Submitted:
12 April 2025
Posted:
15 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
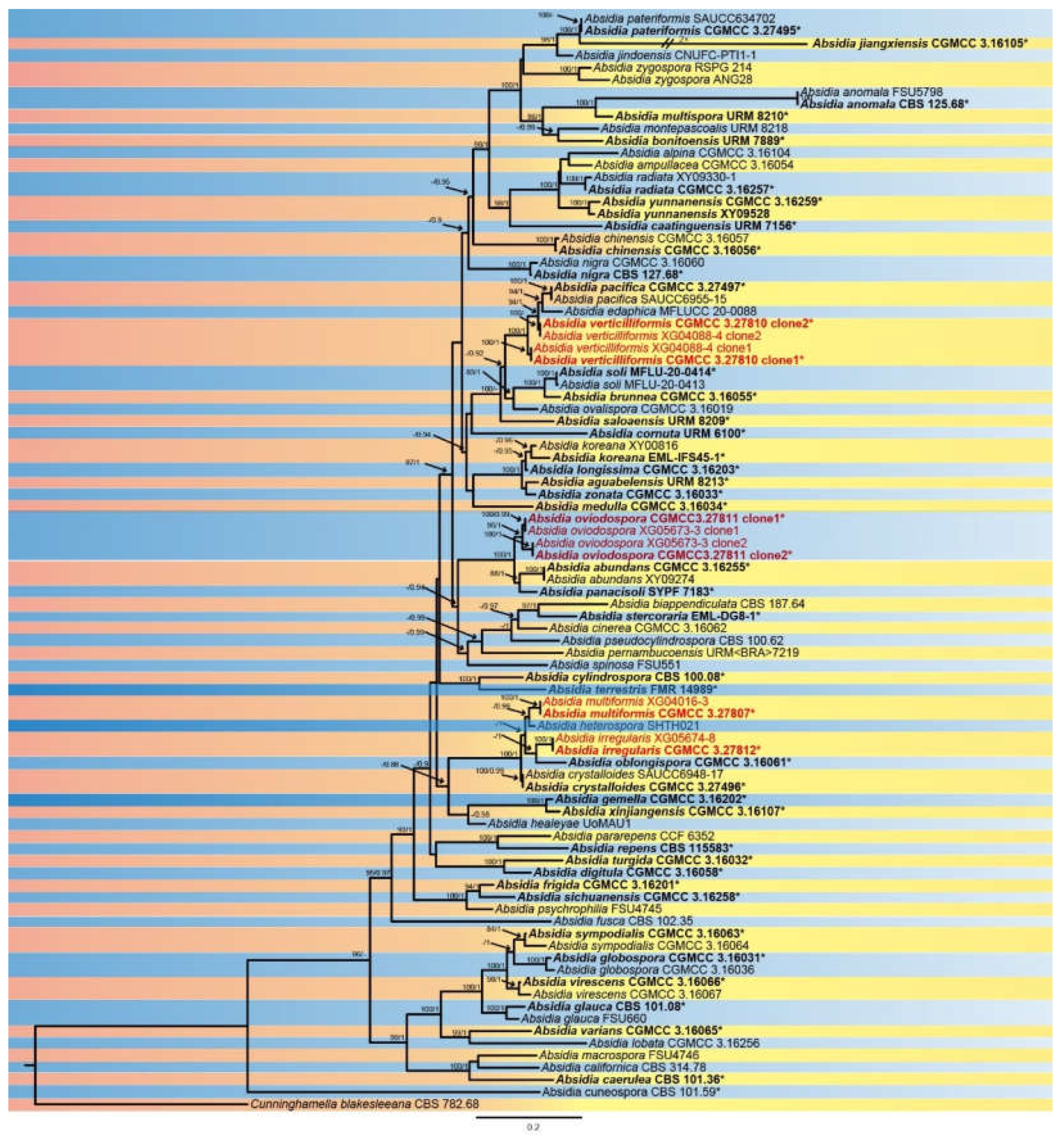
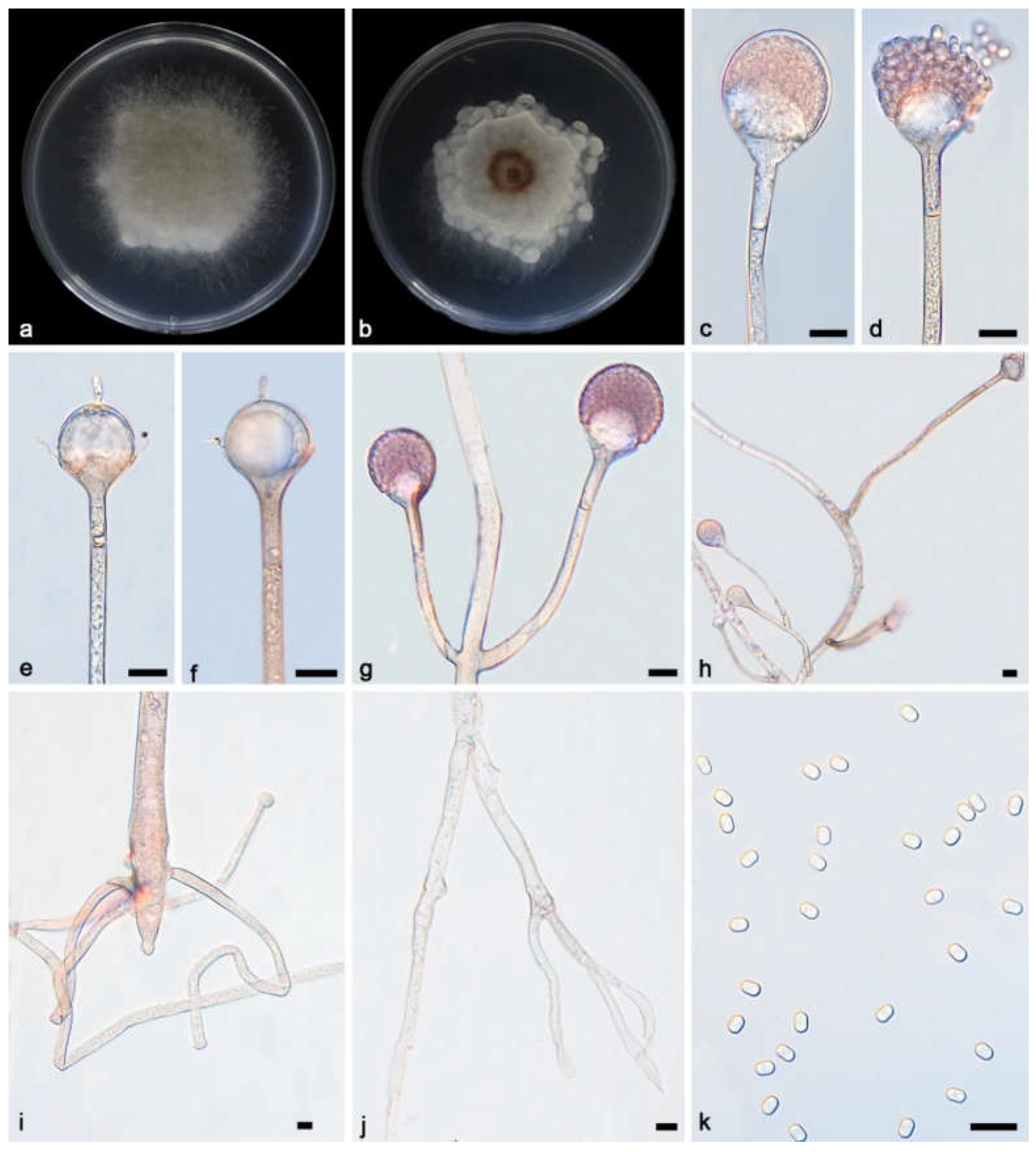
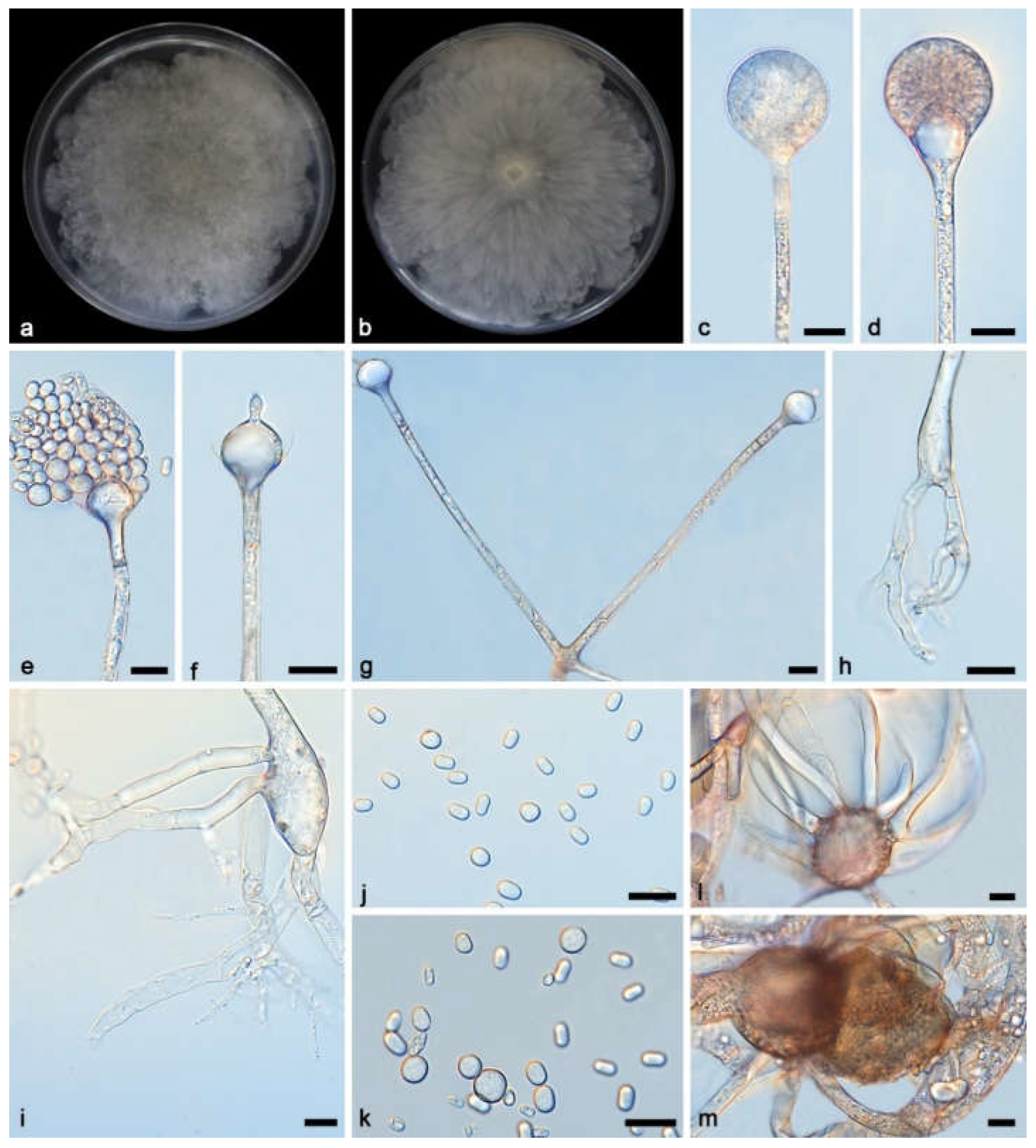
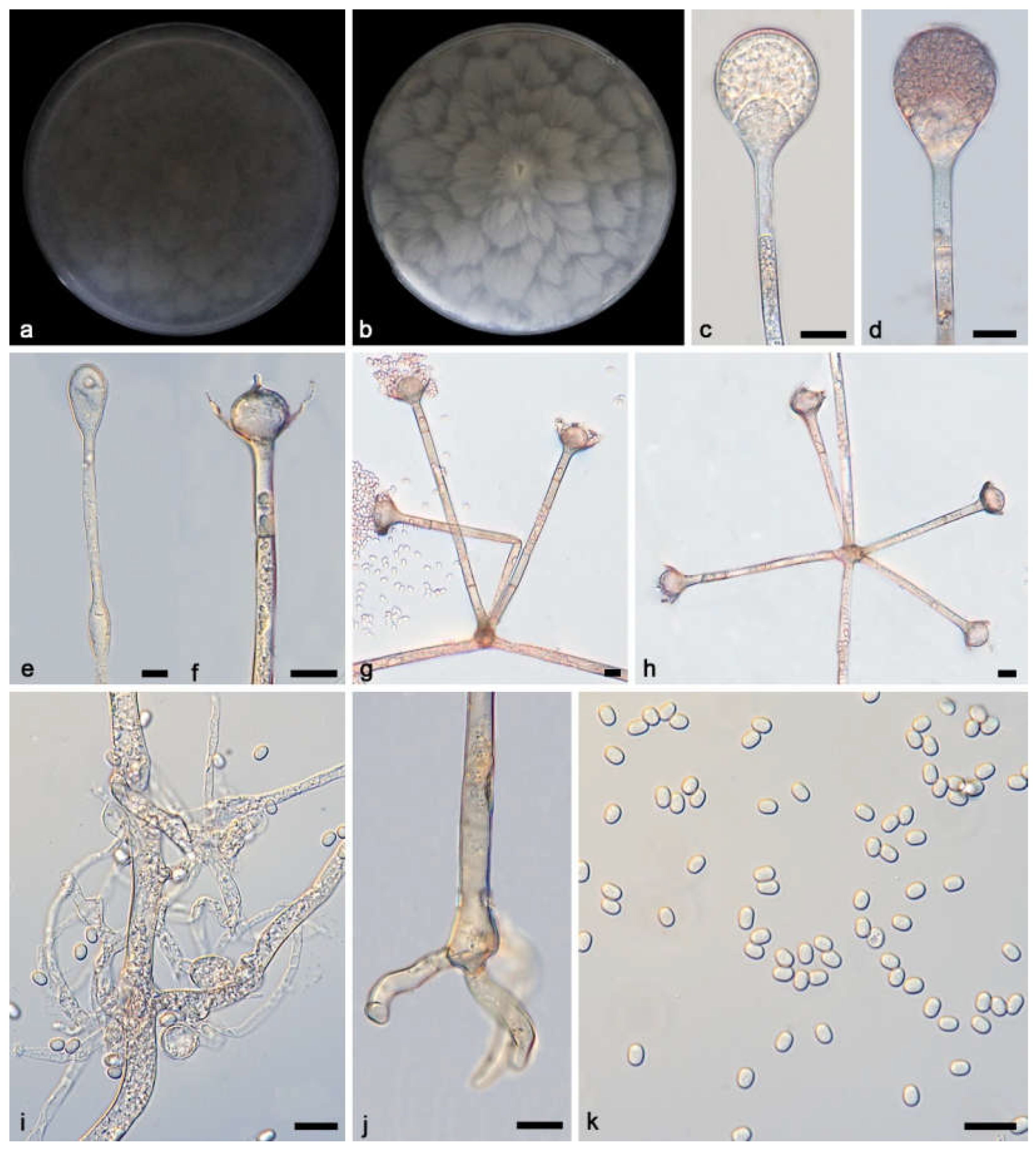
2.1. Isolation and Morphology
2.2. DNA Extraction and Amplification
3. Results
3.1. Phylogenetic Analyses
3.2. Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van Tieghem, P. Troisieme memoire sur les Mucorinees. Ann Sci Nat Bot Ser 1876, 4, 312–398. [Google Scholar]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Diversity 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Arx, J. A. v. On Mucoraceae s. str. and other families of the Mucorales. Sydowia 1982, 35, 10–26. [Google Scholar]
- Benny, G.; Humber, R.; Morton, J.: Zygomycota: Zygomycetes. 2001; 113–146.
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Ainsworth and Bisby's Dictionary of the Fungi (10th edition). Reference Reviews 2009, 23, 42–42. [Google Scholar]
- Hoffmann, K. Identification of the genus Absidia (Mucorales, Zygomycetes): A comprehensive taxonomic revision. 2010; 439–460.
- Hoffmann, K.; Discher, S.; Voigt, K. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycological Research 2007, 111, 1169–1183. [Google Scholar] [CrossRef]
- Hoffmann, K.; Voigt, K. Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biology 2009, 11, 537–554. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Tongkai, Z.; Wang, Y.-J.; Wang, M.; Dai, Y.-C.; Liu, X.-Y. Species diversity and ecological habitat of Absidia (Cunninghamellaceae, Mucorales) with emphasis on five new species from forest and grassland soil in China. Journal of Fungi 2022, 8, 471. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Zong, T.; Dai, Y.; Liu, X. Three new species of Absidia (Mucoromycota) from China based on phylogeny, morphology and physiology. Diversity 2022, 14, 132. [Google Scholar] [CrossRef]
- Braga, A. R.; Nunes, M. C.; Raymundo, A. The experimental development of emulsions enriched and stabilized by recovering matter from spirulina biomass: Valorization of residue into a sustainable protein source. Molecules. [CrossRef] [PubMed]
- Liu, X.; Qiao, L.; Xie, D.; Zhang, Y.; Zou, J.; Chen, X.; Dai, J. Microbial transformation of ginsenoside-Rg1 by Absidia coerulea and the reversal activity of the metabolites towards multi-drug resistant tumor cells. Fitoterapia 2011, 82, 1313–1317. [Google Scholar] [CrossRef]
- Tao, M.-F.; Ding, Z.-Y.; Wang, Y.-X.; Zhang, Z.-X.; Zhao, H.; Meng, Z.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China II: Three new species of Absidia (Cunninghamellaceae, Mucoromycota) from Hainan Province. MycoKeys 2024, 110, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-X.; Zhao, H.; Jiang, Y.; Liu, X.-Y.; Tao, M.-F.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China III: Six new species and a new record of Gongronella (Cunninghamellaceae, Mucoromycota). MycoKeys 2024, 110, 287–317. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Huang, B.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China I: three new species of Backusella (Backusellaceae, Mucoromycota). MycoKeys 2024, 109, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.-Y.; Ji, X.-Y.; Tao, M.-F.; Jiang, Y.; Liu, W.-X.; Wang, Y.-X.; Meng, Z.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China IV: Four new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys (under review).
- Ji, X.-Y.; Ding, Z.-Y.; Nie, Y.; Zhao, H.; Wang, S.; Huang, B.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China V: Five new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys (under review).
- Li, W.; Wei, Y.; Zou, Y.; Liu, P.; Li, Z.; Gontcharov, A.; Stephenson, S.; Wang, Q.; Zhang, S.; li, y. Dictyostelid cellular slime molds from the russian far east. Protist 2020, 171, 125756. [Google Scholar] [CrossRef]
- Zou, Y.; Hou, J.; Guo, S.; Li, C.; Li, Z.; Stephenson, S. L.; Pavlov, I. N.; Liu, P.; Li, Y. Diversity of dictyostelid cellular slime molds, including two species new to science, in forest soils of Changbai Mountain, China. Microbiology Spectrum 2022, 10, e0240222. [Google Scholar] [CrossRef]
- Corry, J. E. L., Curtis, G. D. W., Baird, R. M., Eds.; Elsevier, Rose Bengal Chloramphenicol (RBC) agar. In Progress in Industrial Microbiology 1995, 34, 431–433.
- Wang, Y.-X.; Zhao, H.; Ding, Z.-Y.; Ji, X.-Y.; Zhang, Z.; Wang, S.; Zhang, X.; Liu, X.-Y. Three new species of Gongronella (Cunninghamellaceae, Mucorales) from soil in Hainan, China based on morphology and molecular phylogeny. Journal of Fungi 2023, 9, 1182. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, R.; Liu, S.; Mu, T.; Zhang, X.; Xia, J. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 2022, 88, 171–192. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Voigt, K.; Wostemeyer, J. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiological research 2000, 155, 179–195. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J.: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. 1990; Vol. 31; pp 315–322.
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L. M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O'Donnell, K.; Lutzoni, F.; Ward, T.; Benny, G. Evolutionary relationships among mucoralean fungi (Zygomycota): Evidence for family polyphyly on a large scale. Mycologia 2001, 93, 286–296. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest V2. Program distributed by the author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef]
- Huelsenbeck, J. P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic biology 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Nie, Y.; Zong, T.-K.; Wang, K.; Lv, M.-L.; Cui, Y.-J.; Tohtirjap, A.; Chen, J.-J.; Zhao, C.-L.; Wu, F.; Cui, B.-K.; Yuan, Y.; Dai, Y.-C.; Liu, X.-Y. Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Diversity 2023, 123, 49–157. [Google Scholar] [CrossRef]
- Hesseltine, C. W. J. J. E. The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia 1964, 4, 568–601. [Google Scholar] [CrossRef]
- Zhang, T.-Y.; Yu, Y.; Zhu, H.; Yang, S.-Z.; Yang, T.-M.; Zhang, M.-y.; Zhang, Y.-X. Absidia panacisoli sp. nov., isolated from rhizosphere of Panax notoginseng. International Journal of Systematic and Evolutionary Microbiology 2018, 68, 2468–2472. [Google Scholar] [CrossRef]
- Hurdeal, V. G.; Gentekaki, E.; Lee, H. B.; Jeewon, R.; Hyde, K. D.; Tibpromma, S.; Mortimer, P. E.; Xu, J.-c. Mucoralean fungi in Thailand: Novel species of Absidia from tropical forest soil. Cryptogamie, Mycologie 2021, 42, 39–61. [Google Scholar] [CrossRef]
- Ariyawansa, H.; Hyde, K.; Jayasiri, S.; Buyck, B.; Kandawatte, T.; Dai, D.-Q.; Dai, Y.; Daranagama, D.; Jayawardena, R.; Lücking, R.; Ghobad-Nejhad, M.; Niskanen, T.; Thambugala, K.; Voigt, K.; Zhao, R.-L.; Li, G.-J.; Doilom, M.; Boonmee, S.; Yang, Z.; Chen, X.-H. Fungal Diversity Notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal diversity 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Li, G. J.; Hyde, K. D.; Zhao, R. L.; Hongsanan, S.; Abdel-Aziz, F. A.; Abdel-Wahab, M. A.; Alvarado, P.; Alves-Silva, G.; Ammirati, J. F.; Ariyawansa, H. A.; Baghela, A.; Bahkali, A. H.; Beug, M.; Bhat, D. J.; Bojantchev, D.; Boonpratuang, T.; Bulgakov, T. S.; Camporesi, E.; Boro, M. C.; Ceska, O.; Chakraborty, D.; Chen, J. J.; Chethana, K. W. T.; Chomnunti, P.; Consiglio, G.; Cui, B. K.; Dai, D. Q.; Dai, Y. C.; Daranagama, D. A.; Das, K.; Dayarathne, M. C.; De Crop, E.; De Oliveira, R. J. V.; de Souza, C. A. F.; de Souza, J. I.; Dentinger, B. T. M.; Dissanayake, A. J.; Doilom, M.; Drechsler-Santos, E. R.; Ghobad-Nejhad, M.; Gilmore, S. P.; Góes-Neto, A.; Gorczak, M.; Haitjema, C. H.; Hapuarachchi, K. K.; Hashimoto, A.; He, M. Q.; Henske, J. K.; Hirayama, K.; Iribarren, M. J.; Jayasiri, S. C.; Jayawardena, R. S.; Jeon, S. J.; Jerônimo, G. H.; Jesus, A. L.; Jones, E. B. G.; Kang, J. C.; Karunarathna, S. C.; Kirk, P. M.; Konta, S.; Kuhnert, E.; Langer, E.; Lee, H. S.; Lee, H. B.; Li, W. J.; Li, X. H.; Liimatainen, K.; Lima, D. X.; Lin, C. G.; Liu, J. K.; Liu, X. Z.; Liu, Z. Y.; Luangsa-ard, J. J.; Lücking, R.; Lumbsch, H. T.; Lumyong, S.; Leaño, E. M.; Marano, A. V.; Matsumura, M.; McKenzie, E. H. C.; Mongkolsamrit, S.; Mortimer, P. E.; Nguyen, T. T. T.; Niskanen, T.; Norphanphoun, C.; O’Malley, M. A.; Parnmen, S.; Pawłowska, J.; Perera, R. H.; Phookamsak, R.; Phukhamsakda, C.; Pires-Zottarelli, C. L. A.; Raspé, O.; Reck, M. A.; Rocha, S. C. O.; de Santiago, A. L. C. M. A.; Senanayake, I. C.; Setti, L.; Shang, Q. J.; Singh, S. K. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 2016, 78, 1–237. [Google Scholar] [CrossRef]
- Crous, P.; Luangsa-Ard, J.; Wingfield, M. J.; Carnegie, A.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.; Baseia, I.; Cano, J.; Martín, M.; Morozova, O.; Stchigel, A. M.; Summerell, B.; Brandrud, T. E.; Dima, B.; García, D.; Giraldo López, A.; Guarro, J.; Groenewald, J. Z. Fungal Planet description sheets- 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Wanasinghe, D. N.; Phukhamsakda, C.; Hyde, K. D.; Jeewon, R.; Lee, H. B.; Gareth Jones, E. B.; Tibpromma, S.; Tennakoon, D. S.; Dissanayake, A. J.; Jayasiri, S. C.; Gafforov, Y.; Camporesi, E.; Bulgakov, T. S.; Ekanayake, A. H.; Perera, R. H.; Samarakoon, M. C.; Goonasekara, I. D.; Mapook, A.; Li, W.-J.; Senanayake, I. C.; Li, J.; Norphanphoun, C.; Doilom, M.; Bahkali, A. H.; Xu, J.; Mortimer, P. E.; Tibell, L.; Tibell, S.; Karunarathna, S. C. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Lima, D.; Cordeiro, T.; Fragoso de Souza, C.; Oliveira, R.; Lee, H.; Souza-Motta, C.; Santiago, A. Morphological and molecular evidence for two new species of Absidia from Neotropic soil. Phytotaxa 2020, 446, 61–71. [Google Scholar] [CrossRef]
- Rafhaella, T.; Cordeiro, L.; Thuong, T.; Nguyen, T.; Lima, D.; Silva, S.; Lima, C.; D'Arc, J.; Leitão, A.; Melo, L.; Gurgel, S.; Lee, H.; Luiz, A.; Monteiro, C.; Santiago, A. Two new species of the industrially relevant genus Absidia (Mucorales) from soil of the Brazilian Atlantic Forest. Acta Botanica Brasilica 2020, 34. [Google Scholar]
- Arina, C.; Lima, L.; Diogo, X.; Lima, T.; Rafhaella, L.; Cordeiro, T.; Lee, H.; Thuong, T.; Nguyen, T.; Melo, L.; Gurgel, S.; Santiago, A. Absidia bonitoensis (Mucorales, Mucoromycota),a new species isolated from the soil of an uplandAtlantic forest in Northeastern Brazil. Nova Hedwigia 2021, 112, 241–251. [Google Scholar]
- LeitÃO, J.; Cordeiro, T.; Nguyen, T.; Lee, H.; Gurgel, L.; Santiago, A. Absidia aguabelensis sp. nov.: A new mucoralean fungi isolated from a semiarid region in Brazil. Phytotaxa 2021, 516, 83–91. [Google Scholar] [CrossRef]
- Urquhart, A.; Idnurm, A. Absidia healeyae: a new species of Absidia (Mucorales) isolated from Victoria, Australia. Mycoscience 2021, 62, 331–335. [Google Scholar] [CrossRef]
- Zhao, H.; Zhu, J.; Tongkai, Z.; Liu, X.-L.; Ren, L.-Y.; Lin, Q.; Qiao, M.; Nie, Y.; Zhang, Z.-D.; Liu, X.-Y. Two New Species in the Family Cunninghamellaceae from China. Mycobiology 2021, 49, 1–9. [Google Scholar] [CrossRef]
- Luo, C.; Chen, F.; Phookamsak, R.; Sun, F.; Xu, J.; Jiang, H. Polyphasic taxonomic study of Absidia menglianensis sp. nov. (Cunninghamellaceae, Mucorales) isolated from an avocado plantation in Yunnan, China. Studies in Fungi 2024, 9, e018. [Google Scholar] [CrossRef]
- Nguyen, T. T. T.; Santiago, A.; Hallsworth, J. E.; Cordeiro, T.; Voigt, K.; Kirk, P.; Crous, P. W.; Júnior, M. A. M.; Elsztein, C.; Lee, H. New Mucorales from opposite ends of the world. Studies in Mycology 2024, 109, 273–321. [Google Scholar] [CrossRef] [PubMed]
- Htet, Y. M.; Yasanthika, W. A. E.; Hurdeal, V. G.; Gomes De Farias, A. R. Absidia thailandica sp. nov., an addition to the diversity of soil fungi from Thailand. Phytotaxa 2024, 646, 157–168. [Google Scholar] [CrossRef]
- Hurdeal, V. G.; Jones, E. B. G.; Gentekaki, E. Absidia zygospora (Mucoromycetes), a new species from Nan Province, Thailand.
| Loci | PCR primers | Sequence (5’ – 3’) | PCR cycles | References |
|---|---|---|---|---|
| Act | ACT-512F | ATG TGC AAG GCC GGT TTC GC | 95℃ 3 min; (95℃ 1 min, 55℃ 1 min, 72℃ 1 min) × 30 cycles; 72℃ 10 min | [25] |
| ACT-783R | TAC GAG TCC TTC TGG CCC AT | |||
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | 95℃ 5 min; (95℃ 30 s, 55℃ 30 s, 72℃ 1 min) × 35 cycles; 72℃ 10 min | [26] |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | |||
| LSU | LR0R | GTA CCC GCT GAA CTT AAG C | 95℃ 5 min; (95℃ 50 s, 47℃ 30 s, 72℃ 1.5 min) × 35 cycles; 72℃ 10 min | [27] |
| LR7 | TAC TAC CAC CAA GAT CT | |||
| SSU | NS1 | GTA GTC ATA TGC TTG TCT C C | 95 °C 5 min; (94 °C 60 s, 54 °C 50 s, 72 °C 60 s) × 37 cycles; 72 °C 10 min | [26] |
| NS4 | CTT CCG TCA ATT CCT TTA AG | |||
| TEF1α | EF1-728F | CAT CGA GAA GTT CGA GAA GG | 95℃ 5 min; (95℃ 30 s, 55℃ 60 s, 72℃ 1 min) × 30 cycles; 72℃ 10 min | [28] |
| EF2 | GGA RGT ACC AGT SAT CAT GTT |
| Species | Strains | GenBank accession numbers | ||||
| ITS | LSU | TEF1α | Act | SSU | ||
| Absidia abundans | CGMCC 3.16255* | NR_182590 | ON074683 | n.a. | n.a. | n.a. |
| A. abundans | XY09274 | ON074696 | ON074682 | n.a. | n.a. | n.a. |
| A. aguabelensis | URM 8213* | NR_189383 | NG_241934 | n.a. | n.a. | n.a. |
| A. alpina | CGMCC 3.16104 | OL678133 | n.a. | n.a. | n.a. | n.a. |
| A. ampullacea | CGMCC 3.16054 | MZ354138 | MZ350132 | n.a. | n.a. | n.a. |
| A. anomala | CBS 125.68* | MH859085 | MH870799 | n.a. | n.a. | n.a. |
| A. anomala | FSU5798 | EF030523 | n.a. | n.a. | EF030535 | n.a. |
| A. biappendiculata | CBS 187.64 | MZ354153 | MZ350147 | MZ357420 | MZ357438 | |
| A. bonitoensis | URM 7889* | MN977786 | MN977805 | n.a. | n.a. | n.a. |
| A. brunnea | CGMCC 3.16055* | MZ354139 | MZ350133 | MZ357403 | MZ357421 | n.a. |
| A. caatinguensis | URM 7156* | NR_154704 | NG_058582 | n.a. | n.a. | n.a. |
| A. caerulea | CBS101.36* | MH855718 | MH867230 | n.a. | n.a. | n.a. |
| A. californica | CBS 314.78 | JN205816 | MH872902 | n.a. | n.a. | n.a. |
| A. chinensis | CGMCC 3.16057 | MZ354141 | MZ350135 | n.a. | MZ357422 | n.a. |
| A. chinensis | CGMCC 3.16056* | MZ354140 | MZ350134 | n.a. | n.a. | n.a. |
| A. cinerea | CGMCC 3.16062 | MZ354146 | MZ350140 | MZ357407 | MZ357427 | n.a. |
| A. cornuta | URM 6100* | NR_172976 | MN625255 | n.a. | n.a. | n.a. |
| A. crystalloides | CGMCC3.27496* | PP377803 | PP373736 | PP790574 | PP790582 | PP779723 |
| A. crystalloides | SAUCC693201 | PP377804 | PP373737 | PP790573 | PP790581 | PP779722 |
| A. cuneospora | CBS 101.59* | MH857828 | MH869361 | n.a. | n.a. | n.a. |
| A. cuneospora | FSU5890 | EF030524 | n.a. | n.a. | EF030533 | n.a. |
| A. cylindrospora | CBS 100.08 | JN205822 | JN206588 | n.a. | n.a. | n.a. |
| A. digitula | CGMCC 3.16058* | MZ354142 | MZ350136 | MZ357404 | MZ357423 | n.a. |
| A. edaphica | MFLUCC 20-0088 | NR_172305 | NG_075367 | n.a. | MT410739 | NG_074951 |
| A. frigida | CGMCC 3.16201* | NR_182565 | OM030223 | n.a. | n.a. | n.a. |
| A. fusca | CBS 102.35* | NR_103625 | NG_058552 | n.a. | n.a. | n.a. |
| A. gemella | CGMCC 3.16202* | OM108488 | OM030224 | n.a. | n.a. | n.a. |
| A. glauca | CBS 101.08* | MH854573 | MH866105 | n.a. | n.a. | n.a. |
| A. glauca | FSU660 | AY944879 | EU736302 | EU736248 | EU736225 | EU736275 |
| A. globospora | CGMCC 3.16031* | NR_189829 | MW671544 | MZ357412 | MZ357431 | n.a. |
| A. globospora | CGMCC 3.16036 | MW671539 | MW671546 | MZ357414 | MZ357433 | n.a. |
| A. healeyae | UoMAU1 | n.a. | MT436027 | n.a. | n.a. | n.a. |
| A. heterospora | SHTH021 | JN942683 | JN982936 | n.a. | n.a. | JQ004928 |
| A.irregularis | CGMCC 3.27812* | PQ306325 | PQ289020 | PV126019 | PQ807209 | PQ799254 |
| A.irregularis | XG05674-7 | PQ306326 | PQ289021 | PV126020 | PQ807210 | PQ799255 |
| A. jiangxiensis | CGMCC 3.16105* | OL678134 | PP780377 | n.a. | n.a. | n.a. |
| A. jindoensis | CNUFC-PTI1-1 | MF926622 | MF926616 | MF926513 | MF926510 | MF926626 |
| A. koreana | EML-IFS45-1* | KR030062 | KR030056 | KR030060 | KR030058 | KT321298 |
| A. koreana | XY00816 | OL620083 | ON123771 | n.a. | n.a. | n.a. |
| A. lobata | CGMCC 3.16256 | ON074690 | ON074679 | n.a. | n.a. | n.a. |
| A. longissima | CGMCC 3.16203* | NR_182566 | OM030225 | n.a. | n.a. | n.a. |
| A. macrospora | FSU4746 | AY944882 | EU736303 | EU736249 | AY944760 | EU736276 |
| A. medulla | CGMCC 3.16034 | NR_189832 | MW671549 | MZ357417 | MZ357436 | n.a. |
| A. montepascoalis | URM 8218 | NR_172995 | n.a. | n.a. | n.a. | n.a. |
| A.multiformis | CGMCC 3.27807* | PQ306319 | PQ803168 | PV126021 | PQ807203 | PQ799260 |
| A.multiformis | XG04016-3 | PQ306320 | PQ803169 | PV126022 | PQ807204 | PQ799261 |
| A. multispora | URM 8210* | MN953780 | MN953782 | n.a. | n.a. | n.a. |
| A. nigra | CBS 127.68* | NR_173068 | MZ350146 | MZ357419 | MZ357437 | n.a. |
| A. nigra | CGMCC 3.16060 | MZ354144 | MZ350138 | MZ357406 | MZ357425 | n.a. |
| A. oblongispora | CGMCC 3.16061 | MZ354145 | MZ350139 | n.a. | MZ357426 | n.a. |
| A. ovalispora | CGMCC 3.16019 | NR_176748 | MW264131 | n.a. | n.a. | n.a. |
| A.ovoidospora | CGMCC 3.27811 clone1* | PQ306327 | PQ803164 | PV126015 | PQ807207 | PQ799256 |
| A. ovoidospora | CGMCC 3.27811 clone2* | PV069753 | PQ803165 | PV126017 | PV126023 | PQ799257 |
| A. ovoidospora | XG05673-3 clone1 | PQ306328 | PQ803166 | PV126016 | PQ807208 | PQ799258 |
| A. ovoidospora | XG05673-3 clone2 | PV069754 | PQ803167 | PV126018 | PV126024 | PQ799259 |
| A. pacifica | CGMCC3.27497* | PP377802 | PP373735 | PP839793 | PP790579 | PP779720 |
| A. pacifica | SAUCC413601 | PP377801 | PP373734 | PP839794 | PP790580 | PP779721 |
| A. panacisoli | SYPF 7183* | MF522181 | MF522180 | MF624251 | n.a. | MF522179 |
| A. pararepens | CCF 6352 | MT193669 | MT192308 | n.a. | n.a. | n.a. |
| A. pateriformis | CGMCC3.27495* | PP377805 | PP373738 | PP790575 | PP790583 | PP779724 |
| A. pateriformis | SAUCC634702 | PP377806 | PP373739 | PP790576 | PP790584 | PP779725 |
| A. pernambucoensis | URM<BRA>7219 | MN635568 | MN635569 | n.a. | n.a. | n.a. |
| A. pseudocylindrospora | EML-FSDY6-2 | KU923817 | KU923814 | n.a. | KU923815 | KU923819 |
| A. pseudocylindrospora | CBS 100.62* | NR_145276 | MH869688 | n.a. | n.a. | n.a. |
| A. psychrophilia | FSU4745 | AY944874 | EU736306 | EU736252 | AY944762 | EU736279 |
| A. radiata | CGMCC 3.16257 | ON074698 | ON074684 | n.a. | n.a. | n.a. |
| A. radiata | XY09330-1 | ON074699 | ON074685 | n.a. | n.a. | n.a. |
| A. repens | CBS 115583* | NR_103624 | NG_058551 | n.a. | n.a. | n.a. |
| A. saloaensis | URM 8209* | MN953781 | MN953783 | n.a. | n.a. | n.a. |
| A. sichuanensis | CGMCC 3.16258* | NR_182589 | ON074688 | n.a. | n.a. | n.a. |
| A. soli | MFLU-20-0414* | MT396373 | MT393988 | n.a. | n.a. | MT394049 |
| A. soli | MFLU 20-0413 | MT396371 | MT393985 | n.a. | n.a. | MT394046 |
| A. spinosa | FSU551 | AY944887 | EU736307 | EU736253 | EU736227 | EU736280 |
| A. stercoraria | EML-DG8-1* | KU168828 | KT921998 | KT922002 | KT922000 | NG_065640 |
| A. sympodialis | CGMCC 3.16063* | MZ354147 | MZ350141 | n.a. | n.a. | n.a. |
| A. sympodialis | CGMCC 3.16064 | MZ354148 | MZ350142 | MZ357408 | n.a. | n.a. |
| A. terrestris | FMR 14989* | LT795003 | LT795005 | n.a. | n.a. | n.a. |
| A. turgida | CGMCC 3.16032* | NR_189830 | NG_241931 | MZ357415 | MZ357434 | n.a. |
| A. varians | CGMCC 3.16065* | MZ354149 | MZ350143 | MZ357409 | MZ357428 | n.a. |
| A.verticilliformis | CGMCC 3.27810 clone1* | PQ306315 | PQ803170 | PV126011 | PQ807205 | PQ799262 |
| A.verticilliformis | CGMCC 3.27810 clone2* | PV069755 | PQ803171 | PV126013 | PV126025 | PQ799263 |
| A.verticilliformis | XG04088-4 clone1 | PQ306316 | PQ803172 | PV126012 | PQ807206 | PQ799264 |
| A.verticilliformis | XG04088-4 clone2 | PV069756 | PQ803173 | PV126014 | PV126026 | PQ799265 |
| A. virescens | CGMCC 3.16066* | MZ354150 | MZ350144 | MZ357410 | MZ357429 | n.a. |
| A. virescens | CGMCC 3.16067 | MZ354151 | MZ350145 | MZ357411 | MZ357430 | n.a. |
| A. xinjiangensis | CGMCC 3.16107* | OL678136 | n.a. | n.a. | n.a. | n.a. |
| A. yunnanensis | XY09528 | ON074701 | ON074686 | n.a. | n.a. | n.a. |
| A. yunnanensis | CGMCC 3.16259* | NR_182591 | NG_149054 | n.a. | n.a. | n.a. |
| A. zonata | CGMCC 3.16033* | NR_189831 | MW671548 | MZ357416 | MZ357435 | n.a. |
| A. zygospora | RSPG 214 | KC478527 | n.a. | n.a. | n.a. | n.a. |
| A. zygospora | ANG28 | DQ914420 | n.a. | n.a. | n.a. | n.a. |
| Cunninghamella blakesleeana | CBS 782.68 | JN205869 | MH870950 | n.a. | n.a. | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





