Submitted:
12 March 2025
Posted:
13 March 2025
You are already at the latest version
Abstract
The Dof gene family represents a class of plant-specific transcription factors that play crucial regulatory roles in various biological processes, including plant growth, development, and responses to abiotic stress. However, genome-wide identification and functional characterization of the Dof gene family remain unexplored in Dendrobium officinale. In this study, we performed a genome-wide identification and functional analysis of the DoDof gene family. A total of 28 Dof family members were identified and named DoDof1-28 based on genome annotation data. Phylogenetic analysis classified these genes into four major groups (A-D) and further subdivided them into nine subfamilies. Gene structure analysis revealed that most DoDofs lack introns, with no distinct specificity observed among different subfamilies and considerable diversity within the same subfamily. Sequence alignment analysis demonstrated that all DoDof proteins contain a conserved Dof domain consisting of 52 amino acids, which includes a C2-C2 zinc finger motif and a DNA-binding domain. MEME analysis revealed that the conserved motif composition exhibits a certain degree of conservation among DoDof proteins, but significant differences exist across subfamilies. Expression pattern analysis demonstrated that DoDofs have exhibited diverse expression profiles across different developmental stages, tissues, and under abiotic stresses (such as low temperature, salinity, and drought) in D. officinale, suggesting their potential roles in plant development and stress responses. Subcellular localization analysis indicated that DoDof15, DoDof22, and DoDof24 are localized exclusively in the nucleus. Yeast one-hybrid assays revealed that DoDof22 binds to the promoter of the ABA receptor DoPYL9, while DoDof15 and DoDof24 bind to the promoter of the bHLH transcription factor DobHLH68. These results suggest that DoDof proteins may regulate the growth, development, and stress response processes of D. officinale by binding to the promoters of target genes. This study provides critical insights into the functional roles of Dof transcription factors in Orchidaceae family and establishes a theoretical foundation for molecular breeding and stress resistance improvement in D. officinale.
Keywords:
1. Introduction
2. Results
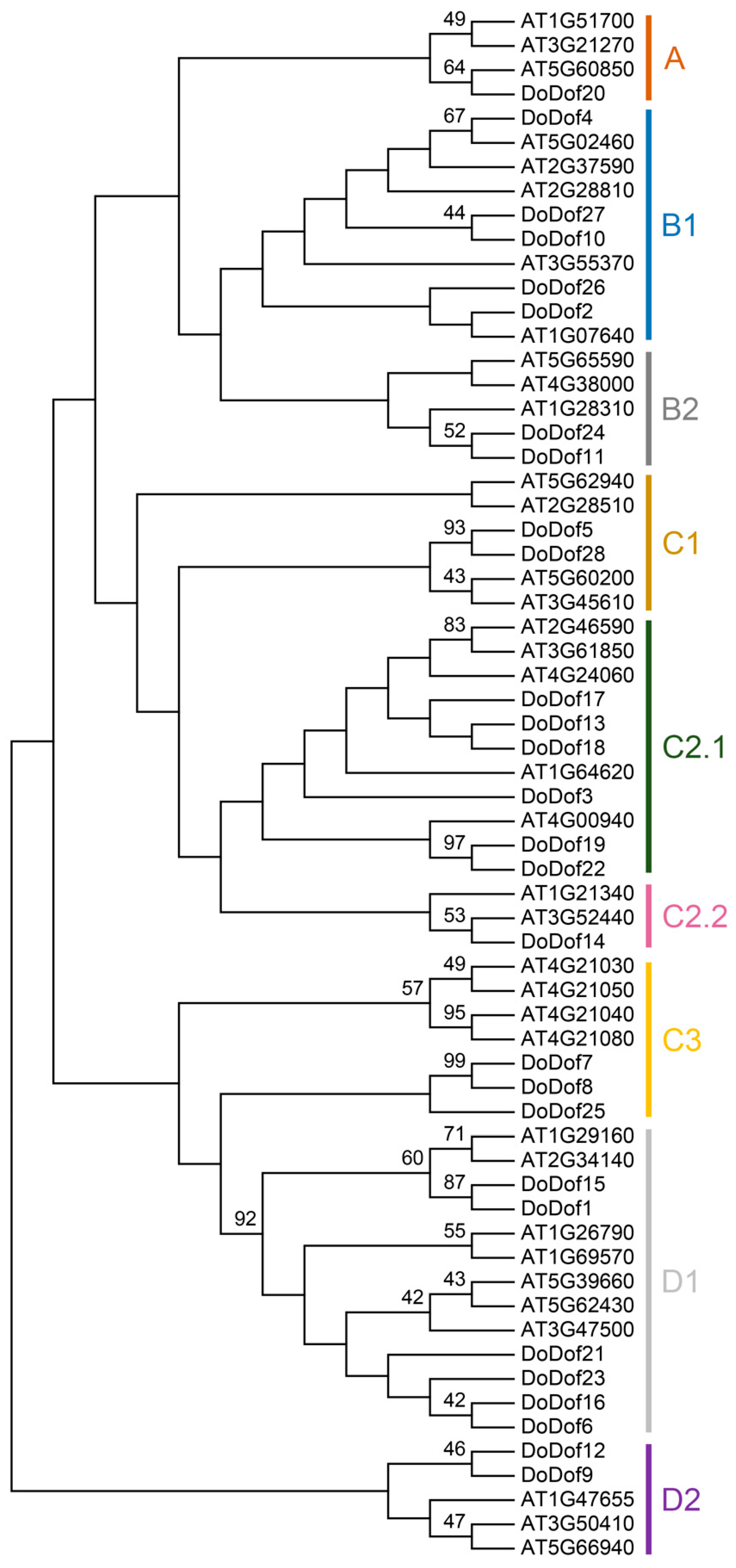
2.1. Identification and Classification of Dof Family in D. officinale
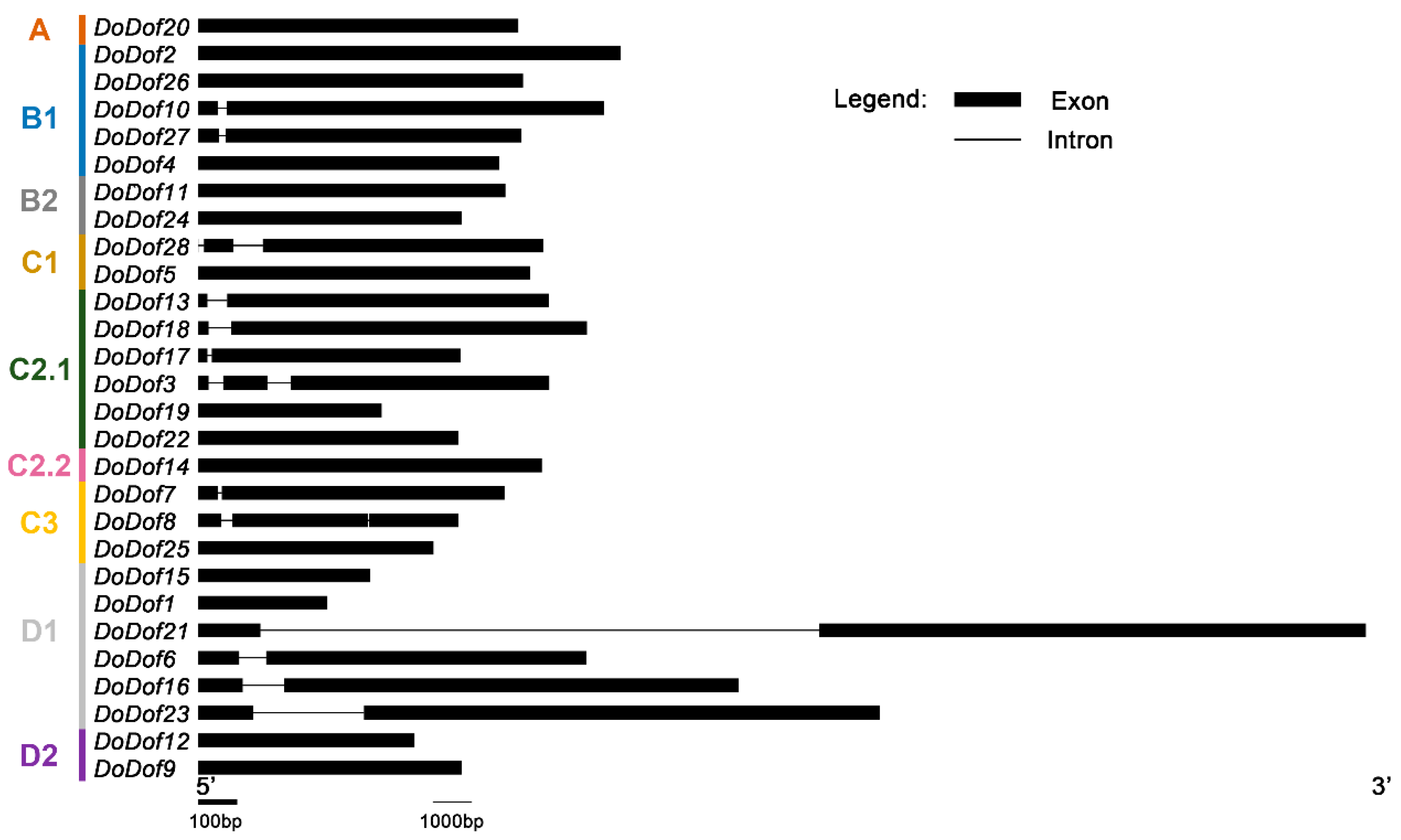
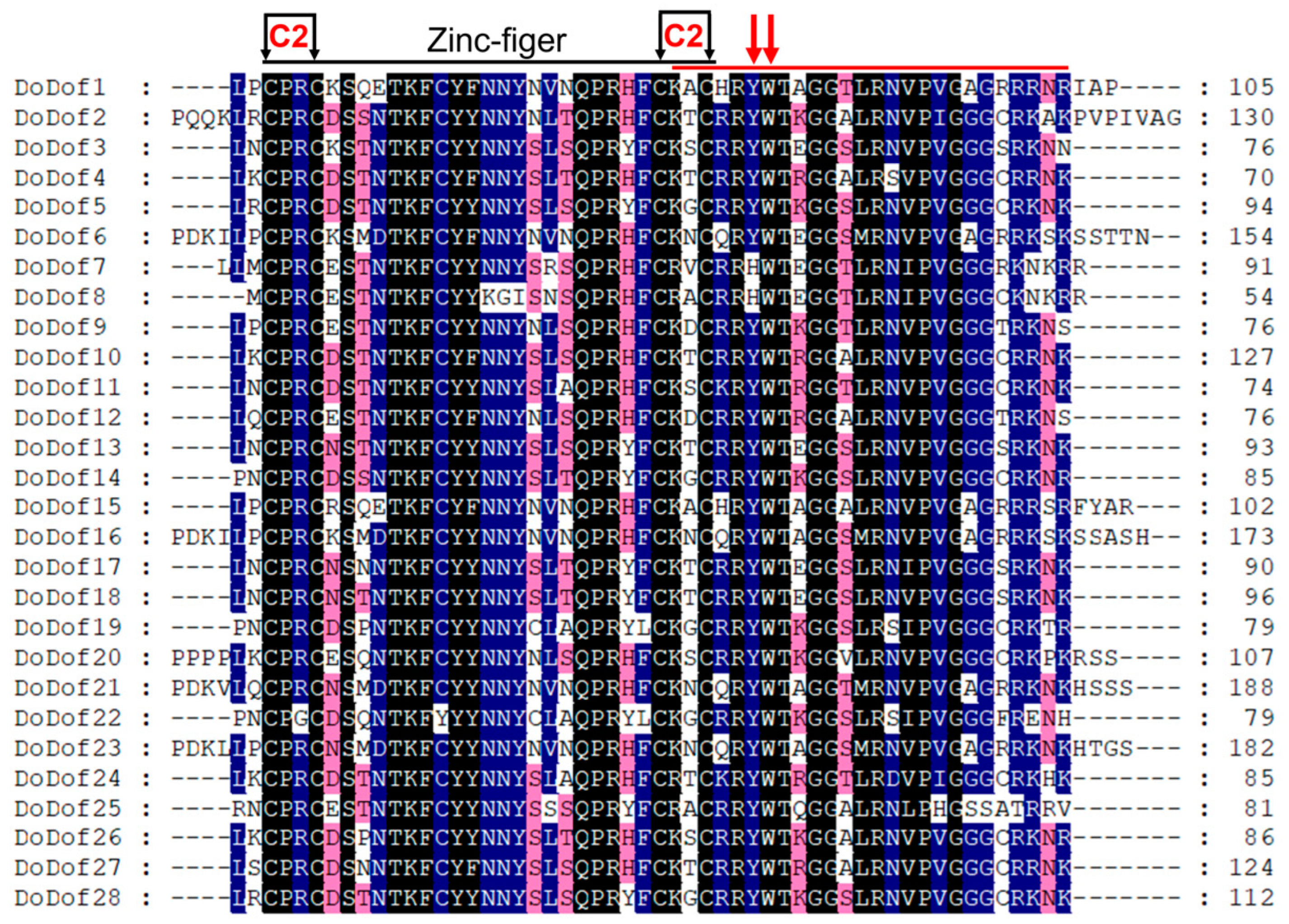
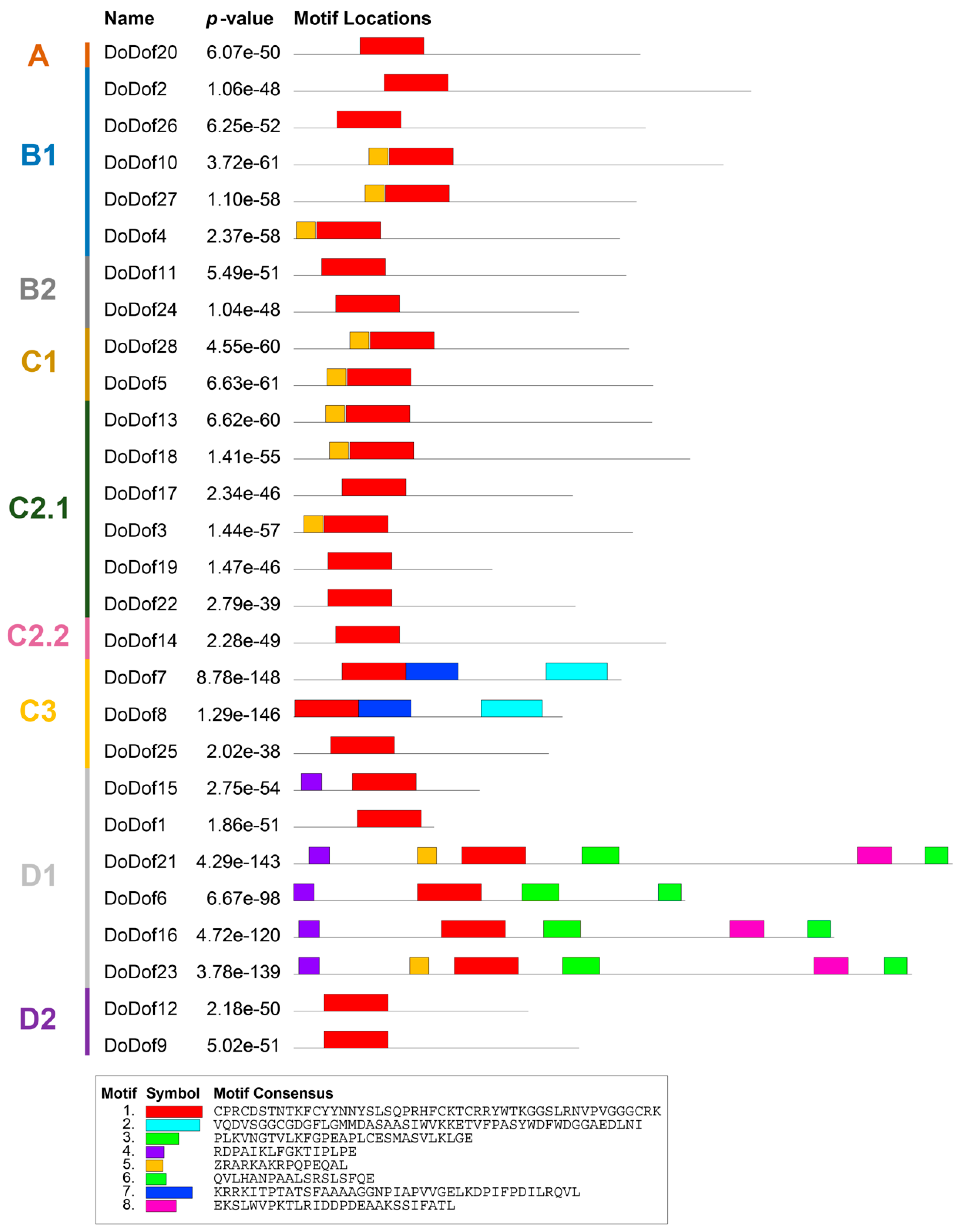
2.2. Sequence Alignment and Gene Structure Analysis of DoDof Proteins
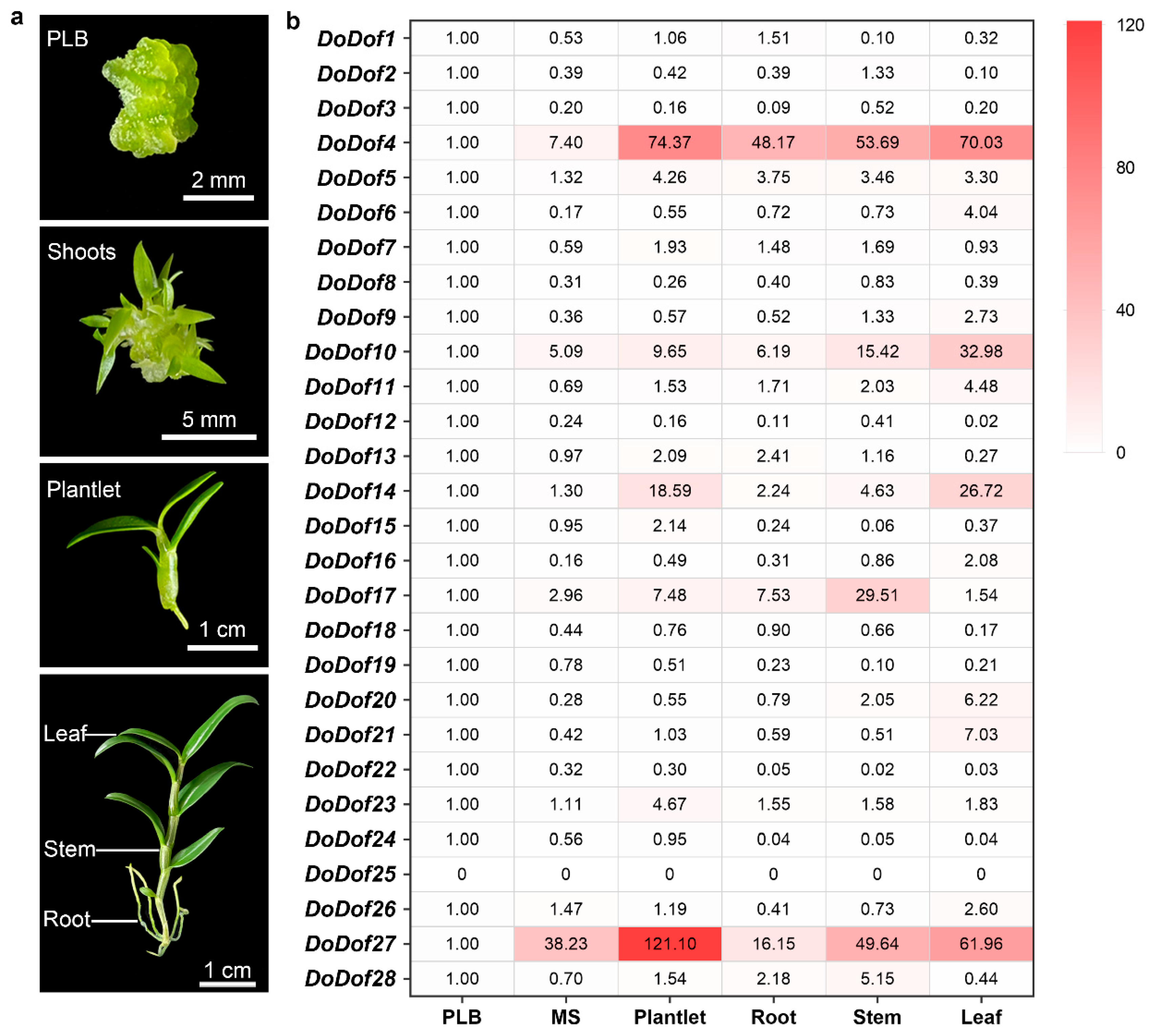
2.3. Expression Patterns of DoDof Genes in Different Developmental Stages and Tissues
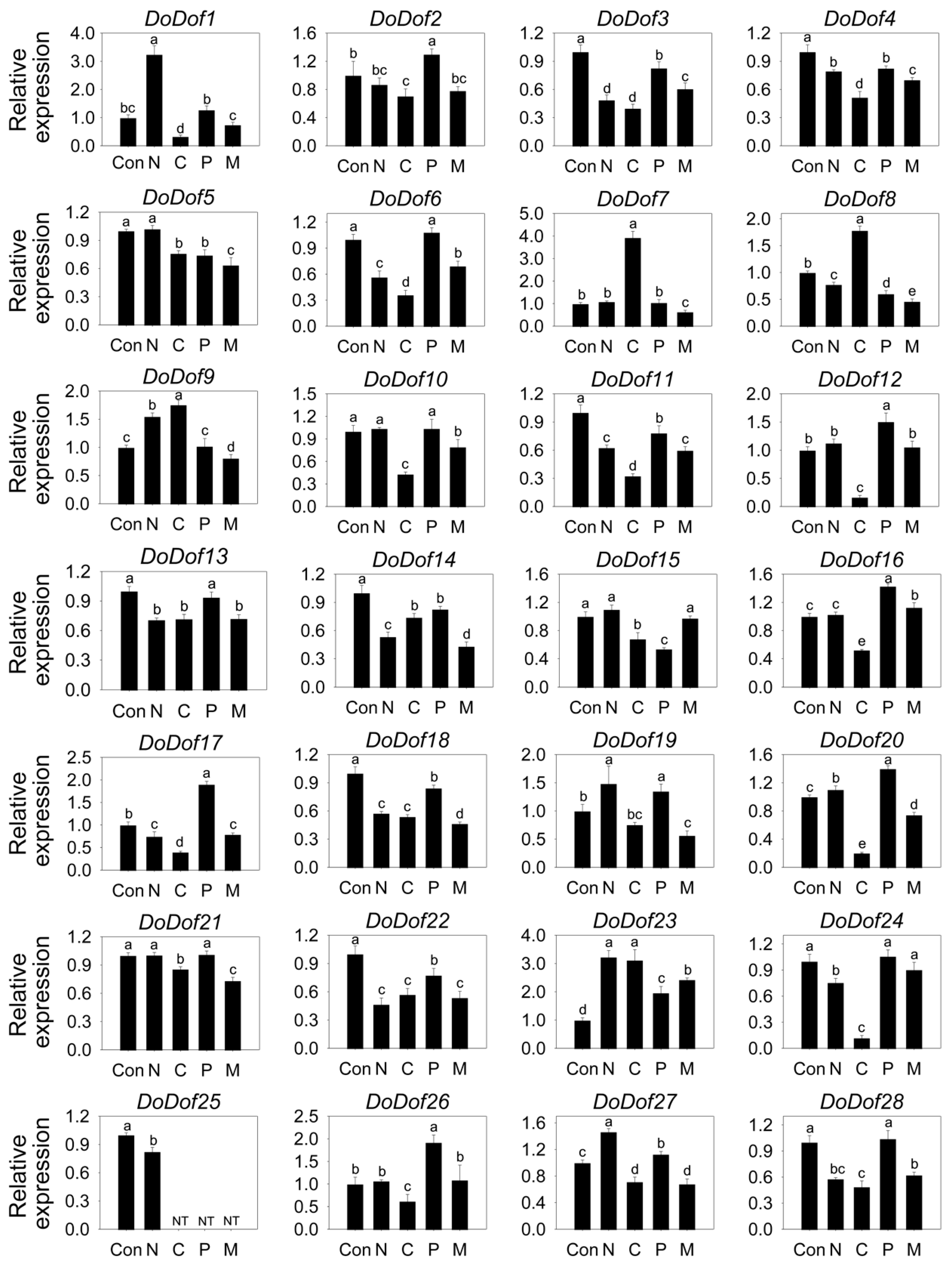
2.4. Expression Patterns of DoDof Genes Under Different Stress Treatments
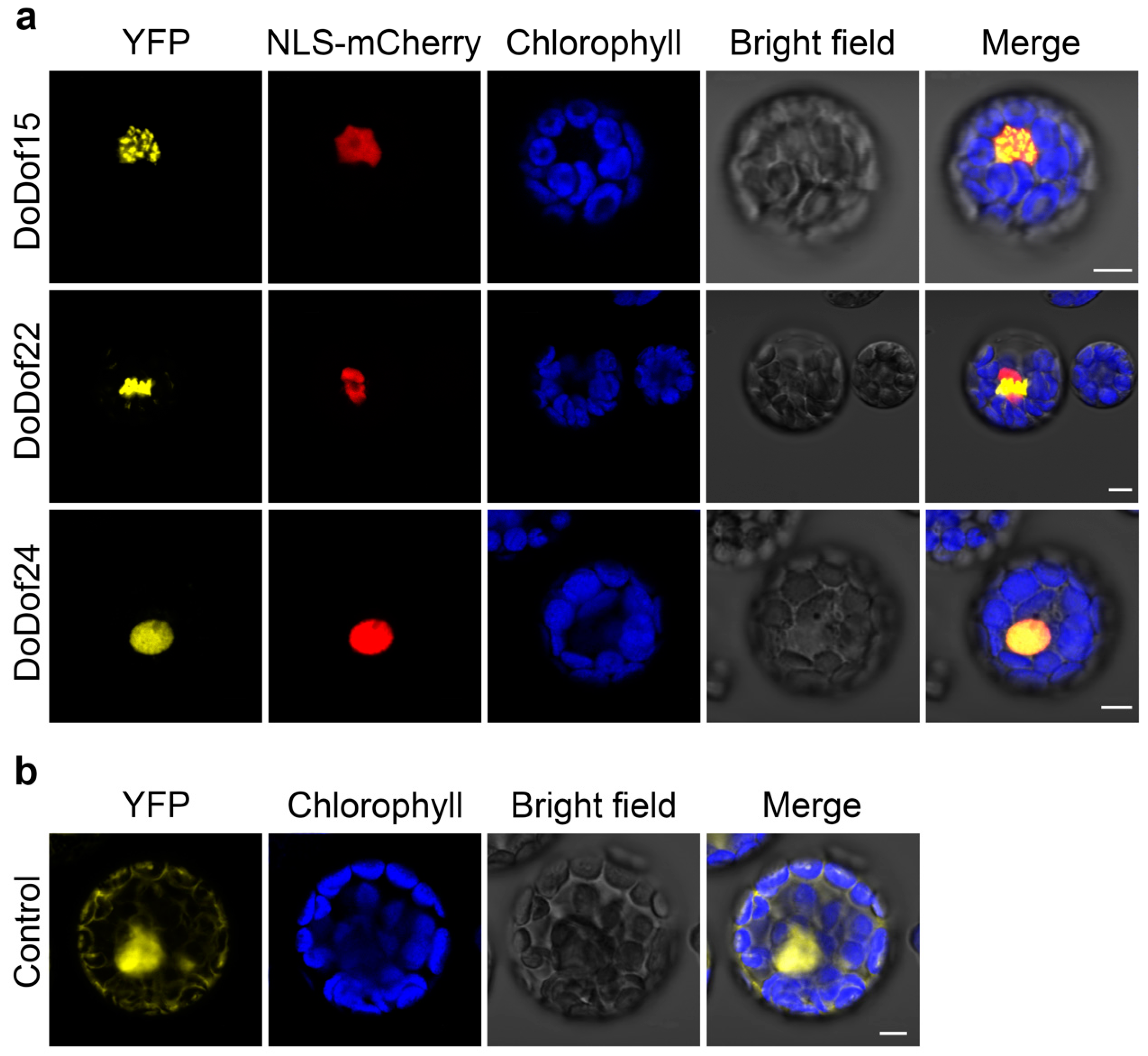
2.5. Subcellular Localization Analysis of DoDof15, DoDof22, and DoDof24
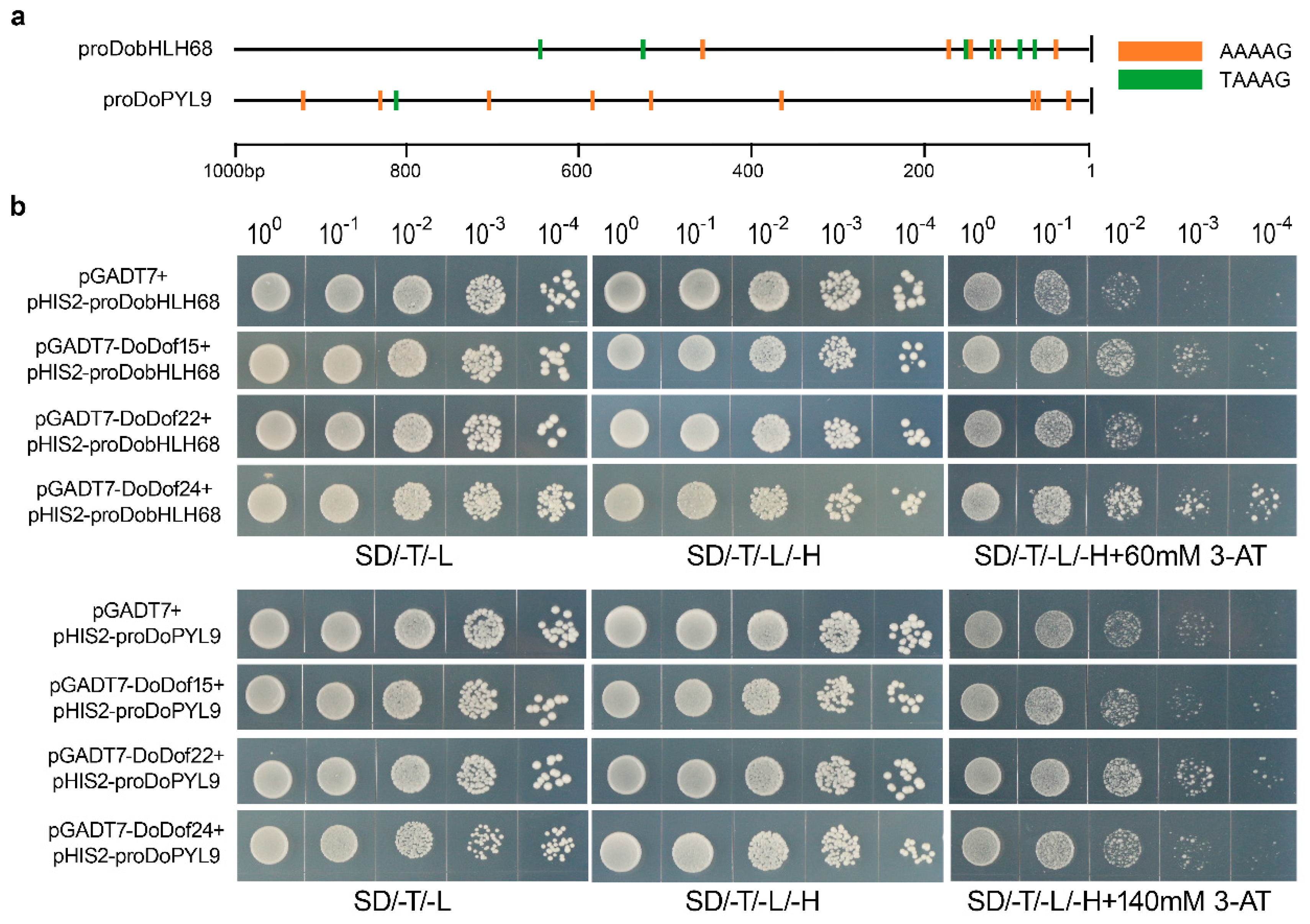
2.6. Binding Analysis of Candidate Target Genes for DoDof15, DoDof22, and DoDof24
3. Discussion
3.1. Dof Transcription Factors Are Ubiquitous in Plants and Possess a Conserved Zinc Finger Domain
3.2. Dof Transcription Factors Regulate Leaf Development in Plant
3.3. Dof Transcription Factors Respond to Abiotic Stress in Plants
3.4. Dof Transcription Factors Show Diversity in Recognizing and Binding to Core Sequences
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of DoDof Transcription Factors and Construction of Phylogenetic Tree
4.3. Bioinformatics Analysis of DoDof Proteins
4.4. RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
4.5. Subcellular Localization Analysis
4.6. Analysis of the Core Sequence 5′-(T/A)AAAG-3′ in the Promoter of D. officinale
4.7. Promoter Self-Activation Detection and Yeast One-Hybrid Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamaguchi-Shinozaki, K.; Shinozaki, K., Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803.
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N. C.; Singh, V. K.; Yadav, D., Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562.
- Shu, Y. J.; Song, L. L.; Zhang, J.; Liu, Y.; Guo, C. H., Genome-wide identification and characterization of the Dof gene family in Medicago truncatula. Genet. Mol. Res. 2015, 14, 10645–10657.
- Song, A.; Gao, T.; Li, P.; Chen, S.; Guan, Z.; Wu, D.; Xin, J.; Fan, Q.; Zhao, K.; Chen, F., Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 199.
- Gupta, S.; Pathak, R. K.; Gupta, S. M.; Gaur, V. S.; Singh, N. K.; Kumar, A., Identification and molecular characterization of Dof transcription factor gene family preferentially expressed in developing spikes of Eleusine coracana L. 3 Biotech 2018, 8, 82.
- Yanagisawa, S., The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560.
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J., Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17.
- Shigyo, M.; Tabei, N.; Yoneyama, T.; Yanagisawa, S., Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 2007, 48, 179–185.
- Zou, X.; Sun, H., DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918.
- Yanagisawa, S.; Schmidt, R. J., Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214.
- Yanagisawa, S., Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004, 45, 386–391.
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M., The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749.
- Noguero, M.; Atif, R. M.; Ochatt, S.; Thompson, R. D., The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45.
- Venkatesh, J.; Park, S. W., Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85.
- Cavalar, M.; Möller, C.; Offermann, S.; Krohn, N. M.; Grasser, K. D.; Peterhänsel, C., The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry 2003, 42, 2149–2157.
- Yanagisawa, S.; Sheen, J., Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 1998, 10, 75–89.
- Nonogaki, H.; Bassel, G. W.; Bewley, J. D., Germination—Still a mystery. Plant Sci. 2010, 179, 574–581.
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C., Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415.
- Gabriele, S.; Rizza, A.; Martone, J.; Circelli, P.; Costantino, P.; Vittorioso, P., The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010, 61, 312–323.
- Santopolo, S.; Boccaccini, A.; Lorrai, R.; Ruta, V.; Capauto, D.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P., DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1. BMC Plant Biol. 2015, 15, 72.
- Ravindran, P.; Verma, V.; Stamm, P.; Kumar, P. P., A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 2017, 10, 1307–1320.
- Jiang, J.; Ding, A. B.; Liu, F.; Zhong, X., Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 2020, 71, 5179–5190.
- Gao, H.; Song, W.; Severing, E.; Vayssières, A.; Huettel, B.; Franzen, R.; Richter, R.; Chai, J.; Coupland, G., PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation. Nat. Plants 2022, 8, 1082–1093.
- Ramachandran, V.; Tobimatsu, Y.; Masaomi, Y.; Sano, R.; Umezawa, T.; Demura, T.; Ohtani, M., Plant-specific Dof transcription factors VASCULAR-RELATED DOF1 and VASCULAR-RELATED DOF2 regulate vascular cell differentiation and lignin biosynthesis in Arabidopsis. Plant Mol. Biol. 2020, 104, 263–281.
- Xu, J.; Dai, H., Brassica napus Cycling Dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 2016, 80, 315–322.
- Liu, X.; Liu, Z.; Hao, Z.; Chen, G.; Qi, K.; Zhang, H.; Jiao, H.; Wu, X.; Zhang, S.; Wu, J.; Wang, P., Characterization of Dof family in Pyrus bretschneideri and role of PbDof9.2 in flowering time regulation. Genomics 2020, 112, 712–720.
- Zang, D.; Wang, L.; Zhang, Y.; Zhao, H.; Wang, Y., ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 2017, 94, 495–507.
- Corrales, A. R.; Nebauer, S. G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R. V.; Medina, J., Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012.
- Latchman, D. S., Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312.
- Ward, J. M.; Cufr, C. A.; Denzel, M. A.; Neff, M. M., The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 2005, 17, 475–485.
- Kurai, T.; Wakayama, M.; Abiko, T.; Yanagisawa, S.; Aoki, N.; Ohsugi, R., Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 2011, 9, 826–837.
- He, L.; Su, C.; Wang, Y.; Wei, Z., ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie 2015, 110, 17–24.
- Ahanger, M. A.; Siddique, K. H. M.; Ahmad, P., Understanding drought tolerance in plants. Physiol. Plant. 2021, 172, 286–288.
- Sanchez-Olvera, M.; Martin-Vasquez, C.; Mayordomo, C.; Illescas-Miranda, J.; Bono, M.; Coego, A.; Alonso, J.; Hern´andez-Gonz´alez, M.; Jim´enez-Arias, D.; Forment, J.; Albert, A.; Granell, A.; Borges, A. A.; Rodriguez, P. L., ABA-receptor agonist iSB09 decreases soil water consumption and increases tomato CO2 assimilation and water use efficiency under drought stress. Environ. Exp. Bot. 2024, 225, 105847.
- Fan, W.; Zhao, M.; Li, S.; Bai, X.; Li, J.; Meng, H.; Mu, Z., Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 2016, 16, 99.
- Qi, L.; Liu, S.; Li, C.; Fu, J.; Jing, Y.; Cheng, J.; Li, H.; Zhang, D.; Wang, X.; Dong, X.; Han, R.; Li, B.; Zhang, Y.; Li, Z.; Terzaghi, W.; Song, C. P.; Lin, R.; Gong, Z.; Li, J., PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol. Plant 2020, 13, 414–430.
- Wang, Y.; Zhang, G.; Zhou, H.; Yin, S.; Li, Y.; Ma, C.; Chen, P.; Sun, L.; Hao, F., GhPYL9-5D and GhPYR1-3 A positively regulate Arabidopsis and cotton responses to ABA, drought, high salinity and osmotic stress. BMC Plant Biol. 2023, 23, 310.
- Yang, J.; Wang, M.; Zhou, S.; Xu, B.; Chen, P.; Ma, F.; Mao, K., The ABA receptor gene MdPYL9 confers tolerance to drought stress in transgenic apple (Malus domestica). Environ. Exp. Bot. 2022, 194, 104695.
- Yanagisawa, S.; Izui, K., Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036.
- Guo, Y.; Qiu, L. J., Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS One 2013, 8, e76809.
- Ma, J.; Li, M. Y.; Wang, F.; Tang, J.; Xiong, A. S., Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 2015, 16, 33.
- Wen, C. L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y., Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072.
- Sun, S.; Wang, B.; Jiang, Q.; Li, Z.; Jia, S.; Wang, Y.; Guo, H., Genome-wide analysis of BpDof genes and the tolerance to drought stress in birch (Betula platyphylla). PeerJ 2021, 9, e11938.
- Lucas-Reina, E.; Romero-Campero, F. J.; Romero, J. M.; Valverde, F., An evolutionarily conserved DOF-CONSTANS module controls plant photoperiodic signaling. Plant Physiol. 2015, 168, 561–574.
- Diaz, I.; Martinez, M.; Isabel-LaMoneda, I.; Rubio-Somoza, I.; Carbonero, P., The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 2005, 42, 652–662.
- Oguchi, R.; Onoda, Y.; Terashima, I.; Tholen, D., Leaf anatomy and function. In The leaf: A platform for performing photosynthesis, Adams Iii, W. W.; Terashima, I., Eds. Springer International Publishing: Cham, 2018; pp 97–139.
- Du, Q.; Liu, T.; Jiao, X.; Song, X.; Zhang, J.; Li, J., Leaf anatomical adaptations have central roles in photosynthetic acclimation to humidity. J. Exp. Bot. 2019, 70, 4949–4961.
- Falquetto-Gomes, P.; Silva, W. J.; Siqueira, J. A.; Araújo, W. L.; Nunes-Nesi, A., From epidermal cells to functional pores: Understanding stomatal development. J. Plant Physiol. 2024, 292, 154163.
- Hetherington, A. M.; Woodward, F. I., The role of stomata in sensing and driving environmental change. Nature 2003, 424, 901–908.
- Negi, J.; Moriwaki, K.; Konishi, M.; Yokoyama, R.; Nakano, T.; Kusumi, K.; Hashimoto-Sugimoto, M.; Schroeder, J. I.; Nishitani, K.; Yanagisawa, S.; Iba, K., A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 2013, 23, 479–484.
- Konishi, M.; Yanagisawa, S., Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol. Biochem. 2007, 45, 623–629.
- Kim, H. S.; Kim, S. J.; Abbasi, N.; Bressan, R. A.; Yun, D. J.; Yoo, S. D.; Kwon, S. Y.; Choi, S. B., The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 2010, 64, 524–535.
- Konishi, M.; Yanagisawa, S., Transcriptional repression caused by Dof5.8 is involved in proper vein network formation in Arabidopsis thaliana leaves. J. Plant. Res. 2015, 128, 643–652.
- Xu, P.; Chen, H.; Cai, W., Transcription factor CDF4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep. 2020, 21, e48967.
- Zhuo, M.; Sakuraba, Y.; Yanagisawa, S., A jasmonate-activated MYC2-Dof2.1-MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell 2020, 32, 242–262.
- Liu, J.; Meng, Q.; Xiang, H.; Shi, F.; Ma, L.; Li, Y.; Liu, C.; Liu, Y.; Su, B., Genome-wide analysis of Dof transcription factors and their response to cold stress in rice (Oryza sativa L.). BMC Genomics 2021, 22, 800.
- Corrales, A. R.; Carrillo, L.; Lasierra, P.; Nebauer, S. G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R. V.; Vicente-Carbajosa, J.; Medina, J., Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764.
- Boccaccini, A.; Lorrai, R.; Ruta, V.; Frey, A.; Mercey-Boutet, S.; Marion-Poll, A.; Tarkowská, D.; Strnad, M.; Costantino, P.; Vittorioso, P., The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. Plant Cell Environ. 2016, 16, 198.
- Mittler, R.; Zandalinas, S. I.; Fichman, Y.; Van Breusegem, F., Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679.
- He, M.; Zhang, X.; Ma, Y.; Zhang, X.; Chen, S.; Zhu, Y.; Wang, Y.; Liu, L.; Ma, Y.; Wang, L.; Xu, L., RsCDF3, a member of Cycling Dof Factors, positively regulates cold tolerance via auto-regulation and repressing two RsRbohs transcription in radish (Raphanus sativus L.). Plant Sci. 2023, 337, 111880.
- Seki, H.; Nakamura, N.; Marutani, M.; Okabe, T.; Sanematsu, S.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Yamada, T.; Ichinose, Y., Molecular cloning of cDNA for a novel pea Dof protein, PsDof1, and its DNA-binding activity to the promoter of PsDof1 gene. Plant Biotechnol. 2002, 19, 251–260.
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J., ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2017, 93, 7–20.
- Diaz, I.; Vicente-Carbajosa, J.; Abraham, Z.; Martínez, M.; Isabel-La Moneda, I.; Carbonero, P., The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 2002, 29, 453–464.
- Murashige, T.; Skoog, F., A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497.
- He, C.; Zeng, S.; Teixeira da Silva, J. A.; Yu, Z.; Tan, J.; Duan, J., Molecular cloning and functional analysis of the phosphomannomutase (PMM) gene from Dendrobium officinale and evidence for the involvement of an abiotic stress response during germination. Protoplasma 2017, 254, 1693–1704.
- Yoo, S. D.; Cho, Y. H.; Sheen, J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572.
- Zhang, G. Q.; Xu, Q.; Bian, C.; Tsai, W. C.; Yeh, C. M.; Liu, K. W.; Yoshida, K.; Zhang, L. S.; Chang, S. B.; Chen, F.; Shi, Y.; Su, Y. Y.; Zhang, Y. Q.; Chen, L. J.; Yin, Y.; Lin, M.; Huang, H.; Deng, H.; Wang, Z. W.; Zhu, S. L.; Zhao, X.; Deng, C.; Niu, S. C.; Huang, J.; Wang, M.; Liu, G. H.; Yang, H. J.; Xiao, X. J.; Hsiao, Y. Y.; Wu, W. L.; Chen, Y. Y.; Mitsuda, N.; Ohme-Takagi, M.; Luo, Y. B.; Van de Peer, Y.; Liu, Z. J., The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029.
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M. R.; Appel, R. D.; Bairoch, A., Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, Walker, J. M., Ed. Humana Press: Totowa, NJ, 2005; pp 571–607.
- Larkin, M. A.; Blackshields, G.; Brown, N. P.; Chenna, R.; McGettigan, P. A.; McWilliam, H.; Valentin, F.; Wallace, I. M.; Wilm, A.; Lopez, R.; Thompson, J. D.; Gibson, T. J.; Higgins, D. G., Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948.
- Saitou, N.; Nei, M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425.
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549.
- Nicholas, K. B.; Nicholas, H. B.; Deerfield, D. W. In GeneDoc: Analysis and visualization of genetic variation, 1997.
- Hu, B.; Jin, J.; Guo, A. Y.; Zhang, H.; Luo, J.; Gao, G., GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297.
- Bailey, T. L.; Elkan, C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings International Conference on Intelligent Systems for Molecular Biology 1994, 2, 28–36.
- He, C.; Zhang, J.; Liu, X.; Zeng, S.; Wu, K.; Yu, Z.; Wang, X.; Teixeira da Silva, J. A.; Lin, Z.; Duan, J., Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 2015, 88, 219–231.
- Livak, K. J.; Schmittgen, T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt.Method. Methods 2001, 25, 402–408.
- Citovsky, V.; Lee, L. Y.; Vyas, S.; Glick, E.; Chen, M. H.; Vainstein, A.; Gafni, Y.; Gelvin, S. B.; Tzfira, T., Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 2006, 362, 1120–1131.
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H. R.; Frank, M. H.; He, Y.; Xia, R., TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




