1. Introduction
The
GASA gene family, widely distributed in plants, is a cysteine-rich gene family regulated by gibberellins (GA) , playing important roles in plant growth and development, and response to biotic or abiotic stress [
1]. The GASA protein generally contains three parts: an N-terminal signal peptide with the characteristics of an extracellular secreted protein, composed of 18 to 29 amino acid residues; a variable hydrophilic segment composed of 7 to 31 polar amino acid residues, in which there are significant variations among different GASA proteins; a C-terminal conserved domain of 60 amino acids, known as GASA domain, containing 12 fully conserved cysteine residues [
2]. Generally, a
GASA gene contains only one GASA domain, in which the deletions or changes of key amino acids can make the gene lose its function [
3,
4]. Therefore, this domain was considered to be the key region for maintaining the spatial structure and function of GASA proteins.
In plants,
GASAs are usually strongly expressed in young and vigorously growing tissues and organs, and most members are regulated by gibberellins [
5,
6]. Analysis of
GASAs sequences showed that the promoter region was enriched with cis-acting elements related to development, phytohormones, and stress, indicating that
GASAs not only involved in plant growth and development but also plant responses to stress. As various plant
GASAs had been identified, it was demonstrated that
GASAs played important regulatory roles in plant growth and development, including seed germination [
7], flower and fruit development [
8], lateral root formation [
9], hormone signal transduction [
10], and response to biotic or abiotic stress [
11,
12].
AtGASA4 was rapidly up-regulated under short-term heat treatment, and the heat tolerance of
Arabidopsis plants were enhanced by overexpressing
AtGASA4 [
13]. In
AtGASA5-overexpressing plants,
AtGASA5 gene acted as a negative regulator in heat-tolerance by regulating both SA signaling and heat shock-protein accumulation [
14].
SmGASA4 isolated from
Salvia miltiorrhiza, improved
Arabidopsis resistance to salt, drought, and paclobutrazol (PBZ) [
15].
AtGASA14 regulated the resistance to abiotic stress by modulating the accumulation of reactive oxygen species [
16].
OsGASR1 enhanced plant tolerance to salt stress via the reactive oxygen species scavenging system in
Oryza sativa [
17]. In
Glycine soja,
GsGASA1 was involved in inhibiting root growth via the accumulation of DELLA proteins under chronic cold stress [
18].
The rubber tree (
Hevea brasiliensis) is an important tropical cash crop and the primary source of natural rubber all over the world [
19]. Natural rubber is an essential raw material, widely used in medical and health care, transportation, aerospace, and other fields [
20]. The rubber tree, originating in the high-temperature, high-humidity, and windless Amazon River basin, is a typical tropical rainforest species with weak resistance to low temperature. China's rubber-planting areas are located between 18°N and 24°N, belonging to non-traditional rubber-planting areas, in which the rubber tree often suffered from abiotic stresses like low temperature, seasonal drought, typhoons [
21]. Among these stresses, low temperature is the most severe disasters affecting rubber tree growth. Therefore, identification of genes associated with cold stress is very important for cold-stress-resistant molecular breeding of rubber tree.
In the present work, the GASA genes of Hevea brasiliensis were genome-wide identified, meanwhile, the structure characteristics and expression patterns of all the HbGASA genes were investigated. Furthermore,the responses of HbGASA genes to low temperature were detected. Our research provided some clues to elucidate the functions of GASA proteins in the the biological functions and regulatory mechanisms of GASAs.
2. Results
2.1. Identification and characteristics of HbGASA genes
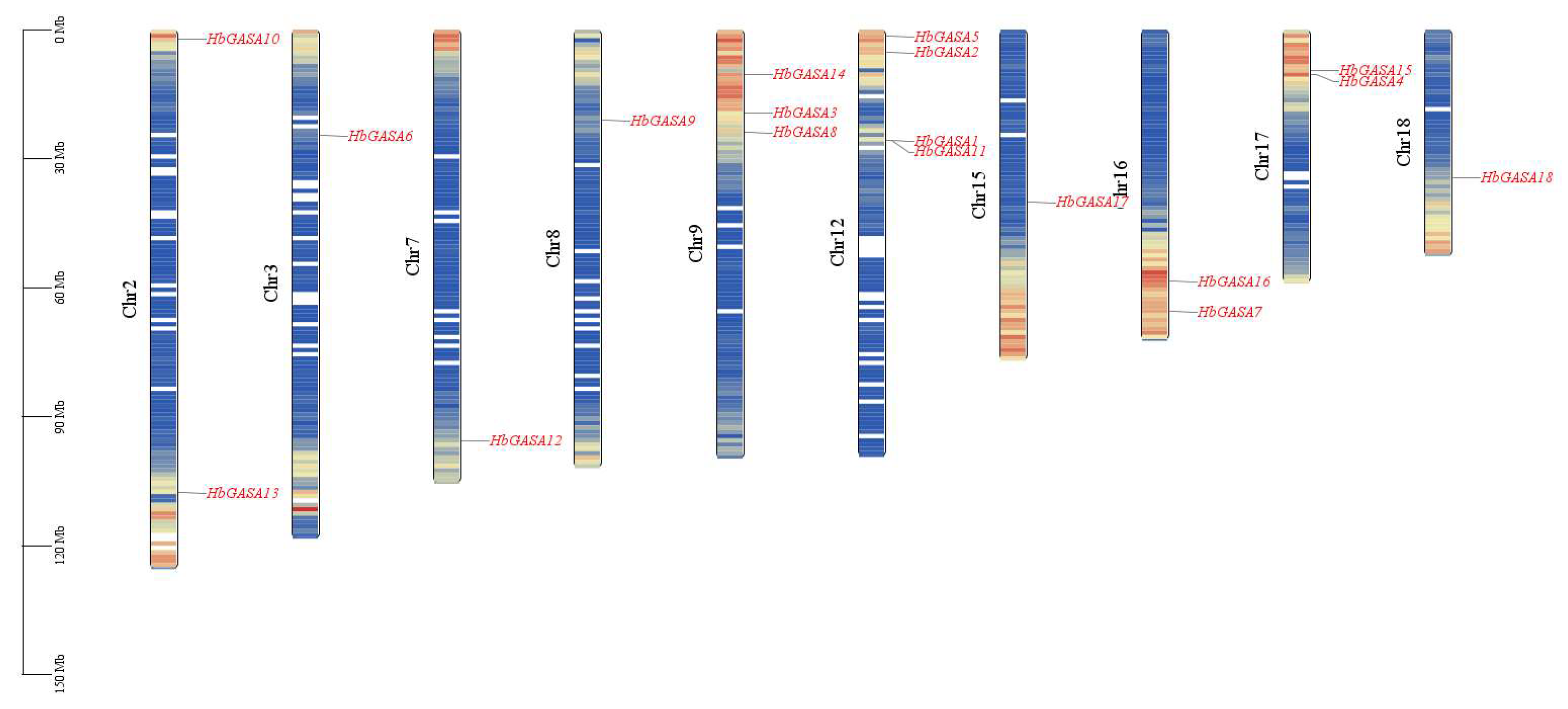
In the rubber tree, a comprehensive exploration of GASA - domain encoding genes was carried out. Employing the HMMER tool, a total of eighteen genes, namely
HbGASA -
HbGASA18, were successfully identified (
Table 1). Chromosomal localization analysis revealed that these 18
HbGASAs were distributed across 10 chromosomes in a rather dispersed manner (
Figure 1). On Chromosome 12, a gene cluster composed of
HbGASA1 and
HbGASA11 was formed. Notably, Chromosome 12 harbored the highest number of
HbGASA genes, with a count of 4, surpassing the quantity on other chromosomes. Sequence analysis further unveiled detailed characteristics of the corresponding proteins. The lengths of the 18 HbGASA proteins spanned from 88 to 253 amino acids. Specifically, HbGASA18 encoded the largest number of amino acids, reaching 253aa, while HbGASA2 and HbGASA9 encoded the fewest, with only 88aa. In terms of relative molecular weight, it ranged from 9680.57 to 26546.86 Da. HbGASA18 exhibited the largest molecular weight, and HbGASA2 the smallest. Regarding the isoelectric point, it varied from 7.46 to 9.98. HbGASA5 had the lowest isoelectric point, and HbGASA18 the highest. Significantly, the isoelectric points of all 18 proteins were above 7.0, indicating that they all belonged to the category of alkaline proteins.Analysis of the instability index showed that five proteins had an instability index less than 40, while the other 13 proteins were unstable, with an instability index greater than 40. In terms of hydrophobicity, except for HbGASA5 and HbGASA14, which encoded hydrophobic proteins, the remaining 16 HbGASAs encoded hydrophilic proteins.
2.2. Protein and gene structure of HbGASA genes
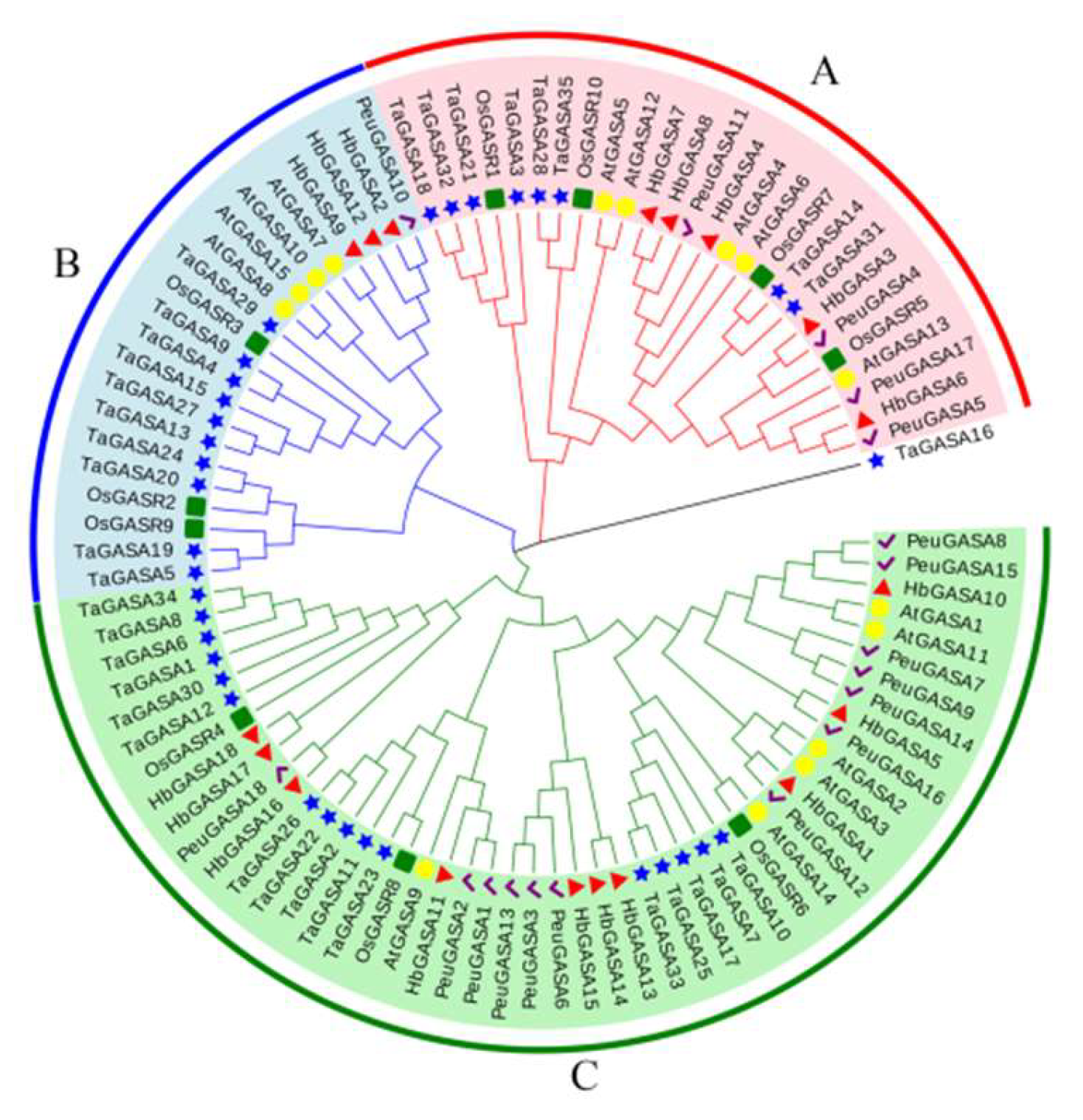
The protein sequences of HbGASAs were aligned with those of GASA proteins from
Arabidopsis, poplar, rice, and wheat. The alignment results revealed that all GASA proteins, with the exception of TaGASA16, were grouped into three subfamilies. Each subfamily encompassed homologous proteins originating from five distinct plant species (
Figure 2). Specifically, within the rubber tree's GASA proteins, there were three pairs of homologous genes. HbGASA7-HbGASA8 belonged to subfamily A, while HbGASA13- HbGASA14 and HbGASA17-HbGASA18 were part of subfamily C. Overall, in the phylogenetic analysis, the GASA proteins of the rubber tree exhibited a closer evolutionary relationship with the homologous proteins in poplar compared to those in the other plant species examined.
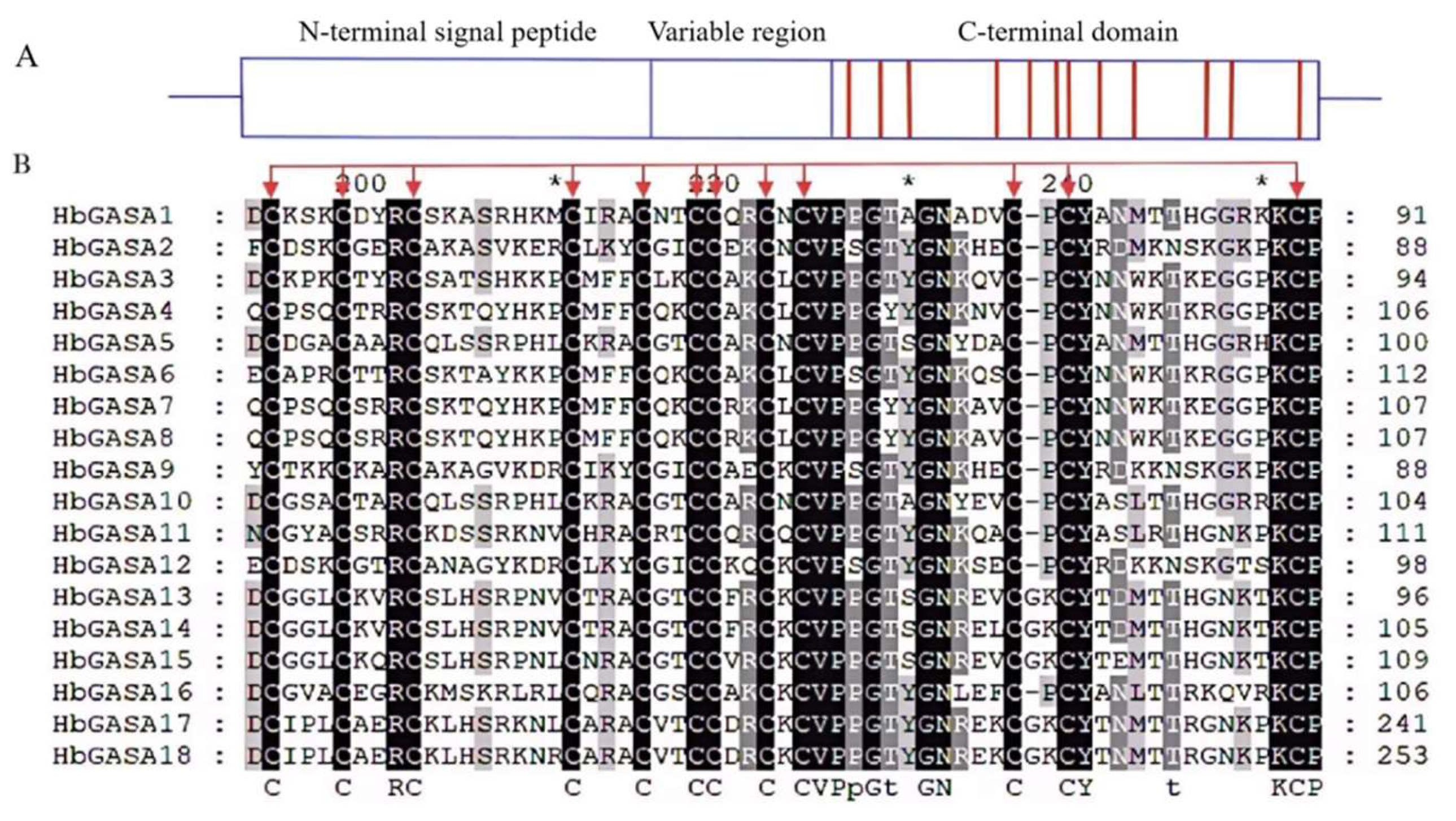
To thoroughly clarify the characteristics of HbGASA proteins, the full-length HbGASA protein sequences were aligned by means of GeneDoc software. The results demonstrated that all 18 HbGASA proteins exhibited a comparable structure. Each of them contained an N-terminal signal peptide ranging from 18 to 30 amino acids and a highly conserved C-terminal domain. Comprising approximately 60 amino acids, this C- terminal domain harbored 12 cysteine residues (
Figure 3,
Figure 4B).
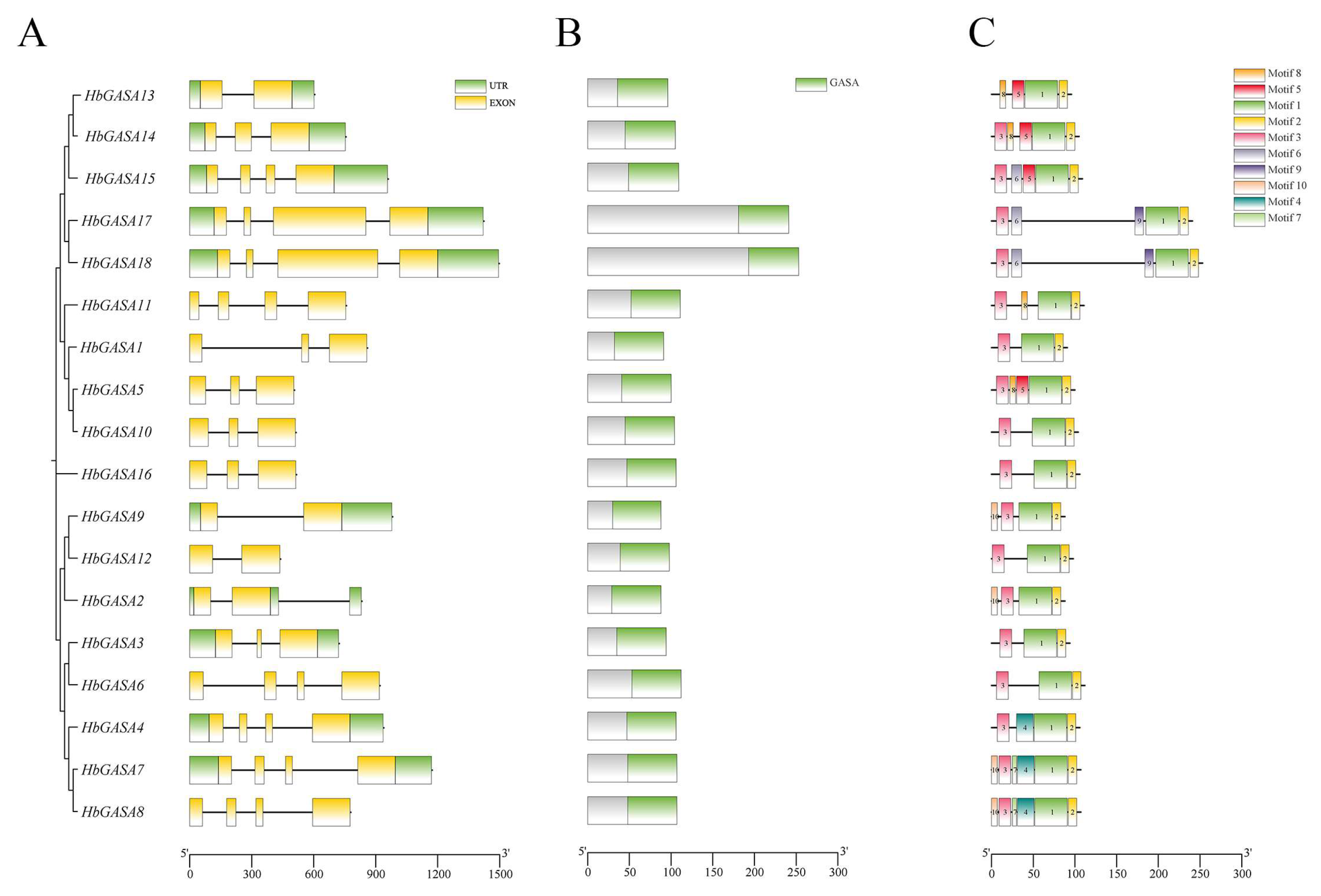
Gene structure analysis unveiled that all 18
HbGASAs were split genes, with exons being separated by introns. Each gene displayed structural disparities to different extents. The coding regions of
HbGASAs encompassed 1 to 3 introns (
Figure 4A). A total of 10 conserved motifs were detected in the 18 HbGASA proteins (
Figure 4C). Notably, all HbGASA proteins harbored Motif 1 and Motif 2, which together form the conserved structural domain of GASA. Moreover, all HbGASA proteins except HbGASA13 contained Motif 3. In contrast, Motif 7 was exclusively present in HbGASA7 and HbGASA8, and Motif 9 was only found in HbGASA17 and HbGASA18, respectively.
2.3. Promoter region analysis of HbGASA genes
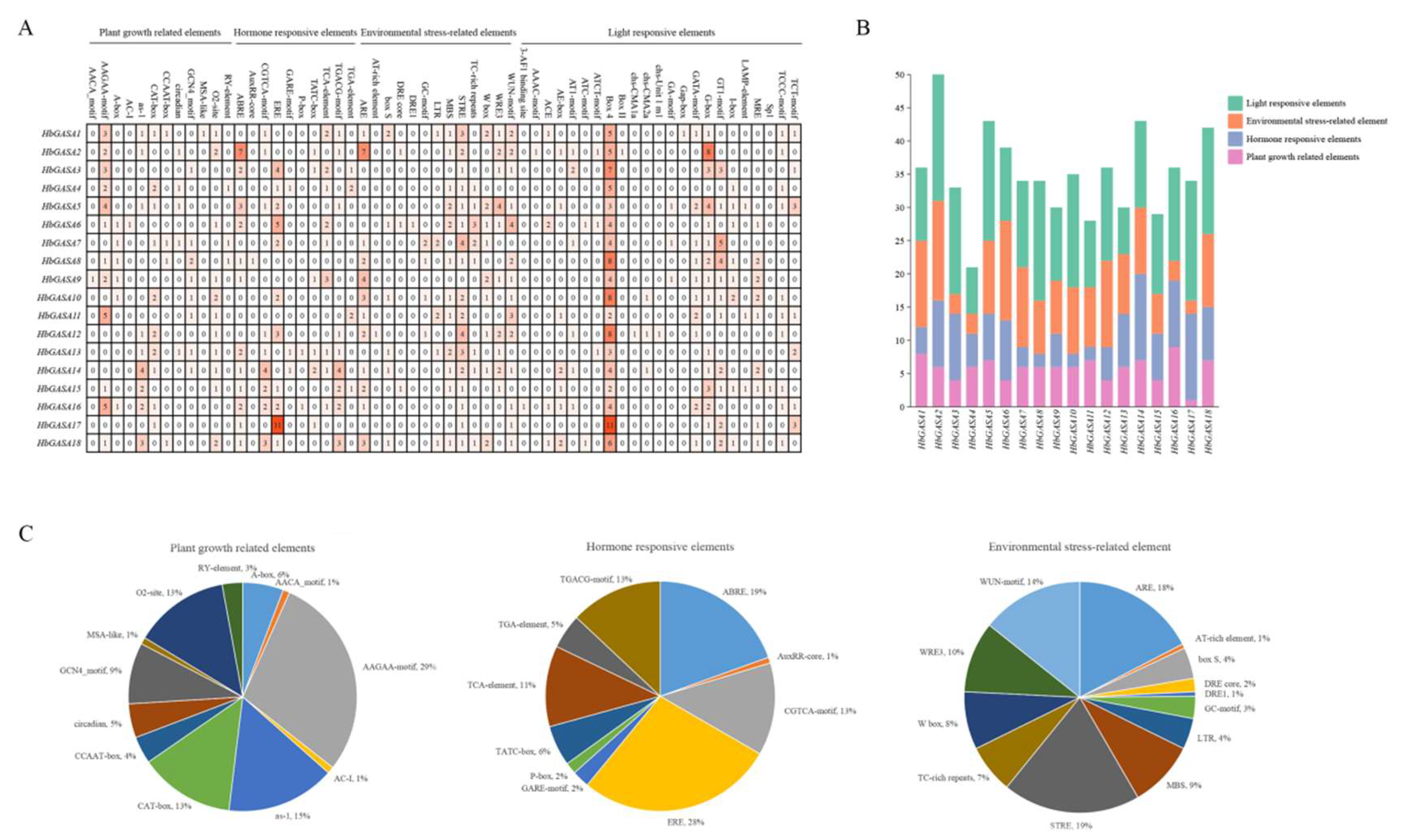
Analysis of cis-acting elements revealed that the promoters of
HbGASA genes harbored various elements associated with growth, stress responses, and phytohormone responses (
Figure 5). A relatively large number of cis-acting elements related to ethylene, abscisic acid (ABA), methyl jasmonate (MeJA), gibberellic acid (GA), and salicylic acid (SA) were detected. Besides these, elements related to cold, drought, and disease were identified in promoter regions of the majority of
HbGASA genes. Notably, the promoter of
HbGASA13 contained the most diverse array of cis-acting elements. These included elements associated with environmental signals like light, low temperature, and disease, as well as those related to hormone responses such as GA, MeJA, and auxin. This implies that
HbGASA13 might play a pivotal role in plant growth and responses to abiotic stress.
2.4. Tissue-specific expression patterns of HbGASA genes
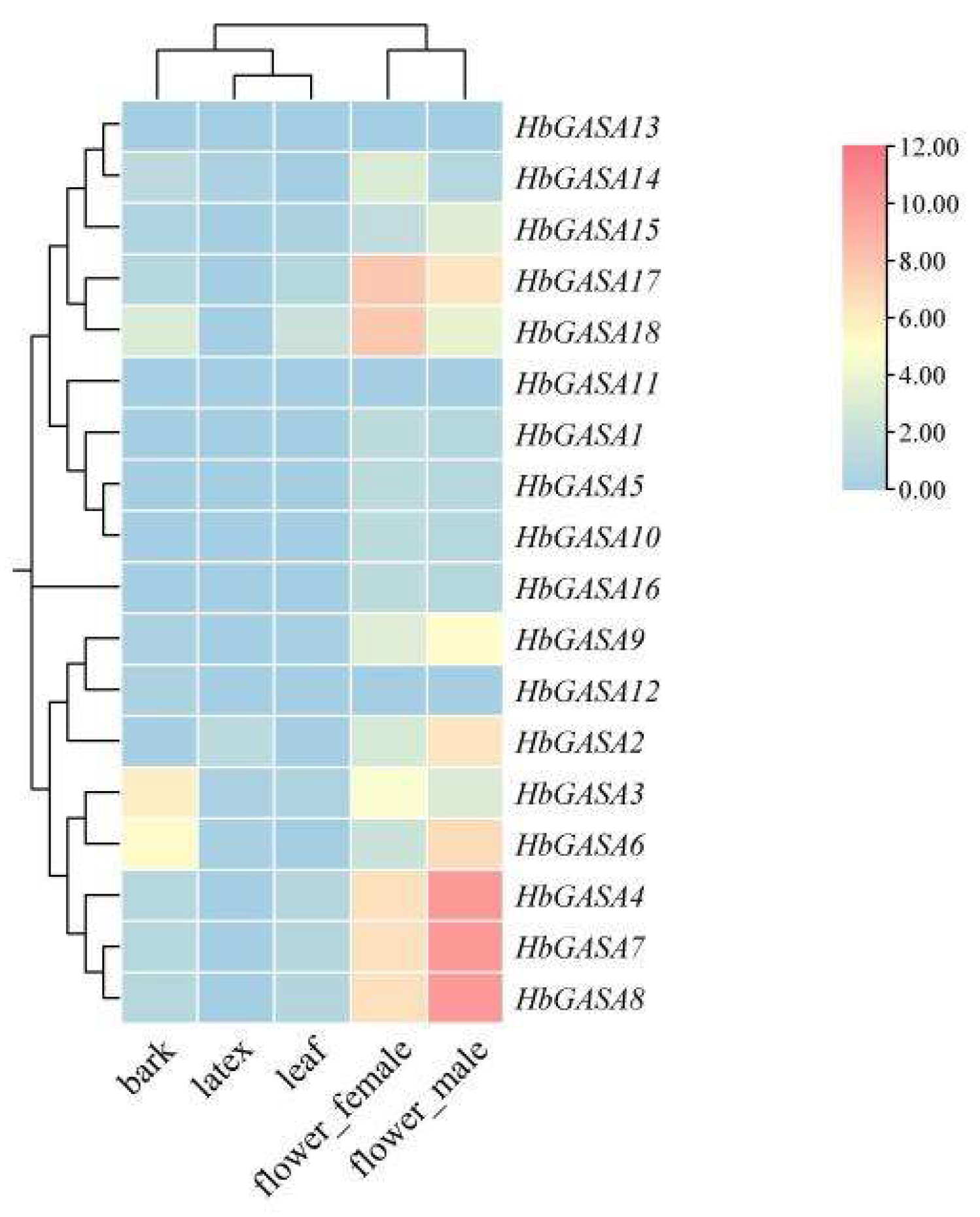
Based on the published RNA-seq data of the rubber tree, remarkable differences in the expression levels of the 18
HbGASAs were evident across various tissues (
Figure 6). To further validate the tissue-specific expression of
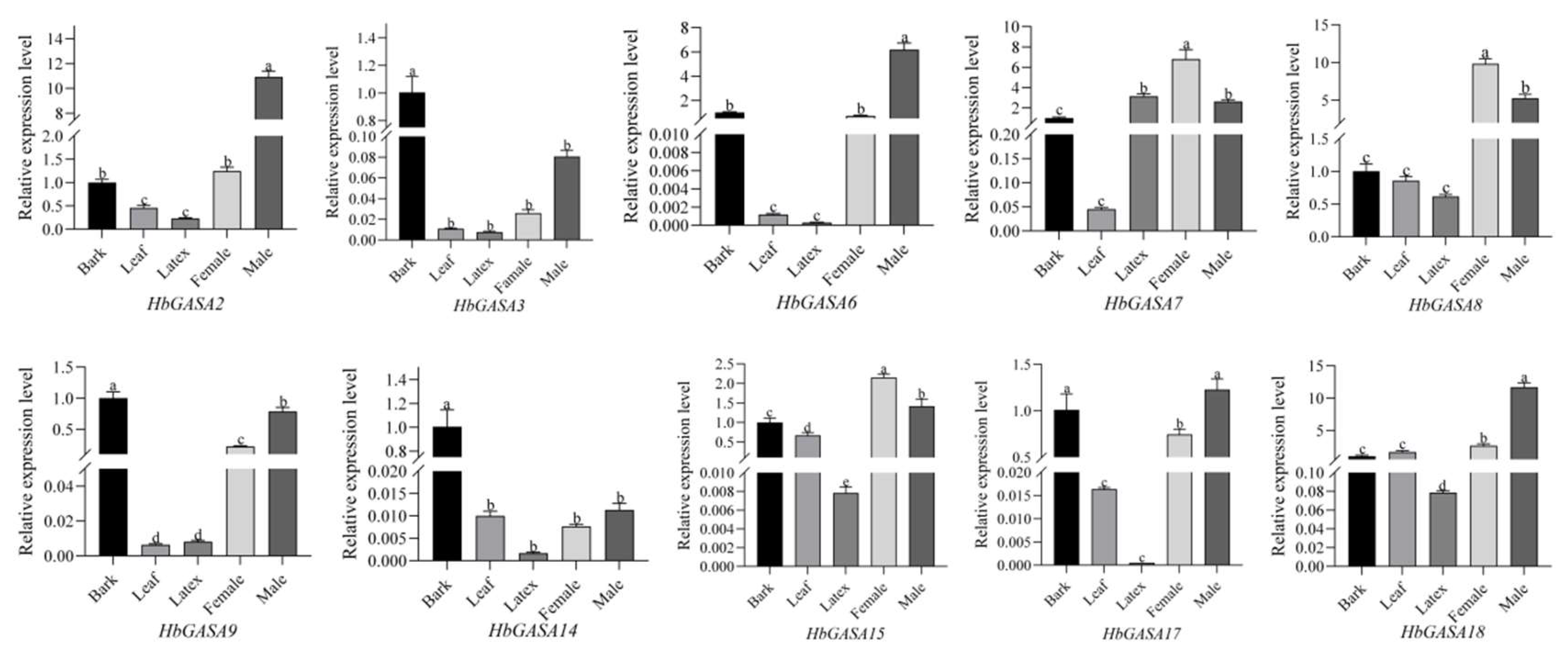
HbGASAs using qRT-PCR, 10 genes with relatively high expression levels, as determined from the expression heatmap, were selected. Among five distinct tissues of the rubber tree, viz., bark, leaves, latex, female flowers, and male flowers, the expression of these ten genes demonstrated tissue- specific characteristics (
Figure 7). Specifically,
HbGASA2,
HbGASA6, and
HbGASA18 exhibited higher expression levels in male flowers.
HbGASA3,
HbGASA9, and
HbGASA14 were more highly expressed in the bark. Moreover,
HbGASA7,
HbGASA8, and
HbGASA15 had elevated expression levels in female flowers.
2.5. Expression profiles of HbGASAs under cold stress
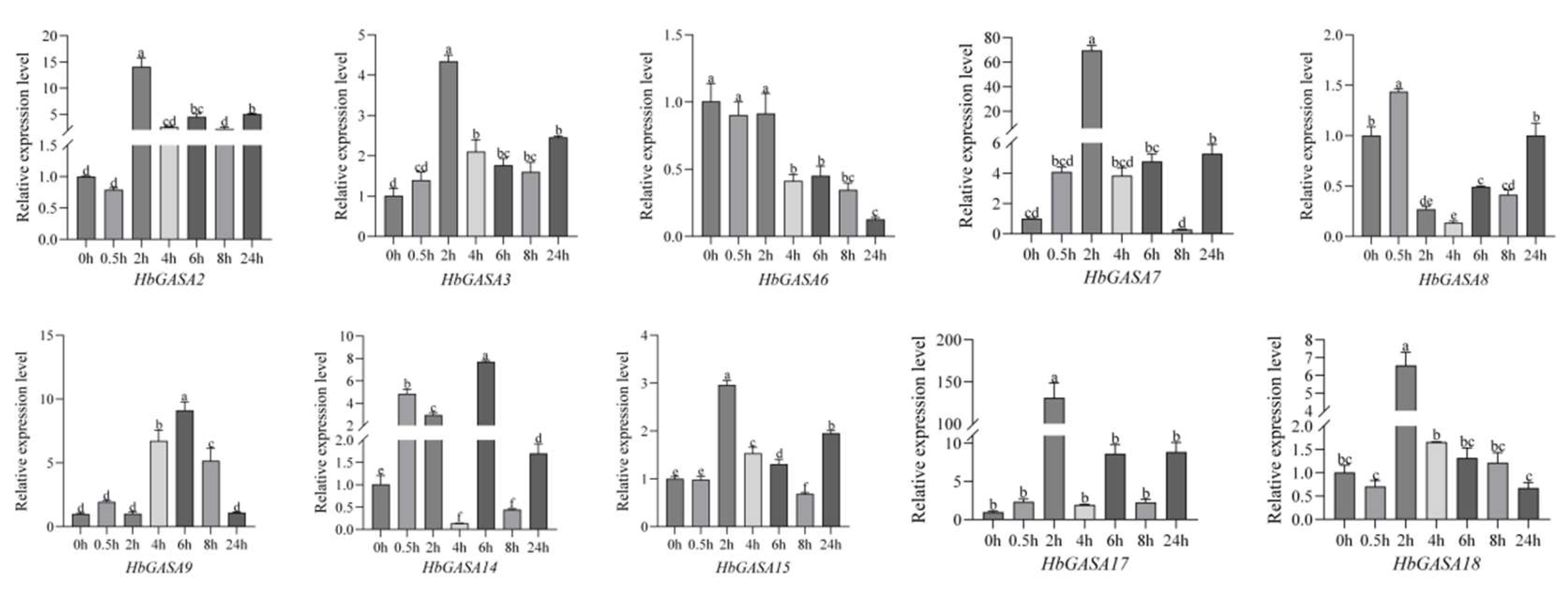
Analysis of the promoter cis-acting elements of
HbGASAs indicated that the promoter regions harbored low-temperature response elements, suggesting that the transcription of
HbGASAs was likely to be regulated by low temperature. The expression of ten selected
HbGASAs was analyzed via qRT-PCR under cold stress. The expression levels of most
HbGASAs exhibited a pattern of initial up-regulated expression followed by down-regulated (
Figure 8). Specifically,
HbGASA2,
HbGASA3,
HbGASA7,
HbGASA15,
HbGASA17, and
HbGASA18 responded rapidly to cold stress. Their expression levels reached the highest points merely 2 hours after the onset of cold stress. Meanwhile,
HbGASA9 and
HbGASA14 reached their peak expression levels 6 hours after the start of cold stress treatment. Among them,
HbGASA8 demonstrated the most rapid response to cold stress during the low-temperature treatment. Its expression level peaked after only 0.5 hours of cold stress exposure. In contrast, the expression level of
HbGASA6 decreased significantly, indicating that it was negatively regulated by cold stress.
3. Discussion
The GASA gene family, a plant-specific gene family regulated by gibberellins, has attracted extensive research attention in recent years. In the present study, a total of 18
HbGASA genes were identified, which is more than reported in the previous study[
22]. This increase can be attributed to the high-quality reference genome that has been published in recent years. In
Arabidopsis, the GASA genes were dispersed across all chromosomes. In contrast, in
Hevea brasiliensis, akin to plants such as rice [
23], grapevine [
24], and cotton [
25], the 18
GASA genes (
HbGASA1-
HbGASA18) were unevenly distributed among 18 chromosomes. This non-random distribution might be associated with the evolutionary history of the rubber tree and the functional specialization of different chromosomal regions. For instance, the gene cluster on chromosome 12, which contains
HbGASA1 and
HbGASA11, could be the outcome of gene duplication events. These events may have led to the functional divergence or sub-functionalization of these genes.
The proteins encoded by
HbGASA genes are all small polypeptides, which is consistent with the findings in
Arabidopsis [
26] and potato [
27]. This conservation during evolution implies that the GASA gene family plays fundamental and universal roles in basic physiological processes of plants. Motifs, which are structural aggregates within protein molecules, have the capacity to determine the specific functions of proteins or form specific structural domains within proteins. Common motifs in protein families play pivotal roles in both the functional expression and structural stability of these proteins [
4].
In this study, a comprehensive motif analysis was carried out on the 18 members of the GASA protein family in Hevea brasiliensis. The conserved motifs and gene structures observed among HbGASAs provided valuable insights into their potential functions. Notably, the presence of Motif 1 and Motif 2 in all 18 HbGASA proteins, which together constituted the conserved GASA domain, underscored their essential role in maintaining the structure and function of GASA proteins. Furthermore, the additional motifs, such as Motif 4 found in HbGASA4, HbGASA7, and HbGASA8, might confer unique functions to these proteins, especially considering their higher expression levels in male flowers. It is thus hypothesized that Motif 4 could be intricately involved in regulating the reproductive development of the rubber tree.
The tissue-specific expression patterns of HbGASA genes vividly illustrated their remarkable functional diversity. Distinct HbGASA genes exhibited preferential expression in particular tissues, including bark, leaves, latex, female flowers, and male flowers. This tissue-specific expression is likely intrinsically linked to the diverse physiological functions of these tissues. For instance, genes that were highly expressed in the bark might be engaged in processes such as pathogen defense or the regulation of latex production. In contrast, genes expressed in flowers were probably pivotal for reproductive development. Gaining a deep understanding of the regulatory mechanisms governing tissue-specific expression is of utmost importance for accurately deciphering the precise functions of HbGASA genes. It will provide key insights into how these genes contribute to the overall growth, development, and survival strategies of the rubber tree in different tissue-specific contexts.
The promoter analysis of
HbGASA genes uncovered the existence of numerous cis- acting elements associated with hormone and stress responses. The prevalence of elements responsive to ethylene, GA, MeJA, ABA, and SA strongly implied that
HbGASA genes were probably intricately involved in complex hormonal regulatory networks. The identification of cold-related elements in the promoters of the majority of the genes were consistent with the observed regulation of multiple
HbGASA genes under low-temperature stress. Additionally,
HbGASA genes have been reported to be regulated by fungal pathogens [
22]. Notably, the wide array of cis-acting elements presented in the HbGASA13 promoter indicated its crucial role in coordinating plant growth and responses to abiotic stress. Nevertheless, the precise mechanisms through which these elements interact with transcription factors and regulate gene expression still await further in-depth investigation. Understanding these regulatory mechanisms is essential for fully elucidating the functions of
HbGASA genes in plant development and stress adaptation.
4. Materials and Methods
4.1. Plant materials and stress treatment
One-year-old bud-grafted seedlings of rubber clone Reyan73397 were supplied by the National Rubber Germplasm Repository of China. To investigate the expression patterns of HbGASA genes under cold stress, these seedlings were placed in a plant growth room at 0℃, with a 16-hour photoperiod and a relative humidity of 80 ± 5%. The treatment duration was 24 hours. Leaves of the plantlets were sampled at 0, 0.5, 2, 4, 6, 8, and 24 hours after the onset of stress treatment. After harvested, the leaves were immediately frozen in liquid nitrogen and stored at −80℃ for RNA extraction. For each sampling time point, three replicate samples were collected.
4.2. Identification and chromosomal distribution of HbGASA genes
The genome sequences, protein sequences, and annotation information of the rubber tree were downloaded from the NCBI database (
https://www.ncbi.nlm.nih.gov/). The Hidden Markov Model (HMM) profile of the GASA domain (PF02704), retrieved from the protein families database (Pfam;
http://pfam-legacy.xfam.org/), was employed to search for GASA proteins in the rubber tree genome using the HMMER program. To verify the presence of the complete GASA domain and extract it from the obtained protein sequences, the NCBI Conserved Domain Database (
https://www.ncbi.nlm.nih. gov/Structure/cdd/cdd.shtml) and the Simple Modular Architecture Research Tool (SMART;
http://smart.embl - heidelberg.de/) were utilized. After eliminating redundant sequences, all non-redundant putative protein sequences containing a conserved GASA domain were selected for further analysis. TBtools was used to visually analyze the chromosomal locations of
HbGASA genes in the rubber tree and generate a chromosomal location map.
4.3. Physicochemical properties of HbGASA proteins
The fundamental physicochemical properties of HbGASA proteins, such as the theoretical relative molecular weight (MW), theoretical isoelectric point (pI), instability index (II), and grand average of hydropathicity (GRAVY), were analyzed using Protparam (
https://web.expasy.org/protparam/). Additionally, the secondary structure of HbGASA proteins was predicted by utilizing SOPMA (
https://npsa-prabi.ibcp.fr/cgi - bin/npsa_automat.pl?page=npsa_sopma.html).
4.4. Analysis of conserved motifs and gene structures
Conserved domain analysis was carried out by NCBI's CD-search tool (https://
www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Meanwhile, MEME (
http://memesuite. org/tools/meme) was utilized to predict the conserved motifs of HbGASA proteins, and visual files were saved for the purpose of creating conserved structure analysis maps and motif structure maps. In addition, the exon–intron study of the
HbGASA genes was conducted using TBtools.
4.5. Analysis of promoters and phylogeny of HbGASA genes
The 2Kb upstream sequences of the start codon ATG of
HbGASA genes were extracted from the rubber tree genome data and served as promoter sequences. The PlantCARE tool (
http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was employed to predict and analyze the characteristics of cis-acting elements in the promoters. Subsequently, TBtools was used to create cis-acting element analysis maps. The HbGASA protein sequences, along with GASA protein sequences from different species including
Arabidopsis, poplar, rice, and wheat, which were downloaded from Phytozome (
https://phytozome-next.jgi.doe.gov) and the NCBI database (
https://www. ncbi.nlm.nih.gov/), were aligned using the Clustal W algorithm in MEGA11 software. In MEGA11, the Neighbor-joining method was utilized to construct a phylogenetic tree with 1000 bootstrap replicates. Finally, the phylogenetic tree was generated using the online software Evolview (
https://www.evolgenius.info/evolview-v2/#login).
4.6. Tissue-specific expression profiles of HbGASA genes
Using the published rubber tree RNA-seq data available at HeveaDB database (
http://hevea.catas.cn), the FPKM (Fragments Per Kilobase of exon per Million reads mapped) values of
HbGASA genes were retrieved and normalized. Based on the normalized FPKM values, the tissue-specific expression profiles of
HbGASA genes in five distinct tissues, namely bark, leaves, latex, female flowers, and male flowers, were acquired. Subsequently, TBtools was employed to construct an expression heatmap of
HbGASA genes. To validate the tissue-specific expression patterns, quantitative real- time polymerase chain reaction (qRT-PCR) was carried out on selected
HbGASA genes in these five different tissues.
4.7. RNA isolation and qRT-PCR reaction
Total RNA was extracted from different tissues (bark, leaves, latex, female flowers, and male flowers) as well as leaves exposed to various cold stress conditions. The extraction was performed using the RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China) following the manufacturer's instructions. The concentration and integrity of the extracted RNA were evaluated using the NanoDrop One spectrophotometer (Thermo Fisher Scientific Inc., USA) and agarose gel electrophoresis. Subsequently, the quanlified RNA samples were reverse-transcribed into cDNA with the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). The cDNA was diluted five-fold. All qRT-PCR assays were conducted using the 2×Q3 SYBR qPCR Master Mix (TOLOBIO, China) according to the manufacturer's protocol. The qRT-PCR primers, designed with Primer 3 software, were listed in
Table 2.
HbYLS8 was selected as the reference gene. The relative expression levels were calculated using the 2
-ΔΔCt method. Each assay included three biological replicates and three technical replicates.
5. Conclusions
A total of eighteen HbGASA genes were identified. All of the identified HbGASA genes included a conserved GASA domain, and can be classified into three groups. Additional analyses of the promoter regions indicated that HbGASA genes were involved in plant growth, development, and stress responses. Analysis of tissue-specific expression patterns demonstrated that, compared to in latex, most HbGASA genes exhibited higher expression levels in barks, leaves, female flowers, and male flowers. Moreover, the expression levels of selected HbGASA genes were investigated under cold stress. The resulting data indicated that HbGASA genes might be involved in the response to cold stress in rubber tree. In summary, the data presented in this study offered fundamental and relevant information of HbGASA genes, which will serve as a valuable foundation for future research.
Author Contributions
Conceptualization, Y.C. and Z.A.; methodology, Y.C.; software, Y.Z.; validation, Z.D. and T.W.; formal analysis, Y.C.; investigation, Y.C.; resources, Y.H. and W.W.; data curation, Y.C.; writing—original draft preparation, Y.C.; writing—review and editing, Z.A.; visualization, Y.Z.; supervision, Z.A.; project administration, Z.D.; funding acquisition, Z.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China, grant number 2023YFD1200204; the Natural Science Foundation of Hainan Province, China, grant number 322RC782 and 323RC527; the Project of National Key Laboratory for Tropical Crop Breeding, grant number NKLTCB2023030.
Acknowledgments
We acknowledge National Rubber Germplasm Repository of China for providing the seedlings of rubber tree.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ABA |
Abscisic acid |
| MeJA |
Methyl jasmonate |
| GA |
Gibberellic acid |
| SA |
Salicylic acid |
| GASA |
Gibberellic Acid-Stimulated in Arabidopsis |
| qRT-PCR |
quantitative real time PCR |
References
- Bouteraa, M.T.; Ben Romdhane, W.; Baazaoui, N.; Alfaifi, M.Y.; Chouaibi, Y.; Ben Akacha, B.; Ben Hsouna, A.; Kacániová, M.; Cavar Zeljkovic, S.; Garzoli, S.; Ben Saad R. GASA proteins: review of their unctions in plant environmental stress tolerance. Plants 2023, 12, 2045. [CrossRef]
- De la Fuente, J. I.; Amaya, I.; Castillejo, C.; Sánchez-Sevilla, J. F.; Quesada, M. A.; Botella, M. A.; Valpuestaet V. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 2006, 10, 2401-2411. [CrossRef]
- Roxrud, I.; Lid, S. E.; Fletcher, J. C.; Schmidt, Ed D.L.; Opsahl-Sorteberg, H.-G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48(3), 471-483.
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [CrossRef]
- Ben-Nissan, G.; Lee, J. Y.; Borohov, A.; Weiss, D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and flowering transition. Plant J. 2004, 37(2), 229-238. [CrossRef]
- Li, W.; Zhen, W.; Xu, Y.; Joo, S.-H.; Kim, S.-K.; Xue Z.; Xu, Z.; Wang Z.; Chong K. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009, 57(3), 498-510.
- Zhong, C.; Xu, H.; Ye, S.; Wang, S.; Li, L.; Zhang, S.; Wang, X. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 2015, 169, 2288-2303.
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009, 69, 745-759. [CrossRef]
- Zimmermann, R.; Sakai, H.; Hochholdinger, F. The gibberellic acid stimulated-like gene family in maize and its role in lateral root development. Plant Physiol. 2010, 152, 356-365.
- Bouteraa, M.T.; Romdhane, W.B.; Ben Hsouna, A.; Amor, F.; Ebel, C.; Ben Saad, R. Genome-wide characterization and expression profiling of GASA gene family in Triticum turgidum ssp. durum (desf.) husn. (Durum wheat) unveils its involvement in environmental stress responses. Phytochemistry 2023, 206, 113544.
- Cheng, X.; Wang, S.; Xu, D.; Liu, X.; Li, X.; Xiao, W.; Cao, J.; Jiang, H.; Min, X.; Wang, J.; Zhang, H.; Chang, C.; Lu, J.; Ma, C. Identification and analysis of the GASR gene family in common wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination. Front. Genet. 2019, 10, 980.
- Herbel, V.; Sieber-Frank, J.; Wink, M. The antimicrobial peptide snakin-2 is up-regulated in the defense response of tomatoes (Solanum lycopersicum) as part of the jasmonate-dependent signaling pathway. J. Plant Physiol. 2017, 208, 1–6. [CrossRef]
- Ko, C.-B.; Woo, Y.-M.; Lee, D. J.; Lee M.-C.; Kim C.S. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol. Bioch. 2006, 44, 722-728. [CrossRef]
- Zhang, S.; Wang, X. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J. Plant Physiol. 2011, 168, 2093-2101. [CrossRef]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, W.; Wang, C.; Chen, C. Transcriptome analyses from mutant Salvia miltiorrhiza reveals important roles for SmGASA4 during plant development. Int. J. Mol. Sci. 2018, 19: 2088.
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637-1647. [CrossRef]
- Lee, S.-C.; Hans, S.-K.; Kim, S.-R. Salt- and ABA-inducible OsGASR1 is involved in salt tolerance. J. Plant Biol. 2015, 58, 96-101. [CrossRef]
- Li, K.-L.; Bai, X.; Li, Y.; Cai, H.; Ji, W.; Tang, L.-L.; Wen, Y.-D.; Zhu, Y.-M. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153-2160. [CrossRef]
- Priyadarshan, P.M.; Goncalves, P.S. Hevea gene pool for breeding. Genet. Resour. Crop Evol. 2003, 50, 101-114. [CrossRef]
- Priyadarshan, P.M. Refinements to Hevea rubber breeding. Tree Genet. Genomes 2017, 13, 20. [CrossRef]
- Priyadarshan, P.M.; Hoa, T.T.T.; Huasun H.; de Gonçalves, P. S. Yielding potential of rubber (Hevea brasiliensis) in sub-optimal environments. J. Crop Improv. 2005, 14(1-2), 221-247.
- An, B.; Wang, Q.; Zhang, X.; Zhang B.; Luo, H.; He, C. Comprehensive transcriptional and functional analyses of HbGASA genes reveal their roles in fungal pathogen resistance in Hevea brasiliensis. Tree Genet. Genomes 2018, 14: 41.
- Muhammad, I.; Li, W.Q.; Jing, X.Q.; Zhou, M.R.; Shalmani, A.; Ali, M.; Wei, X.Y.; Sharif, R.; Liu, W.T.; Chen, K.M. A systematic in silico prediction of gibberellicacid stimulated GASA family members: a novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 2019, 234-235,117-132.
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-wide characterization and expression profiling of GASA genes during different stages of seed development in Grapevine (Vitis vinifera L.) predict their involvement in seed development. Int. J. Mol. Sci. 2020, 21, 1088.
- Qiao, K.; Ma, C.; Lv, J.; Zhang, C.; Ma, Q.; Fan, S. Identification, characterization, and expression profiles of the GASA genes in Cotton. J. Cotton Res. 2021, 4, 7. [CrossRef]
- Zhang, S.; Wang, X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin. Sci. Bull. 2008, 53, 3839-3846. [CrossRef]
- Nahirnak, V.; Rivarola, M.; de Urreta, M.G.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-wide analysis of the Snakin/GASA gene family in Solanum Tuberosum cv. Kennebec. Am J. Potato Res. 2016, 93(2), 172-188. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).