, A.; Offringa, R.; Heuvelink, E. Auxin-Induced Fruit Set in Capsicum Annuum L. Requires Downstream Gibberellin Biosynthesis. J Plant Growth Regul 2012, 31, 570–578. [Google Scholar] [CrossRef]
- Asahira, T.; Nitsch, J.P. Tubérisation in Vitro: Ullucus Tuberosus et Dioscorea. Bulletin de la Societe Botanique de France 1968, 115, 345–352. [Google Scholar] [CrossRef]
- Shohat, H.; Cheriker, H.; Kilambi, H.V.; Illouz Eliaz, N.; Blum, S.; Amsellem, Z.; Tarkowská, D.; Aharoni, A.; Eshed, Y.; Weiss, D. Inhibition of Gibberellin Accumulation by Water Deficiency Promotes Fast and Long-term ‘Drought Avoidance’Responses in Tomato. New Phytologist 2021, 232, 1985–1998. [Google Scholar] [CrossRef]
- Toyomasu, T.; Kawaide, H.; Sekimoto, H.; von Numers, C.; Phillips, A.L.; Hedden, P.; Kamiya Toyomasu, Y. ; Numers, von Frontier Research Pro-Gram, The Institute of Physical and Chemical Research. 1997.
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An Overview of Gibberellin Metabolism Enzyme Genes and Their Related Mutants in Rice. Plant Physiol 2004, 134, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Ueguchi-Tanaka, M.; Sentoku, N.; Kitano, H.; Matsuoka, M.; Kobayashi, M. Cloning and Functional Analysis of Two Gibberellin 3-Hydroxylase Genes That Are Differently Expressed during the Growth of Rice.
- Han, F.; Zhu, B. Evolutionary Analysis of Three Gibberellin Oxidase Genesin Rice, Arabidopsis, and Soybean. Gene 2011, 473, 23–35. [Google Scholar] [CrossRef]
- Phillips, A.L.; Ward, D.A.; Uknes, S.; Appleford, N.E.J.; Lange, T.; Huttly, A.K.; Gaskin, P.; Graebe, J.E.; Hedden, P. Isolation and Expression of Three Gibberellin 20-Oxidase CDNA Clones from Arabidopsis. Plant Physiol 1995, 108, 1049–1057. [Google Scholar] [CrossRef]
- Sun, H.; Pang, B.; Yan, J.; Wang, T.; Wang, L.; Chen, C.; Li, Q.; Ren, Z. Comprehensive Analysis of Cucumber Gibberellin Oxidase Family Genes and Functional Characterization of CsGA20ox1 in Root Development in Arabidopsis. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Hedden, P.; Phillips, A.L. Gibberellin Metabolism: New Insights Revealed by the Genes. Trends Plant Sci 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E. V The DNA-Repair Protein AlkB, EGL-9, and Leprecan Define New Families of 2-Oxoglutarate-and Iron-Dependent Dioxygenases. 2001. [Google Scholar]
- Huang, Y.; Wang, X.; Ge, S.; Rao, G.Y. Divergence and Adaptive Evolution of the Gibberellin Oxidase Genes in Plants Genome Evolution and Evolutionary Systems Biology. BMC Evol Biol 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science (1979) 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Sémon, M.; Wolfe, K.H. Consequences of Genome Duplication. Curr Opin Genet Dev 2007, 17, 505–512. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.-J.; Tan, G.-F.; Zhou, W.-Q.; Wang, G.-L. Gibberellin and the Plant Growth Retardant Paclobutrazol Altered Fruit Shape and Ripening in Tomato. Protoplasma 2020, 257, 853–861. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.D.; Amasino, R.M. Overexpression of a Novel Class of Gibberellin 2-Oxidases Decreases Gibberellin Levels and Creates Dwarf Plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J.R. Breadfruit (Artocarpus altilis) Gibberellin 2-Oxidase Genes in Stem Elongation and Abiotic Stress Response. Plant Physiology and Biochemistry 2016, 98, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Yamaguchi, S.; Hanada, A.; Kuwahara, A.; Yoshioka, Y.; Kato, T.; Tabata, S.; Kamiya, Y.; Sun, T.P. Distinct and Overlapping Roles of Two Gibberellin 3-Oxidases in Arabidopsis Development. Plant Journal 2006, 45, 804–818. [Google Scholar] [CrossRef]
- Hu, J.; Mitchum, M.G.; Barnaby, N.; Ayele, B.T.; Ogawa, M.; Nam, E.; Lai, W.-C.; Hanada, A.; Alonso, J.M.; Ecker, J.R.; et al. Potential Sites of Bioactive Gibberellin Production during Reproductive Growth in Arabidopsis. Plant Cell 2008, 20, 320–336. [Google Scholar] [CrossRef]
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.D.; Chen, L.-J.; Yu, S.-M. A Novel Class of Gibberellin 2-Oxidases Control Semidwarfism, Tillering, and Root Development in Rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef]
- He, H.; Yamamuro, C. Interplays between Auxin and GA Signaling Coordinate Early Fruit Development. Hortic Res 2022, 9. [Google Scholar] [CrossRef]
- Wei, H.; Chen, J.; Zhang, X.; Lu, Z.; Liu, G.; Lian, B.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Characterization, Expression Pattern, and Function Analysis of Gibberellin Oxidases in Salix matsudana. Int J Biol Macromol 2024, 266, 131095. [Google Scholar] [CrossRef]
- Perglova, I.; Pergl, J.; Pysek, P.; Pyšek, P. Reproductive Ecology of Heracleum mantegazzianum. In Ecology and management of giant hogweed; CAB International: Wallingford, 2007; pp. 55–73. ISBN 1845932064. [Google Scholar]
- Koornneef, M.; Van der Veen, J.H. Induction and Analysis of Gibberellin Sensitive Mutants in Arabidopsis thaliana (L.) Heynh. Theoretical and Applied genetics 1980, 58, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.; Pharis, R.P. Role of Gibberellins in the Development of Floral Organs of the Gibberellin-Deficient Mutant, Ga1-1, of Arabidopsis thaliana. Canadian Journal of Botany 1999, 77, 944–954. [Google Scholar] [CrossRef]
- Pimenta Lange, M.J.; Knop, N.; Lange, T. Stamen-Derived Bioactive Gibberellin Is Essential for Male Flower Development of Cucurbita maxima L. J Exp Bot 2012, 63, 2681–2691. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Nakajima, M.; Asano, K.; Nishiyama, T.; Sakakibara, H.; Kojima, M.; Katoh, E.; Xiang, H.; Tanahashi, T.; Hasebe, M. The GID1-Mediated Gibberellin Perception Mechanism Is Conserved in the Lycophyte Selaginella moellendorffii but Not in the Bryophyte Physcomitrella patens. Plant Cell 2007, 19, 3058–3079. [Google Scholar] [CrossRef]
- Bell, C.R. Breeding Systems and Floral Biology of the Umbelliferae; or, Evidence for Specialization in Unspecialized Flowers. In The biology and chemistry of the Umbelliferae; Academic Press, 1971; pp. 93–107. [Google Scholar]
- Serrani, J.C.; Carrera, E.; Ruiz-Rivero, O.; Gallego-Giraldo, L.; Peres, L.E.P.; García-Martínez, J.L. Inhibition of Auxin Transport from the Ovary or from the Apical Shoot Induces Parthenocarpic Fruit-Set in Tomato Mediated by Gibberellins. Plant Physiol 2010, 153, 851–862. [Google Scholar] [CrossRef]
- Alabadí, D.; Blázquez, M.A.; Carbonell, J.; Ferrándiz, C.; Pérez-Amador, M.A. Instructive Roles for Hormones in Plant Development. International Journal of Developmental Biology 2009, 53, 1597–1608. [Google Scholar] [CrossRef]
- Olimpieri, I.; Siligato, F.; Caccia, R.; Soressi, G.P.; Mazzucato, A.; Mariotti, L.; Ceccarelli, N. Tomato Fruit Set Driven by Pollination or by the Parthenocarpic Fruit Allele Are Mediated by Transcriptionally Regulated Gibberellin Biosynthesis. Planta 2007, 226, 877–888. [Google Scholar] [CrossRef]
- Serrani, J.C.; Ruiz-Rivero, O.; Fos, M.; García-Martínez, J.L. Auxin-Induced Fruit-Set in Tomato Is Mediated in Part by Gibberellins. Plant Journal 2008, 56, 922–934. [Google Scholar] [CrossRef]
- Cong, L.; Yue, R.; Wang, H.; Liu, J.; Zhai, R.; Yang, J.; Wu, M.; Si, M.; Zhang, H.; Yang, C.; et al. 2,4-D-Induced Parthenocarpy in Pear Is Mediated by Enhancement of GA4 Biosynthesis. Physiol Plant 2018. [CrossRef]
- Gillaspy, G.; Ben-David, H.; Gruissem, W. Fruits: A Developmental Perspective. Plant Cell 1993, 5, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- McAtee, P.; Karim, S.; Schaffer, R.; David, K. A Dynamic Interplay between Phytohormones Is Required for Fruit Development, Maturation, and Ripening. Front Plant Sci 2013, 4, 1–7. [Google Scholar] [CrossRef]
- Moravcová, L.; Perglová, I.; Pyšek, P.; Jarošík, V.; Pergl, J. Effects of Fruit Position on Fruit Mass and Seed Germination in the Alien Species Heracleum mantegazzianum (Apiaceae) and the Implications for Its Invasion. Acta Oecologica 2005, 28, 1–10. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs; Oxford University Press, 1997; Vol. 25. [Google Scholar]
- Bailey, T.L.; Elkan, C. The Value of Prior Knowledge in Discovering Motifs with MEME. In Proceedings of the Ismb; 1995; Vol. 3, pp. 21–29. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.-T.; Kong, Y.-Z.; Wang, Q.; Sun, Y.-H.; Gong, D.-P.; Lv, J.; Liu, G.-S. MapGene2Chrom, a Tool to Draw Gene Physical Map Based on Perl and SVG Languages. Yi Chuan 2015, 37, 91–97. [Google Scholar]
- Chou, K.-C.; Shen, H.-B. Plant-MPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS One 2010, 5, e11335. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Olechnovič, K.; Venclovas, Č. VoroMQA: Assessment of Protein Structure Quality Using Interatomic Contact Areas. Proteins: Structure, Function, and Bioinformatics 2017, 85, 1131–1145. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol Biol Evol 2024, 41, msae263. [Google Scholar] [CrossRef]
- De Boer, T.J.; Backer, H.J. A New Method for the Preparation of Diazomethane. Recueil des Travaux Chimiques des Pays-Bas 1954, 73, 229–234. [Google Scholar] [CrossRef]
- Gaskin, P.; MacMillan, J. GC-MS of the Gibberellins and Related Compounds. Methodology and a library of spectra. University of Bristol (Cantock’s Enterprises), Bristol, England, UK 1991.
- Hasegawa, H.; Shinohara, Y.; Hashimoto, T.; Matsuda, R.; Hayashi, Y. Prediction of Measurement Uncertainty in Isotope Dilution Gas Chromatography/Mass Spectrometry. J Chromatogr A 2006, 1136, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Frisse, A.; Pimenta, M.J.; Lange, T. Expression Studies of Gibberellin Oxidases in Developing Pumpkin Seeds. Plant Physiol 2003, 131, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fernald, R.D. Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. Journal of computational biology 2005, 12, 1047–1064. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia electronica 2001, 4, 1. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. 2021.
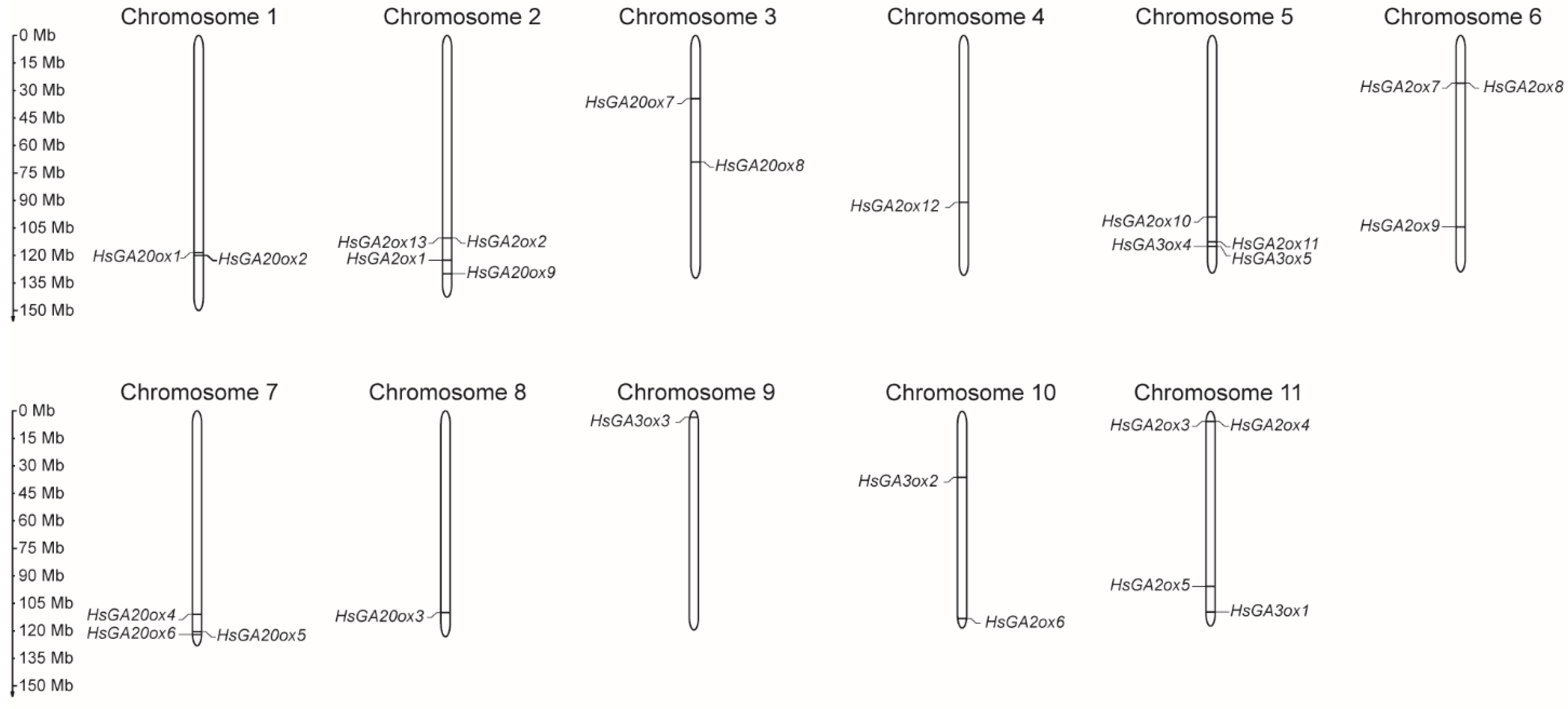
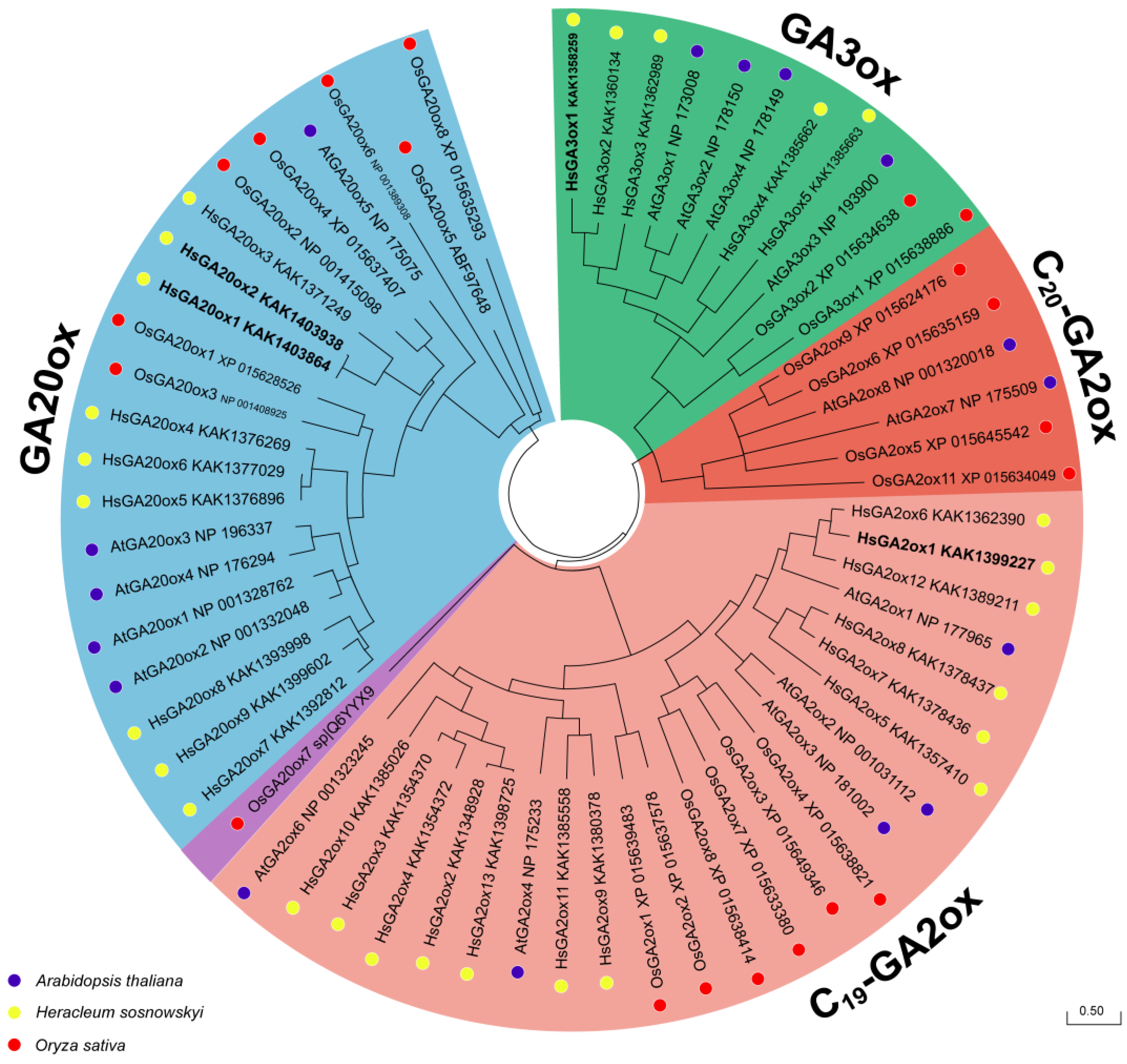
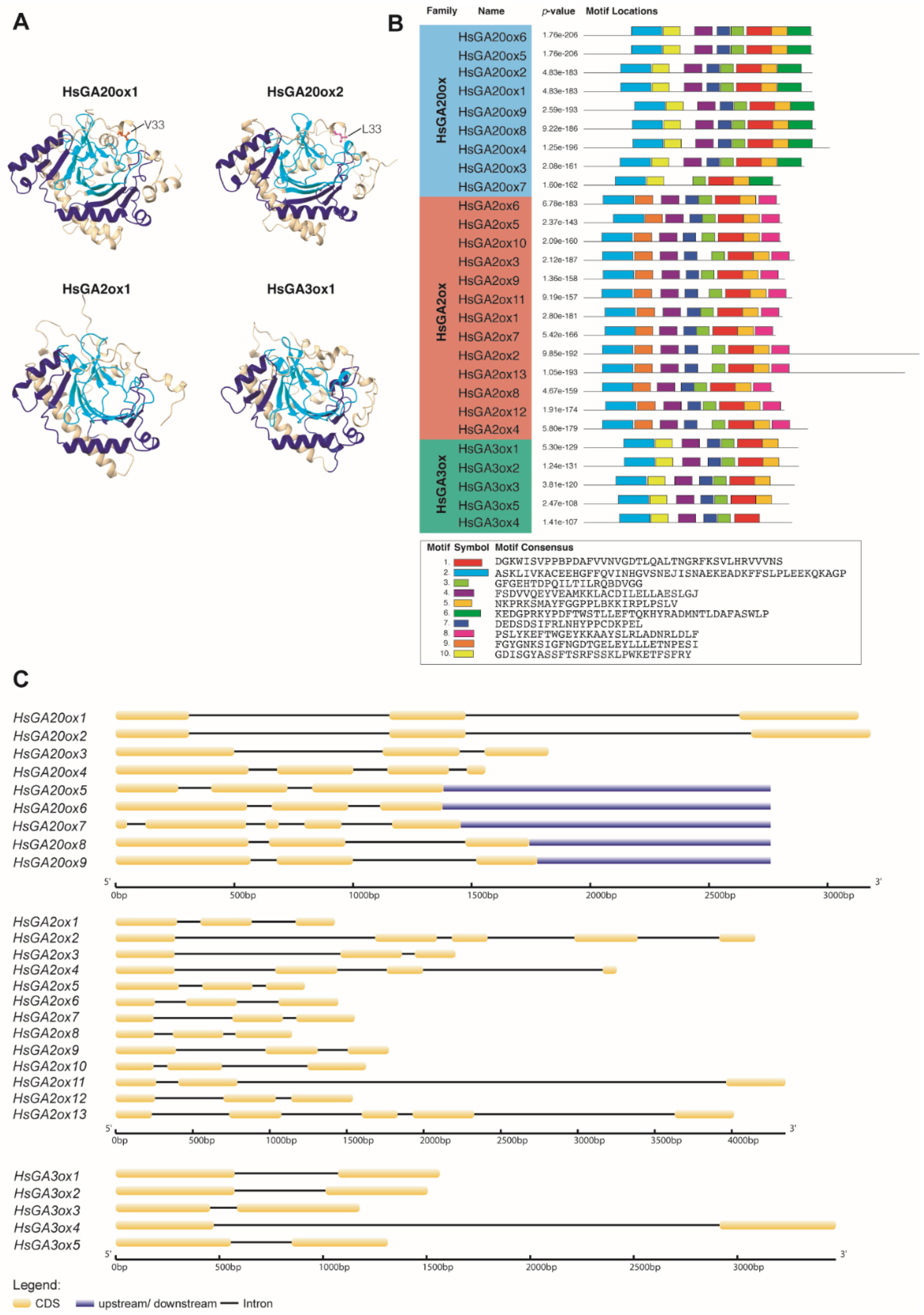
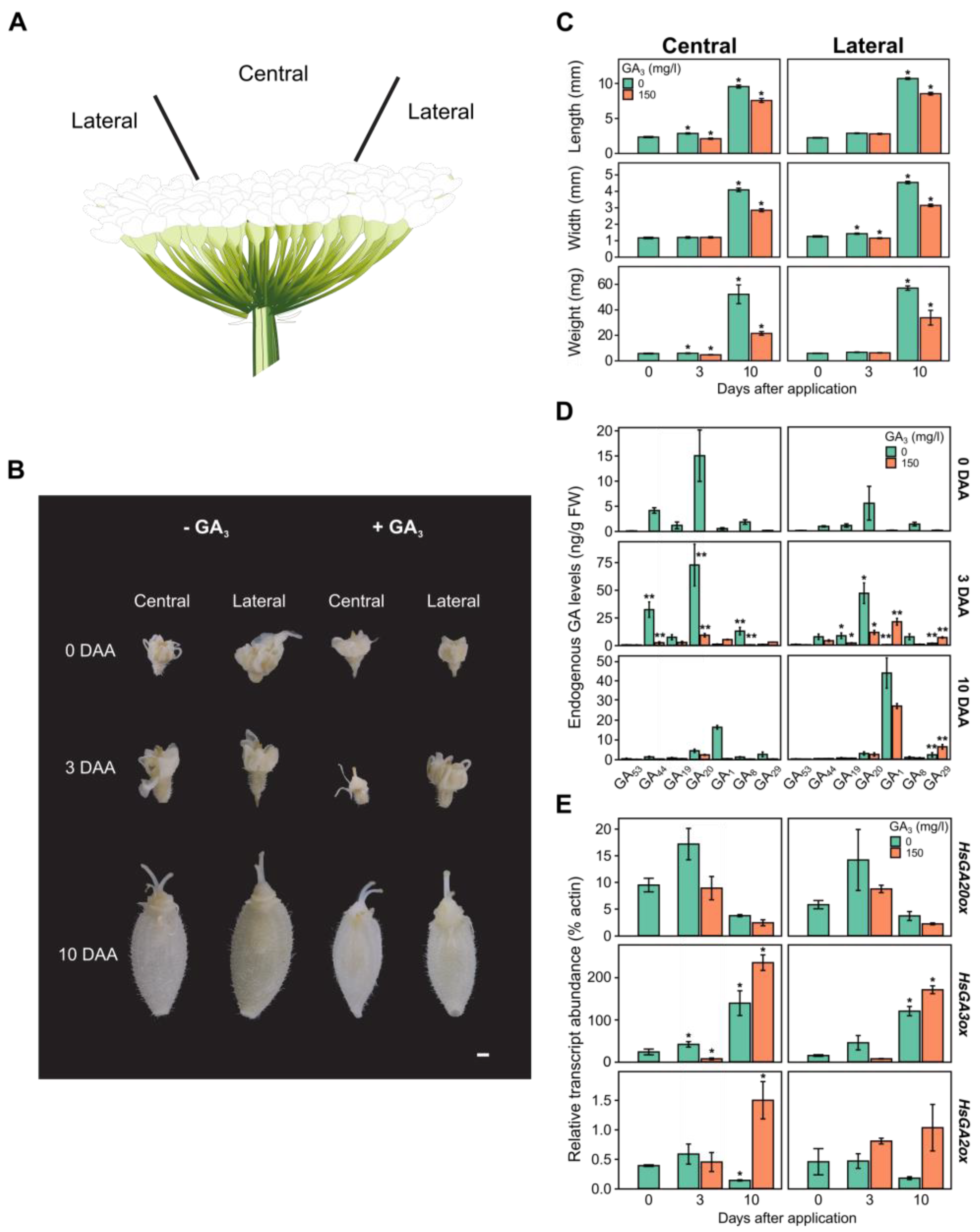
| Protein Name | Accession no. | Gene Name | CDS (nt) | Length (aa) | MW (kDa) | Domain Location | Type | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| HsGA20ox1 | KAK1403938 | HsGA20ox1 | 1134 | 377 | 42.88 | 44-144a 209-306b |
GA20ox | Cytoplasm |
| HsGA20ox2 | KAK1403864 | HsGA20ox2 | 1134 | 377 | 42.89 | 44-144a 209-306b |
GA20ox | Cytoplasm |
| HsGA20ox3 | KAK1371249 | HsGA20ox3 | 1095 | 364 | 41.33 | 43-144a 209-306b |
GA20ox | Cytoplasm |
| HsGA20ox4 | KAK1376269 | HsGA20ox4 | 1221 | 406 | 46.14 | 63-169a 227-325b |
GA20ox | Cytoplasm |
| HsGA20ox5 | KAK1376896 | HsGA20ox5 | 1140 | 379 | 42.96 | 61-167a 225-323b |
GA20ox | Cytoplasm |
| HsGA20ox6 | KAK1377029 | HsGA20ox6 | 1140 | 379 | 43.02 | 61-167a 225-323b |
GA20ox | Cytoplasm |
| HsGA20ox7 | KAK1392812 | HsGA20ox7 | 978 | 325 | 36.81 | 62-164a 227-325b |
GA20ox | Cytoplasm |
| HsGA20ox8 | KAK1393998 | HsGA20ox8 | 1152 | 383 | 43.73 | 34-135a 181-260b |
GA20ox | Cytoplasm |
| HsGA20ox9 | KAK1399602 | HsGA20ox9 | 1149 | 382 | 43.38 | 66-170a 230-328b |
GA20ox | Cytoplasm |
| HsGA2ox1 | KAK1399227 | HsGA2ox1 | 987 | 328 | 36.87 | 26-115a 176-273b |
C19-GA2ox | Cytoplasm |
| HsGA2ox2 | KAK1348928 | HsGA2ox2 | 1665 | 554 | 61.92 | 20-79a 172-291b |
C19-GA2ox | Cytoplasm Nucleus |
| HsGA2ox3 | KAK1354370 | HsGA2ox3 | 1047 | 348 | 38.86 | 20-88a 173-291b |
C19-GA2ox | Cytoplasm |
| HsGA2ox4 | KAK1354372 | HsGA2ox4 | 1113 | 370 | 41.05 | 20-88a 173-292b |
C19-GA2ox | Cytoplasm |
| HsGA2ox5 | KAK1357410 | HsGA2ox5 | 987 | 328 | 36.77 | 40-96a 180-275b |
C19-GA2ox | Cytoplasm |
| HsGA2ox6 | KAK1362390 | HsGA2ox6 | 975 | 342 | 36.31 | 23-110a 173-269b |
C19-GA2ox | Cytoplasm |
| HsGA2ox7 | KAK1378436 | HsGA2ox7 | 954 | 317 | 35.81 | 26-111a 170-265b |
C19-GA2ox | Cytoplasm |
| HsGA2ox8 | KAK1378437 | HsGA2ox8 | 945 | 314 | 35.39 | 22-123a 166-261b |
C19-GA2ox | Cytoplasm |
| HsGA2ox9 | KAK1380378 | HsGA2ox9 | 999 | 332 | 37.21 | 21-80a 174-275b |
C19-GA2ox | Cytoplasm |
| HsGA2ox10 | KAK1385026 | HsGA2ox10 | 981 | 326 | 36.65 | 19-88a 169-276b |
C19-GA2ox | Cytoplasm |
| HsGA2ox11 | KAK1385558 | HsGA2ox11 | 1035 | 344 | 38.19 | 19-84a 172-288b |
C19-GA2ox | Cytoplasm |
| HsGA2ox12 | KAK1389211 | HsGA2ox12 | 996 | 331 | 37.05 | 27-118a 177-276b |
C19-GA2ox | Cytoplasm |
| HsGA2ox13 | KAK1398725 | HsGA2ox13 | 1593 | 530 | 59.09 | 20-79a 172-291b |
C19-GA2ox | Cytoplasm |
| HsGA3ox1 | KAK1358259 | HsGA3ox1 | 1065 | 354 | 39.98 | 56-158a 211-308b |
GA3ox | Cytoplasm |
| HsGA3ox2 | KAK1360134 | HsGA3ox2 | 1068 | 355 | 40.02 | 57-159a 210-309b |
GA3ox | Cytoplasm |
| HsGA3ox3 | KAK1362989 | HsGA3ox3 | 1047 | 348 | 39.05 | 46-145a 196-295b |
GA3ox | Cytoplasm |
| HsGA3ox4 | KAK1385662 | HsGA3ox4 | 1035 | 344 | 38.59 | 48-141a 201-297b |
GA3ox | Cytoplasm |
| HsGA3ox5 | KAK1385663 | HsGA3ox5 | 1020 | 339 | 38.00 | 50-152a 206-302b |
GA3ox | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





