1. Introduction
The brassinosteroids (BRs) is a natural plant hormones similar to animal sterols, it can play an active role in increasing plant development[
1], slow down the aging process [
2], promoting nutrient accumulation and substance synthesis[
3,
4], and enhancing plant stress resistance at very low concentration[
5,
6,
7]. It is considered the sixth-largest plant hormone after auxin, gibberellin, Cytokinin, abscisic acid and ethylene, and plays a crucial role in plant growth and development.
In plants, there are three BR synthesis pathways: the early C-6 oxidation pathway and the late C-6 oxidation pathway, which are dependent on canberylalcohol (CN) , and the early C-22 and C-23 hydroxylation pathways, which are CN-independent[
8]. Based on previous studies of
Arabidopsis thaliana, the CN-dependent pathway is the main way of BR biosynthesis, and the analysis of BR biosynthesis and signal transduction pathway is more in-depth in
A.thaliana[
9], and multiple genes were identified, such as
DWF4[
10],
CPD[
11],
ROT3 and
CYP90D1[
12],
BR6ox1/2[
13],
DET2[
14], and so on.
The
DET2 gene of
A. thaliana, known as
DWF6 gene, is highly consistent with steroid 5α-reductase, which catalyzes the synthesis of dihydrotestosterone in animals and catalyzes the development of the male genitalia and prostate during animal embryonic development[
15]. The
DET2 gene in plants is the rate-limiting gene in three pathways of BRs biosynthesis, and all the intermediates in the BR biosynthetic pathway after
DET2 reaction can be used rescue the
det2 mutant phenotype[
16]. Many studies have speculated that
DET2 affect cell division and growth during plant development by regulating BRs content and then regulating cell wall synthesis and extension. The expression of
CycD3 was significantly increased after treatment with 24-epi-brassinolide in
A. thaliana, which indicated that brassinolide might have some effect on cell division, and in vitro application of brassinolide to the
DET2 mutant of
A. thaliana could increase the number and size of cells in its leaves[
17]. The study of BR-related mutants (
det2-1and
bril-301) in
A. thaliana showed the expression of cellulose synthase gene and the content of cellulose decreased significantly[
18]. When the amino acid Glu240 in
DET2 protein was replaced, the activity of 5α-dehydrogenase was completely lost, resulting in the decrease of BR biosynthesis, the mutant cells obviously reduced, and the plant appeared yellowing and dwarfing[
19]. The overexpression of
DET2 gene showed the elongation of fibroblasts[
20], the improvement of endogenous castasterone (CS) levels, enhance cambium meristem cell activity and xylem (XY) differentiation to promote secondary stem growth[
21].
The fast growth of forest trees is determined by the comprehensive action of many factors of external and endogenous signals[
22,
23].
Populus yunnanensis is an endemic
Tacamahaca and a representative specie of
Populus in low-latitude and high-altitude areas ( 1600 ~ 3200 m above sea level ) of southwest China, and has the characteristics of strong adaptability, fast growth and easy survival of cuttings. It plays an important role in forestry production and environmental protection[
24]. Because of its stronger photosynthetic ability and tissue-life ability than other poplar species, it grows more rapidly. The peak of plant height growth occurs in June-october, even at an altitude of 3,000-4,000 m, the height growth of the tree can also reach 1.0-2.4 m, which has strong fast growth and high development and utilization value[
25,
26].
At present, there are few studies on steroid dehydrogenase family in plants, and studies in other plants have been limited to the cloning of related homologous genes of A. thaliana and the analysis of corresponding mutants. Different plants may have different synthetic pathways, so it is necessary to verify the universality of these pathways or reveal new synthetic pathways through the study of many types of plants in the future. As to whether the BR biosynthesis and signal transduction mechanism of woody plants are identical or specific to A. thaliana, and whether there are similarities and differences in the roles played by DET2, nothing is certain. The study on the relationship between steroid dehydrogenase family and its members in woody plants can provide molecular basis for the rapid growth of P. yunnanensis and speed up the process of forest genetic breeding. The whole genome sequencing in P. yunnanensis is now complete, providing a solid foundation for molecular biology research in P. yunnanensis. The preliminary study showed that the upright and inverted cuttings of P. yunnanensis grew faster in September, and their height growth and stem diameter were significantly higher than those in July. In this study, we firstly identified the steroid dehydrogenase family members in P. yunnanensis and analyze their structure and evolution. The growth of stem is caused by the continuous or periodic cell division of meristem at the end of stem, which makes the stem grow. The differentially expressed genes of this family were screened based on the transcriptome of the stem tip in P. yunnanensis, and the expression and subcellular localization were analyzed to provide a certain reference for the study of rapid growth factor-related genes in P. yunnanensis. We aimed to identify the DET2 family genes involved in BR synthesis regulation, and to elucidate the molecular mechanism of PyDET2e involved in the rapid growth of P. yunnanensis.
2. Materials and Methods
2.1. Plant Materials and Vector
The cuttings of three clones of P. yunnanensis were grown for one year in a greenhouse (Southwest Forestry University, Kunming, China. 102.76 E, 25.06 N). We obtained cuttings of about 15 cm and preserved the plants in the soil in February 2022. The cuttings with different tissue were used in experiments performed in July 2023. We obtained roots, stem, lateral buds, branch, young leaves (1st to 3rd leaves) and tip of stem. All samples were frozen in liquid nitrogen and stored at −80℃ until next use. The plant over-expression vector pSuper1300-GFP was preserved in our laboratory.
2.2. Date Sources
The complete genome sequences for
P. yunnanensis and the RNA-seq data were retrieved from our laboratory group (Southwest Forestry University, Kunming 6500224, China) (BioProject:PRJNA886471). The
DET2 protein sequences of
A. thaliana and
P. trichocarpa were obtained from the phytozome database (
https://phytozome-next.jgi.doe.gov/, accessed on 15 June 2023).
2.3. Genome-Wide Identification of DET2 Gene Members in P. yunnanensis
The HMM of the steroid dehydrogenase domain (PF02544) was obtained from the online server of the Pfam database (
http://pfam.wustl.edu, accessed on 15 June 2023). Then, the protein sequences of the
DET2 gene family members from
A. thaliana and
P. yunnanensis were obtained from the phytozome database (
https://phytozome-next.jgi.doe.gov/, accessed on 15 June 2023) used PF02544 to retrieve. Based on the genome and GFF files of
P. yunnanensis, the proteome files were extracted used TBtools(v2.008)[
27]. First, to classify the
PyDET2 gene family members, the known
AtDET2 protein sequences were used to query the homologous sequences in the
P. yunnanensis genome by performing BLASTp searches (E-value < 10
−14) with TBtools(v2.008). Then, we further identified and screened the conserved domains utilizing the NCBI Conserved Domain Search Tool (CD-Search)(
https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 15 June 2023). After removing incomplete and redundant sequences, the existence of
DET2 gene family of
P. yunnanensis was determined.
2.4. Amino acid Sequence and Phylogenetic Analysis
2.5. Gene Structure , Conserved Motif and Cis-Acting Element Analysis
The conserved motifs of
DET2 proteins were analyzed using the MEME website (
https://meme-suite.org/meme/tools/meme, accessed on 16 June 2023). The upstream 2.0 kb sequence of 14
DET2 genes was defined as the promoter region, and submitted it to the PlantCARE website (
http://bioinformatics.psb.ugent.be/webtools//plantcare/html/, accessed on 16 June 2023). The conserved domains of
DET2 proteins, gene structure, motifs and cis-elements in the
DET2 genes were been visualized using TBtools(v2.008).
2.6. Analysis of PyDET2s Location in Chromosomes and Collinearity
The sequence of the PyDET2 genes to individual chromosomes by analyzing P. yunnanensis genome annotation information and analyze the gene duplication events with default parameters using TBtools(v2.008) was visualized.
2.7. Codon Bias and Influence Factors Analysis
CodonW and Emboss (
http://EMBOSS.bioinformatics.nl/cgi-bin/EMBOSS/cusp, a accessed on 17 June 2023) were used to calculate the codon bias parameters of the coding proteins of the
DET2 genes family of
P. yunnanensis, then analyse the ENc-Plot, PR2-plot and Neutral mapping analysis used R Studio(V3.6.0)[
29].
2.8. Expression Pattern Analysis of PyDET2e
TBtools software was used to analyze the expression pattern by constructing heatmaps of PyDET2s expression based RNA-seq date.
Primers were designed based on CDS sequences for RT-qPCR by using NCBI primer-designing tool (
https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 18 June 2023) (
Table S1). The total RNA was isolated using the E.Z.N.A@ Plant RNA kit (Omega Bio-tek Inc., Norcross, USA) according to its manual. The K5800C (KAIAO, Beijing, China) was been used to detected the quality and purity of RNA. The 500ng RNA of each sample was used for 1st strand cDNA synthesis using Hifair® III Reverse Transcriptase (YEASEN, Shanghai, China). Then, the cDNA was diluted 7-fold for RT-qPCR. The Applied Biosystems 6800 real-time PCR machine and Hieff SYBR Green Master Mix (Yeasen Biotechnology Co.Ltd., Shanghai, China) were used to perform RT-qPCR with procedures according the manufacturer instructions.The reaction mixture (20.0 µL) was prepared with 1.0 µL cDNA template, 0.4 µM primers (F/R), 10 µl mix, and 8.2µL RNase-free water. The PCR thermal cycle conditions were as follows: denaturation at 95 ◦C for 2 min, 45 cycles of 95 ◦C for 10 s, and 56 ◦C for 30 s. Fluorescence intensities were measured for RT-qPCR at the end of each cycle. The relative transcript abundance values were calculated using the 2
−∆∆Ct method. Visualization and statistical analysis of data were performed by GraphPad (v8.0.2). Differences between means of each group were assessed by one-way analysis of variance using SPSS21, and P-values equal to 0.05 was kept statistically significant.
2.9. Subcellular Localization of PyDET2e
The pSuper1300-GFP vector with the CaMV35S promoter was used to identify the localization of PyDET2e by injecting tobacco epidermal cells. For this purpose, Firstly, we designed appropriate primers with restriction sites (Xbal 1 and Hind Ⅲ) according to the CDS sequence of PyDET2e and the multiple cloning site sequence in vector, and cloned this fragment into the pMD-19T vector. Then, the positive bacteria was sent for sequencing (Shanghai Biotechnology Co., Ltd., Shanghai, China) after transformation and spread plate. After the sequencing, the gene fragment was digested with the aforesaid restriction enzymes and ligated into the Xbal 1-Hind Ⅲ digested pSuper1300-GFP vector using T4 ligase (Takara Bio, Shanghai, China). The over-expression vector PyDET2e-pSuper1300-GFP was successfully transformed into tobacco epidermal cells, while pSuper1300-GFP alone was also transformed to use as control. The expression of GFP can be observed with fluorescence confocal microscopy three days after injecting tobacco leaves.
3. Results
3.1. Identification of the DET2 Genes in P. yunnanensis
There are 14
DET2 genes of
P. yunnanensis were identified and named
PyDET2a–
PyDET2n according to their position on chromosomes of
P. yunnanensis (
Table S2). The lengths of amino acids ranged from 253 aa (
PyDET2b) to 351 aa (
PyDET2f). The molecular weight ranged from 20.71 kDa (
PyDET2k) to 40.35 kDa (
PyDET2f), with an average of 31.44 kDa, and the isoelectric point ranged from 8.79 (
PyDET2a) to 9.66 (
PyDET2e), with an average of 9.32. A grand average of hydropathicity (GRAVY) index analysis showed that all proteins were hydrophobic. Most are unstable proteins, and the instability index ranged from 22.63 (
PyDET2l) to 46.84(
PyDET2a). The secondary structure of all proteins was mainly α–helix. The prediction of subcellular localization showed that most of the proteins were mainly located in the cell membrane and chloroplast.
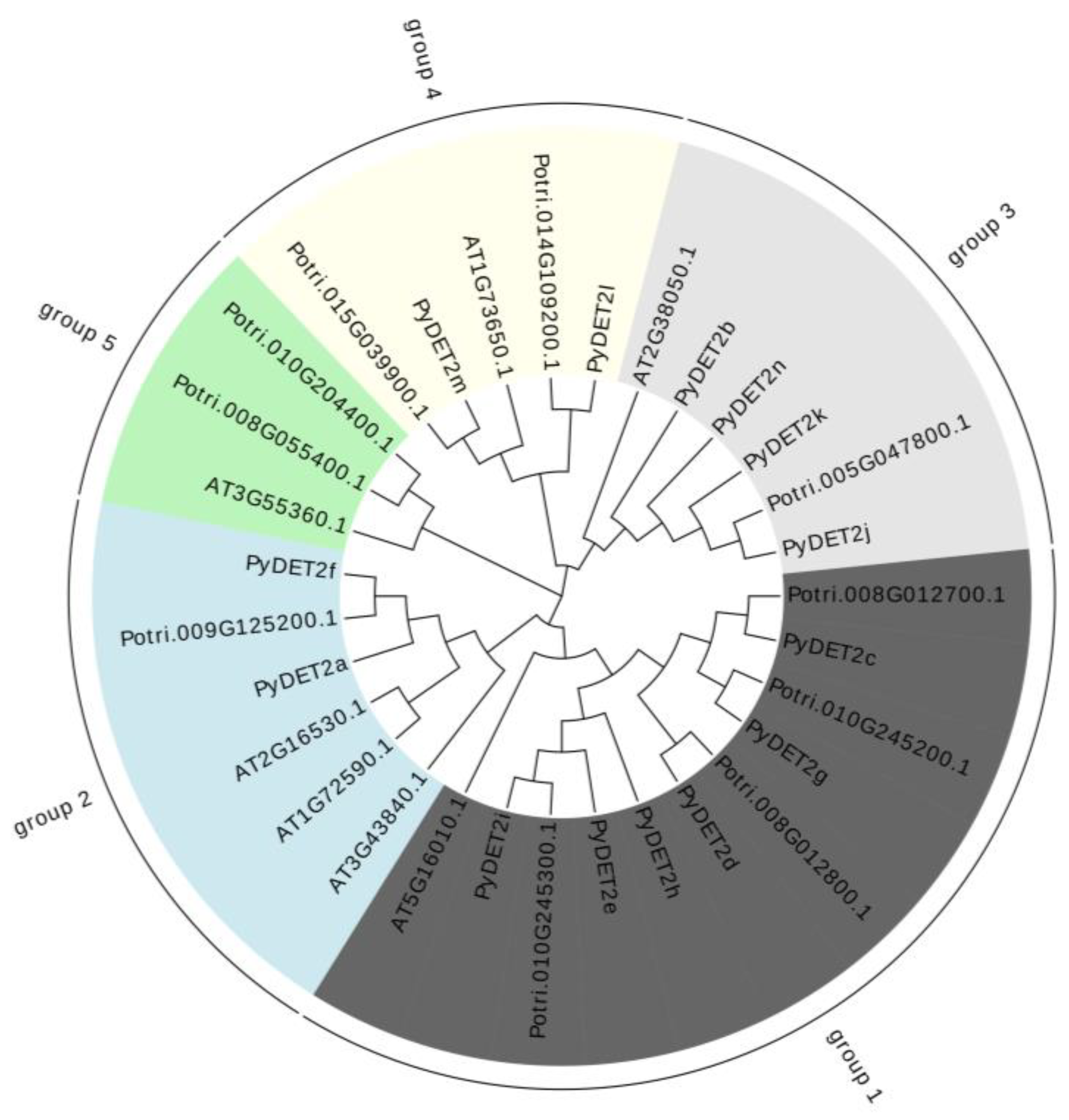
3.2. Comparative Phylogenetic Analysis and Motif Elicitation of PyDET2s
A phylogenetic tree using the protein sequences of members of three
DET2 families was built to understand the evolutionary relationship of the
DET2 genes between different species.These genes including 7
AtDET2s, 14
PyDET2s and 10
PtDET2s were not evenly clustered into 5 subgroups (
Figure 1). We analyzed the exons and introns of the
PyDET2s to determine the structural characteristics of genes, and the gene structure of the same group was similar(
Figure 2).
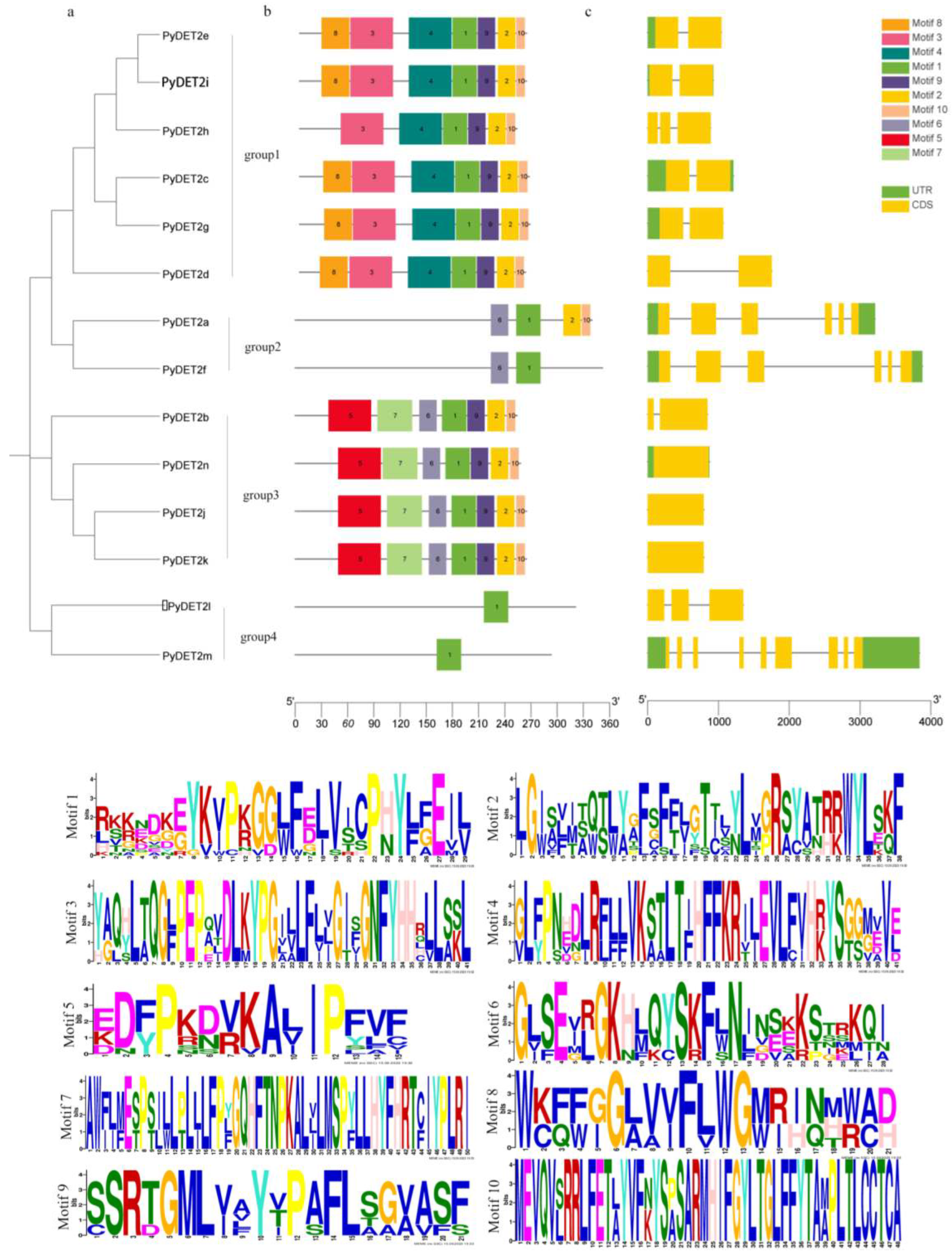
The conserved protein motifs of
DET2 proteins were showed in
Figure 2. Approximately all sequences were found to exhibit one types of highly conserved protein motifs with a similar width of 29 amino acids, which are demonstrated as dark green blocks (motif 1), which is presumed to be conservative in this family, and this motif was present at the N-terminus of all members. The composition of motif is similar in the same subgroup. The first subgroup contained 1,2,3,4,9,10 motif. The second subgroup contains 9,10 motif. The third subgroup contained 1,2,5,6,7,9,10 motif. The fourth subgroup only contains motif 1.
Figure 1.
Phylogenetic tree of 31 DET2 proteins from P. yunnanensis, P. trichocarpa and A. thaliana.
Figure 1.
Phylogenetic tree of 31 DET2 proteins from P. yunnanensis, P. trichocarpa and A. thaliana.
Note: the black block represent the group 1, the blue block represent the group 2, the gray blue block represent the group 3, the yellow block represent the group 4, the green block represent the group 5.
Figure 2.
Analysis of the conserved motifs and gene structure depending on the phylogenetic relationships of DET2 genes in P. yunnanensis.
Figure 2.
Analysis of the conserved motifs and gene structure depending on the phylogenetic relationships of DET2 genes in P. yunnanensis.
Note: (a) The phylogenetic tree using 14 DET2 proteins with the ML method was build. Four subgroups (group 1, group2, group 3 and group 4 ) were shown in different colors; (b)Analysis of conserved motifs of DET2 genes in P. yunnanensis. Gray lines represent sequences of different lengths, and blocks of different colors represent different conserved motifs; (c)Exon/intron structure analysis of DET2 genes P. yunnanensis. Black lines represent introns, yellow boxes represent exons, and green boxes represent untranslated regions (UTR).
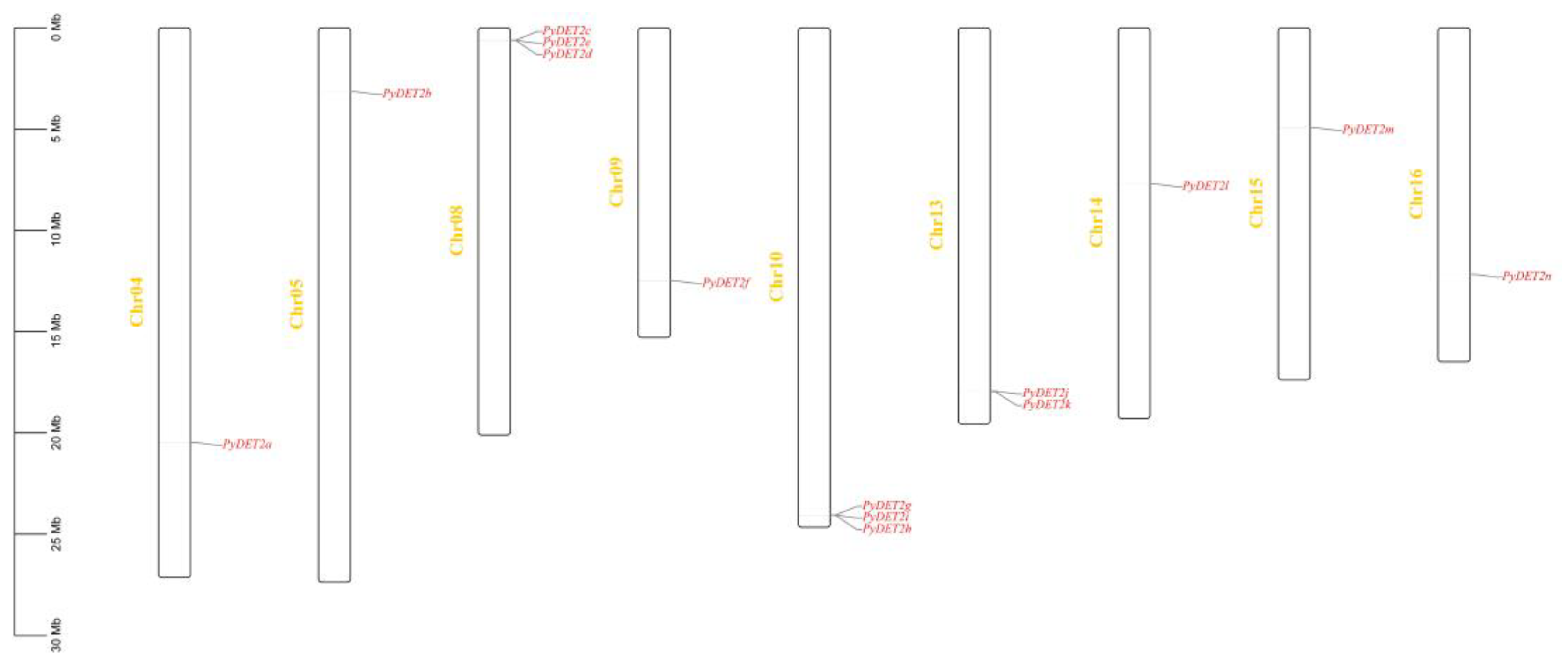
3.3. Localization and Duplication of PyDET2s
The result of chromosome localization is shown in
Figure 3. 14 members were distributed in chromosome 4,5,8,9,10,13,14,15 and chromosome 16, and there are serial replications in the chromosome 8(
PyDET2c,
PyDET2e,
PyDET2d), chromosome 10(
PyDET2g,
PyDET2i,
PyDET2h), and chromosome 13(
PyDET2j, PyDET2k).
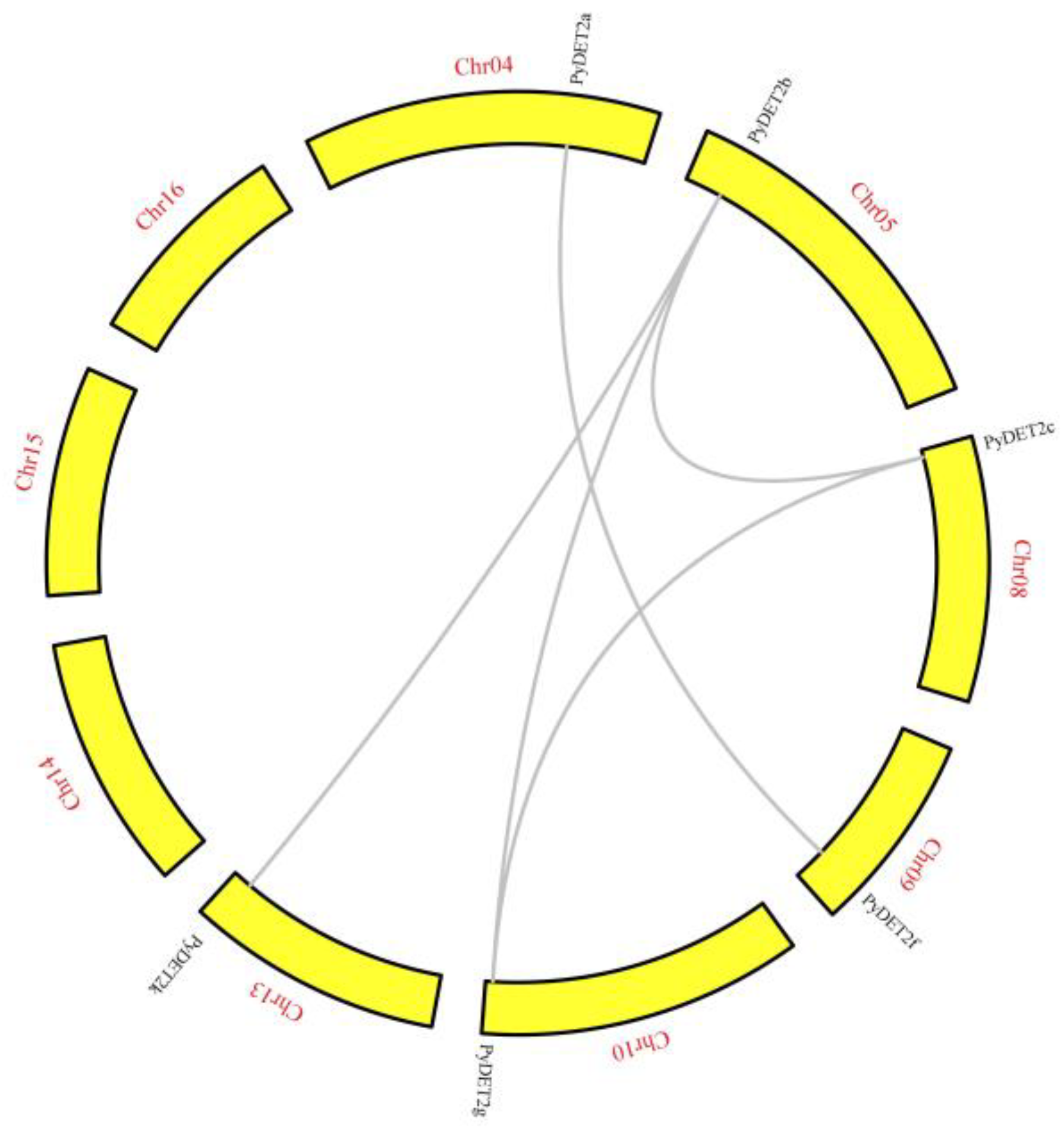
We analyzed the collinear blocks and gene duplication type. Five duplication
PyDET2 gene pairs were observed (
PyDET2a-
PyDET2f,
PyDET2b-
PyDET2c,
PyDET2b-
PyDET2g,
PyDET2b-
PyDET2j,
PyDET2c-
PyDET2g), and located on different chromosomes (chromosomes 4, 5, 8, 9, 10, 13) in
P. yunnanensis(
Figure 4). The duplication events including whole-genome duplications and segmental during the evolutionary process of the
PyDET2s were found. We also found that there were 6 pairs of collinearity genes between
P. yunnanensis and
A. Thaliana, 14 pairs of collinearity genes between
P. yunnanensis and
P. trichocarpa, and no collinearity was found between
PyDET2d,
h,
i and
k (
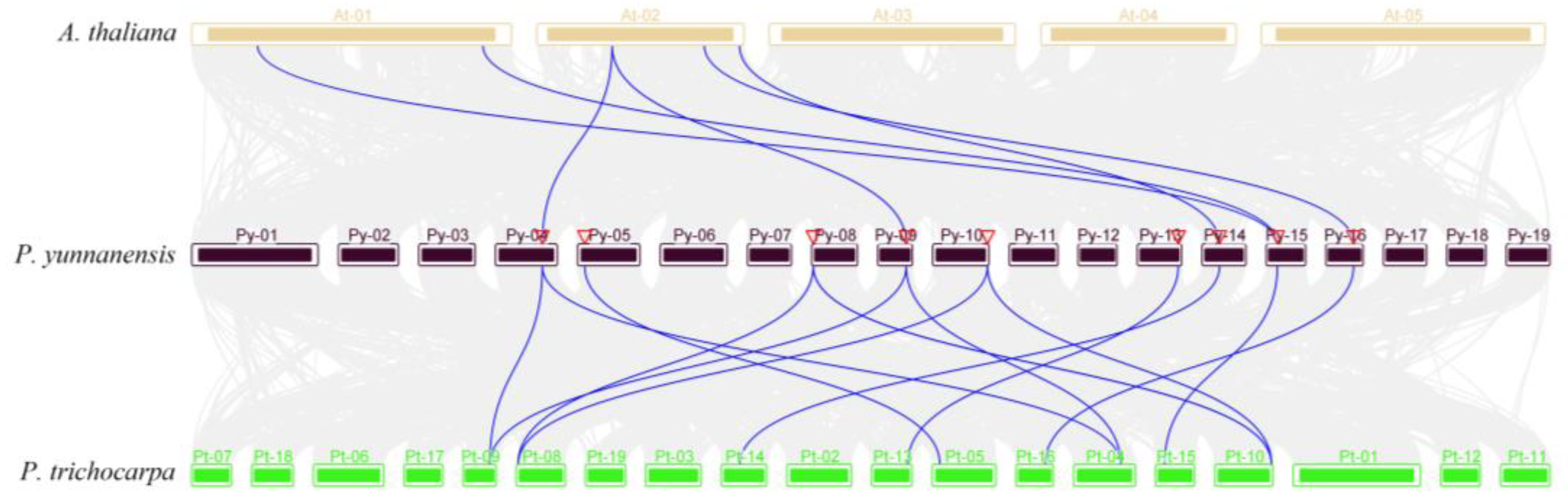
Figure 5).
3.4. Prediction of Cis-Regulatory Elements in the Promoters of PyDET2s
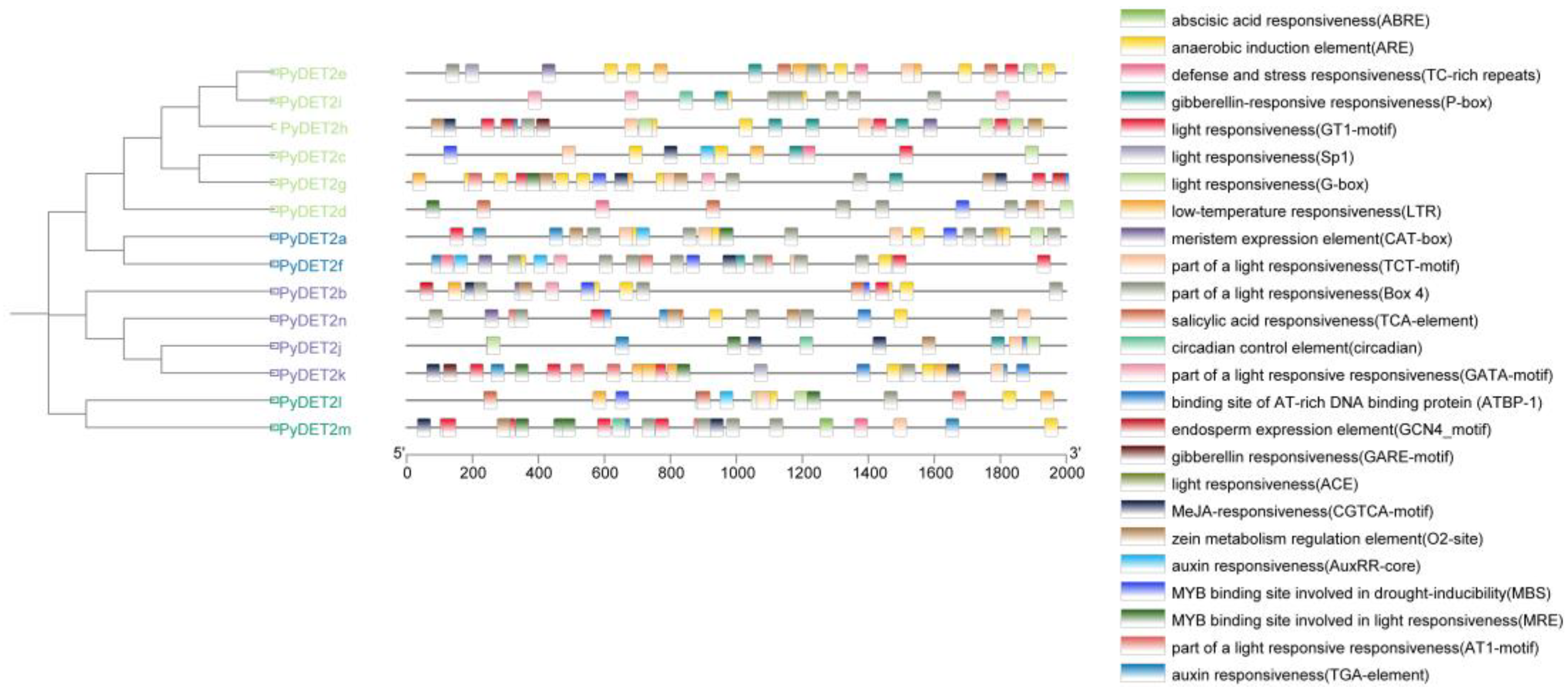
We studied the potential regulation of cis-acting elements in
PyDET2s(
Figure 6). Twenty-five cis-regulatory element were identified, and light responsiveness is the most element. We also found many hormone response and stress elements are abundant in the promoter regions of family members, such as abscisic acid responsiveness (ABRE), anaerobic induction element (ARE), MeJA (CGTCA motif), auxin responsiveness (AuxRR-core, TGA-element), gibberellin responsiveness (P-box), salicylic acid responsiveness(TCA-element), zein metabolism regulation element(O2-site), defense and stress responsiveness(TC-rich repeats), low-tempterature responsiveness(LTR), Therefore, members of this family play an important role in plant hormone signal response and stress. In addition, many elements that regulates plant growth and development were identified, meristem expression element(CAT-box) and endosperm expression element(GCN4-motif). 12 genes contained MYB binding sites, and the secondary binding of MYB transcription factors could regulate the light response and drought stress.
3.5. Analysis of codon preference and its influencing factors
The codon usage bias in the
DET2 family of
P. yunnanensis was investigated by analyzing the related parameters(
Table S3). The codon adaptation index (CAI) of
DET2 family genes ranged from 0.168 to 0.211, with an average of 0.186. The codon preference index (CBI) of
DET2 family genes ranged from -0.142 to 0.045, with an average of -0.05, and the optimal codon usage frequency FOP values ranged from 0.323 to 0.420, with an average of 0.372, and less than 0.5. The content of T3s ranged from 0.3364 to 0.4514, with an average of 0.3967. The content of A3s ranged from 0.1761 to 0.3574, with an average of 0.2851. The content of C3s ranged from 0.1701 to 0.3458, with an average of 0.2674. The content of G3s ranged from 0.2584 to 0.3638, with an average of 0.2803. The frequency of GC1(except
PyDET2m), GC2, and GC3(except
PyDET2n) and the average GC (except
PyDET2n)content were below 50%.These results indicated a preference for A/U-ending codons. Additionally, GC1had a higher frequency than GC2 and GC3, with GC2 showing the lowest frequency(except
PyDET2a).
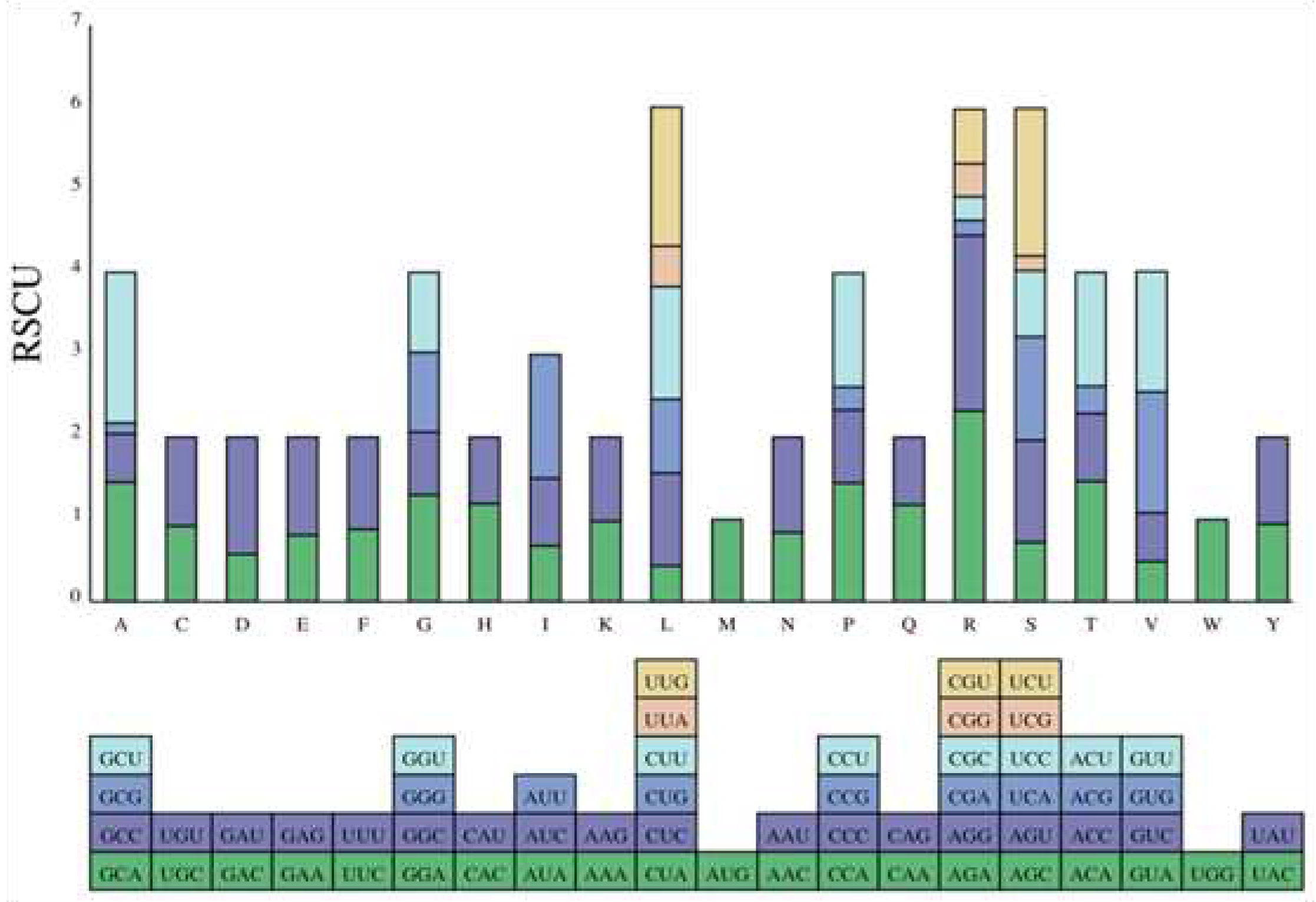
A total of 3933 codons (including stop codons) were found in the 14
DET2 genes of
P. Yunnanensis. The RSCU value analysis of the codons showed that there were 2331 codons with RSCU > 1, and the RSCU of AGA was 2.32 with the highest frequency among the 59 codons (excluding stop codon and start codon). There are 29 high-frequency codons showed obvious A/U preference ending(
Figure 7).
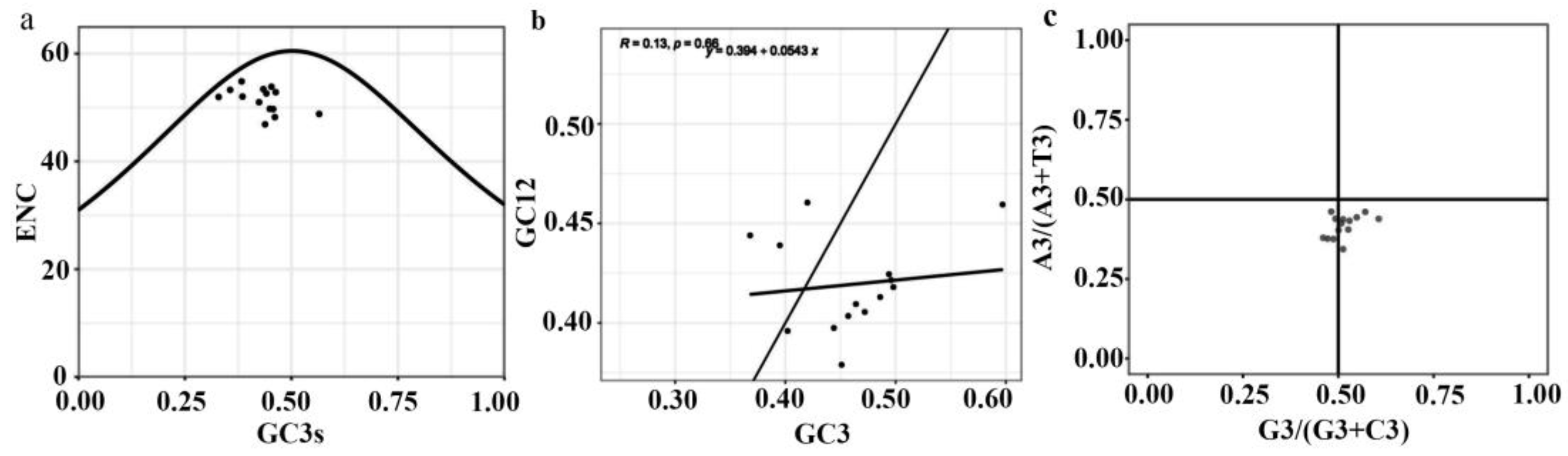
To analyze the codon usage variation in
DET2 family, the ENc-plot, Neutral-plot and PR2-plot analysis was performed as shown in
Figure 8. The
DET2 family members having GC3s distributions ranging from 0.368 to 0.597, with the exception of
PyDET2n having the GC3s content of 0.565(greater than 0.5), and all the points are distributed below the desired curve, which indicated that natural selection played a major role in the formation of codon usage bias. Meanwhile, the distribution of CDSs was not evenly around the center point (A=U/T, G=C), and the formation of codon usage patterns are not only affected by mutation pressure, but also by natural selection. In order to further investigate the degree and extent of mutation pressure and natural selection. The correlation coefficient and regression coefficient were 0.13 and 0.394, respectively, which indicated that the correlation between the base composition at the 1st and 2nd position of codon and that at the 3rd position of codon was weak, and the coding gene of
DET2 gene family was highly conserved, codon preference is strongly influenced by selection pressure. These findings align with the results from the ENc-plot and PR2-plot analyses.
3.6. Gene Expression Analysis
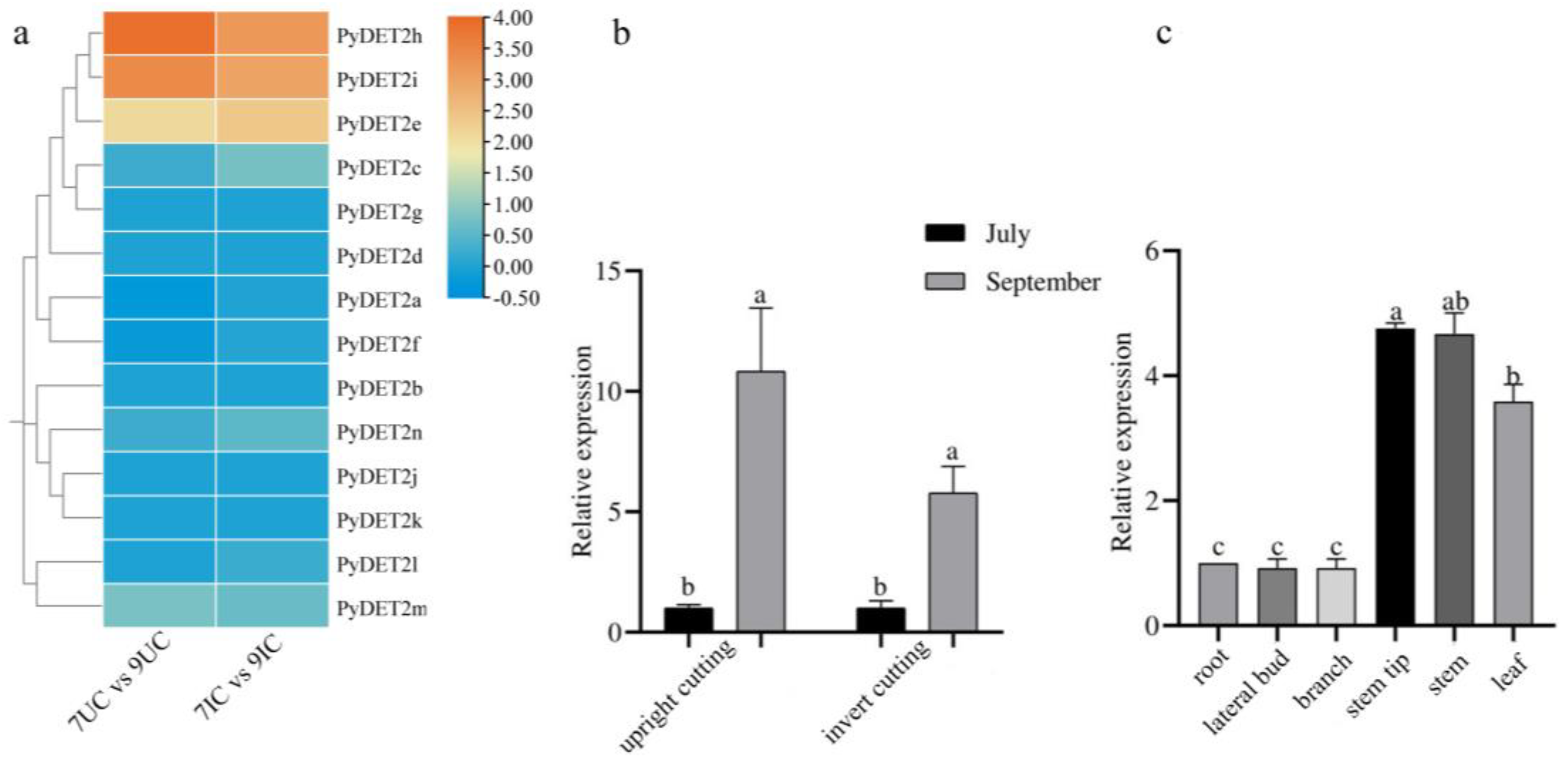
Based on the RNA-seq transcriptome data, the RPKMs of the stem tip in two growth phases (initial and rapid growth stages) and two kinds of cuttings (upright and inverted cuttings) were used to show the expression level of the
DET2 family members in
P. yunnanensis. The result shows that three genes (
PyDET2h,
PyDET2i and
PyDET2e) show higher expression in September than in July both in inserted and upright cuttings (
Figure 9). Therefore, we speculate that these three genes play an important role in high growth of
P. yunnanensis. We selected the
PyDET2e for RT-qPCR. The results showed that the transcriptome data were reliable. Subsequently, we analyzed its tissue specificity and found that its expression level in stem, stem tip and leaf was significantly higher than that in other tissues (
Figure 9).
3.7. Vector construction and subcellular localization of PyDET2e
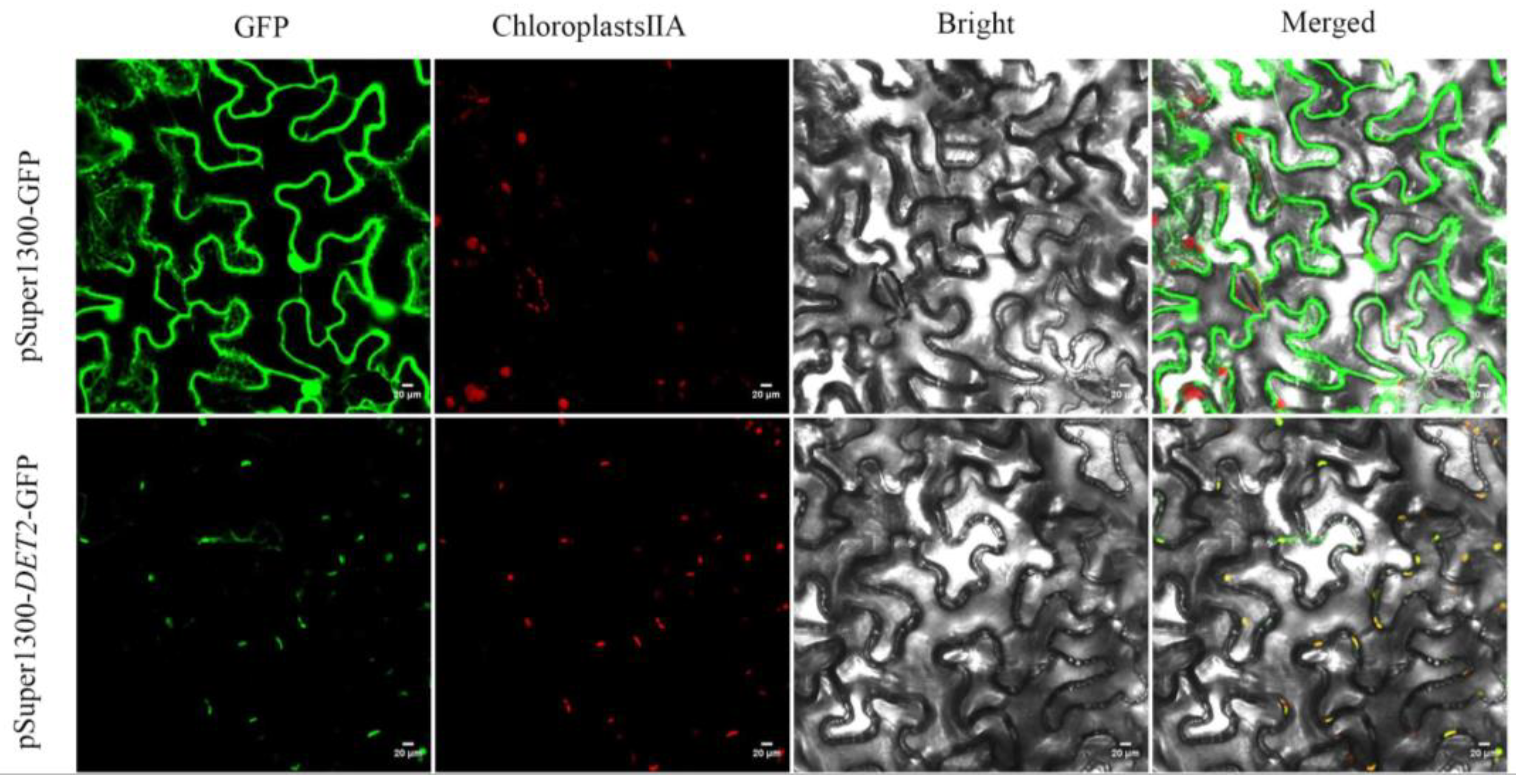
We used the online PSORT website to predict the subcellular localization of
PyDET2e, and the result showed that it was located in chloroplast. In order to determine the location of the
PyDET2e protein in cells, a
GFP vector for
PyDET2e protein subcellular localization analysis was constructed and the empty
GFP fusion protein was used as the control. The position of the fusion protein was observed by fluorescence confocal microscopy three days after injecting tobacco leaves. The results showed that
PyDET2e-GFP was distributed in the chloroplasts and that the
PyDET2e protein was located in the chloroplasts. However, However, the pSuper 1300-
GFP alone was dispersed throughout the cell(
Figure 10).
4. Discussion
BRs has a wide range of physiological functions, such as plant height, plant type, flowering time, plant root growth, stem elongation, leaf extension, microtubule system development, plant morphology in the dark conditions, pollen tube elongation, seed development[
30,
31,
32]. Cell division and cell expansion are the main factors controlling the growth of plant organs, and the expansion of cells determines the growth of an organ. The rice yield had been increased by regulate BRs to change plant height, leaf angle and grain size[
33]. The main signal transduction pathways of BRs have been established and the key gene functions of these pathways have been verified in
A. thaliana, and the
DET2 is a key rate-limiting gene in brassinolide biosynthesis pathway, and its effect is self-evident[
34,
35,
36].
We identified 14
DET2 family genes from the genome of
P. yunnanensis. The
PyDET2s protein has the similar sequence length. The isoelectric point of most
PyDET2s protein are greater than 7, suggesting that
PyDET2s may encode an alkaline protein that plays a biological function in an alkaline subcellular environment[
37]. The 14 genes of
DET2 family were classified into four groups (group 1, group 2, group 3 and group 4). The 14 promoter regions of the
PyDET2 gene also contain various cis-regulatory element, which suggests that proteins encoded by members in
DET2 family are involved in many processes of growth and development.(
Figure 6.). Codon analysis showed that the genes in this family had a significant A/U preference, and natural selection was the main factor affecting their formation(
Figure 8.).
Gene duplication is one of the major forces that act on gene expansion and ultimately drive biological evolution. In total,
14 DET2 genes have evolved repeatedly from WGD and fragments. The two types of gene duplication also contributed to the amplification of
DET2s. The collinearity analysis showed that some genes had no collinearity between
DET2 family and
A. thaliana and
P. trichocarpa, these may be
DET2 members with new function in
P. yunnanensis (
Figure 5.).
The stem tip secretes and accumulates various hormones, and through the growth, the division, the differentiation causes the stem to elongate unceasingly, and forms the stem related structure. In our previous research, we found that height growth and stem diameter of
P. yunnanensis cuttings in September were significantly higher than those in July, whether it is upright or inverted cuttings. Based on the analysis transcriptome of stem-tip and RT-qPCR in
DET2 family, we found that the expression level of
PyDET2h,
PyDET2i and
PyDET2e was significantly higher than that in July. These three genes had similar motif and gene structure, which suggested that they had similar effect on fast-growth of
P. yunnanensis. These genes are located in group1, and their codon composition is quite similar, so it is inferred that they play similar biological functions(
Figure 9.).
The
PyDET2e was selected to clone and construct overexpression vector containing
GFP for subcellular localization. The CDS length of
PyDET2e is 795 bp, encoding 264 amino acids with a molecular weight of 30.02 kDa. It is an unstable hydrophobic protein with no signal peptide and three transmembrane regions composed of α-helices, and presumed that the protein is a non-secretory membrane protein with a hydrophobic index of 0.34, which accords with the characteristics of membrane localization protein. Protein phosphorylation is at the end of the signaling chain and can be achieved by phosphorylation of transcription factors for the purpose of regulating gene, and site predictions indicate that there are 17 sites at which serine may be phosphorylated, it is speculated that the changes in the conformation of
PyDET2e protein may be regulated by the SER phosphorylation site; Whether the predicted phosphorylation sites really exist and what roles they play can be further analyzed by protein phosphorylation modification omics and Western blot methods(
Figure S1). The subcellular localization results showed that
PyDET2e was mainly expressed in chloroplast and a little in cytoplasm. As to which part of the chloroplast it is expressed, it needs further analysis(
Figure 10.). Chloroplasts are the main sites of photosynthesis in plants, and are involved in nitrogen and sulfur assimilation, amino acid, fatty acid, nucleotide, and hormone synthesis. Light directly regulates the expression of hundreds of chloroplast-related genes. Plant hormones such as brassinolide, Cytokinin, auxin and gibberellin are regulated by light and control chloroplast development, especially in the early stages of plant development[
38]. In previous studies, the
det2 (deetiolated2) presented different phenotypes under light and dark conditions, and were identified as being involved in right-regulated development, perhaps related to their localization in chloroplasts. Other results also had provided a mechanism by which BRs modulate photomorphogenesis and chloroplast development[
39]. In plant green tissues, a complex network of transcription factors, light and hormone signals is formed, and
PyDET2e is expressed in which part of the chloroplast, the role of this gene in chloroplast development and BRs synthesis remains to be further studied and demonstrated. The tissue specificity indicated that
PyDET2e was highly expressed not only in stem tips and stems, but also in leaves, suggesting that it also played a role in leaf development, and further research is needed.
5. Conclusions
14 DET2 family genes were identified in P. yunnanensis, and the evolutionary relationships was analyzed with a phylogenetic tree. The gene structure, phylogenetic relationship, cis-acting elements, and collinearity of PyDET2s were analyzed, which increases the understanding of the DET2 gene family in P. yunnanensis. The analysis of cis-acting elements suggested that DET2 genes might be involved in plant growth, development, glucose metabolism, and hormone signal transduction. Tissue-specific analysis of PyDET2e combined with transcriptome analysis revealed that PyDET2e in September was significantly higher than that in July (both in upright and invert cutting), and the expression level of it in stem, tip of stem and leaf was significantly higher than that in other parts, subcellular localization showed that the protein was mainly located in chloroplast.
Supplementary Materials
Figure S1: title; Table S1: Primer sequence of RT-qPCR; Table S2: Identification of DET2 family genes in P. yunnanensis; Table S3: Base composition of codons in the DET2 gene family of P. yunnanensis; Figure S1: Prediction of phosphorylation sites and transmembrane regions of PyDET2e protein.
Author Contributions
Conceptualization, Z.Q.; Data curation, D.Z.,C.H.; Formal analysis, J.L.; Funding acquisition, D.Z.,C.H.; Investigation, X.Z.; Methodology, Z.Q., J.L. and H.G.; Project administration, D.Z.,C.H.; Software, J.L.,X.Z.; Supervision, D.Z.,C.H.; Validation, Z.Q.; Visualization, Z.Q.; Writing—original draft, Z.Q. ; Writing—review and editing, Z.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Postgraduate Fund Project of Yunnan Education Department (2023Y0784) and Youth Talents Special Project of Yunnan Province "Xingdian Talents Support Program"(202101AU070144).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genomic data and RNA-seq date of P. yunnanensis can be obtained by contacting the corresponding author. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sathiyamoorthy, P.; Nakamura, S. In vitro root induction by 24-epibrassinolide on hypocotyl segments of soybean [Glycine max (L.) Merr.]. Plant Growth Regulation. 1990, 73–76. [CrossRef]
- Topping, J.F.; May,V.J.; Muskett, P.R.; Lindsey, K. Mutations in the HYDRA1 gene of Arabidopsis perturb cell shape and disrupt embryonic and seedling morphogenesis. Development. 1997, 124(21):4415-24. [CrossRef]
- Oh, M.H.; Honey, S.H.; Tax, F.E. The Control of Cell Expansion, Cell Division, and Vascular Development by Brassinosteroids: A Historical Perspective. Int J Mol Sci. 2020, 21(5):1743. [CrossRef]
- Mohammadreza, Asghari.; Rana, Rezaei-Rad. 24-Epibrassinolide enhanced the quality parameters and phytochemical contents of table grape. Journal of applied botany and food quality. 2018, 91:226-231.
- Barket, A.; Syed, A.H.; Shamsul, H.; Qaiser, H.; Sangeeta, Y.; Qazi, F.; Aqil, A. A role for brassinosteroids in the amelioration of aluminium stress through antioxidant system in mung bean (Vigna radiata L. Wilczek). Environmental and Experimental Botany. 2008, 62: 153-159.
- Pinhero, R.G.; Rao, M.V.; Paliyath, G.; Murr, D.P.; Fletcher, R.A. Changes in Activities of Antioxidant Enzymes and Their Relationship to Genetic and Paclobutrazol-Induced Chilling Tolerance of Maize Seedlings. Plant Physiol. 1997, 114(2):695-704. [CrossRef]
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000, 42(1):93-113.
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annual Review of Plant Biology. 2003, 54: 137-164. [CrossRef]
- Fujioka, S.; Takatsuto, S.; Yoshida, S. An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiology. 2002, 130(2): 930-939. [CrossRef]
- Si, J.; Sun, Y.; Wang, L.U.; Qin, Y.; Wang, C.; Wang, X. Functional analyses of populus euphratica brassinosteroid biosynthesis enzyme genes DWF4 (PeDWF4) and CPD (PeCPD) in the regulation of growth and development of Arabidopsis thaliana. J Biosci. 2016, 41(4): 727-742. [CrossRef]
- Ohnishi, Y.; Totoki, Y.; Toyoda, A.; Watanabe, T.; Yamamoto, Y.; Tokunaga, K.; Sakaki, Y.; Sasaki, H.; Hohjoh, H. Active role of small non-coding RNAs derived from SINE/B1 retrotransposon during early mouse development. Molecular Biology Reports. 2012, 39(2): 903-909. [CrossRef]
- Ohnishi, T.; Szatmari, A.M.; Watanabe, B.; Fujita. S.; Bancos, S.; Koncz, C.; Lafos, M.; Shibata, K.; Yokota, T.; Sakata, K.; Szekeres, M.; Mizutani, M. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell. 2006, 18(11): 3275-3288.
- Jager, C.E.; Symons, G.M.; Nomura, T.; Yamada, Y.; Smith, J.J.; Yamaguchi, S.; Kamiya. Y.; Weller, J.L.; Yokota, T.; Reid, J.B. Characterization of two brassinosteroid C-6 oxidase genes in pea. Plant Physiology. 2007, 143(4): 1894-1904. [CrossRef]
- Poppenberger, B.; Rozhon, W.; Khan, M.; Husar, S.; Adam, G.; Luschnig, C.; Fujioka, S.; Sieberer, T. CESTA, a positive regulator of brassinosteroid biosynthesis. Embo Journal. 2011, 30(6): 1149-1161. [CrossRef]
- Noguchi, T.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Yoshida, S.; Li, J.; Chory, J. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999, 120(3):833-40. [CrossRef]
- Chory, J.; Nagpal, P.; Peto, C.A. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991, 3(5):445-459.
- Oh, M.H.; Clouse, S.D. Brassinolide affects the rate of cell division in isolated leaf protoplasts of Petunia hybrida. Plant Cell Rep. 1998, 17(12):921-924. [CrossRef]
- Xie, L.; Yang, C.; Wang, X. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J Exp Bot. 2011, 62(13):4495-506. [CrossRef]
- Szekeres, M.; Koncz, C. Biochemical and genetic analysis of brassinosteroid metabolism and function in Arabidopsis. Plant Physiology & Biochemistry. 1998, 36(1):145-155. [CrossRef]
- Luo, M.; Xiao, Y.; Li, X.; Lu, X.; Deng, W.; Li, D.; Hou, L.; Hu, M.; Li, Y.; Pei, Y. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant Journal. 2007, 51(3): 419-430. [CrossRef]
- Wang, Y.; Hao, Y.; Guo, Y.; Shou, H.; Du, J. PagDET2 promotes cambium cell division and xylem differentiation in poplar stem. Front Plant Sci. 2022, 13:923530. [CrossRef]
- Mao, Z.; He, S.; Xu, F.; Wei, X.; Jiang, L.; Liu, Y.; Wang, W.; Li, T.; Xu, P.; Du, S. Photoexcited CRY1 and PhyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis thaliana. New Phytol. 2020, 225, 848–865.
- Liu, G.; Li, Y.; Liu, Y.; Guo, H.; Guo, J. Genome-wide identification and analysis of monolignol biosynthesis genes in Salix matsudana Koidz and their relationship to accelerated growth. For. Res. 2021, 1, 8. [CrossRef]
- Li, X.; Yang, Y.; Sun, X.; Lin, H.; Chen, J.; Ren, J.; Hu, X.; Yang, Y. Comparative Physiological and Proteomic Analyses of Poplar (Populus yunnanensis) Plantlets Exposed to High Temperature and Drought. PLoS ONE 2014, 9, e107605. [CrossRef]
- Luo, J.X.; Zhen, W.; Gu, Y.J.; Cao, X.J. Study on the growth characteristics of Populus yunnanensis. JOURNAL OF SOUTHWEST FORESTRY UNIVERSITY, 2006(06):22-25.
- Liu, Y.Q.; Fu, D.R. Development and Utilization of Sect Ⅲ. Tacamachaca Gene Resources on the Plateau of Western Sichuan. JOURNAL OF CENTRAL SOUTH FORESTRY UNIVERSITY. 2004, (05):129-131.
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; Xia, R. TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Mol Plant. 2023, S1674-2052(23)00281-2.
- Sudhir, K.; Glen, S.; Li, M.; Christina, K.; Koichiro, T. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549.
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2018.
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: a review. Biol Res. 2018, 51(1):46. [CrossRef]
- Wang, H.; Yang, C.; Zhang, C.; Wang, N.; Lu, D.; Wang, J.; Zhang, S.; Wang, Z.X.; Ma, H.; Wang, X. Dual role of BKI1 and 14-3-3s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell. 2011, 21(5):825-34. [CrossRef]
- Bajguz, A.; Piotrowska-Niczyporuk, A. Biosynthetic Pathways of Hormones in Plants. Metabolites. 2023, 13(8):884. [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Matsuoka, M. Brassinosteroids and rice architecture. Journal of Pesticide Science. 2004, 29: 184-188. [CrossRef]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012, 46:701-24. [CrossRef]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang. Y.; Fujioka, S.; Yoshida, S.; Asami, T.; Chory, J. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002, 2(4):505-13. [CrossRef]
- Schumacher, K.; Chory, J. Brassinosteroid signal transduction: still casting the actors. Curr Opin Plant Biol. 2003, (1):79-84. [CrossRef]
- Lyons, G.; Carmichael, E.; Mcroberts, C.; Aubry, A.; Thomson, A.; Reynolds, C.K. Prediction of lignin content in ruminant dietsand fecal samples using rapid analytical techniques. J Agric Food Chem. 2018, 66, 13031–13040. [CrossRef] [PubMed]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: the interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233(5):2000-2016. [CrossRef]
- Zhang, D.; Tan, W.; Yang, F.; Han, Q.; Deng, X.; Guo, H.; Liu, B.; Yin, Y.; Lin, H. A BIN2-GLK1 Signaling Module Integrates Brassinosteroid and Light Signaling to Repress Chloroplast Development in the Dark. Dev Cell. 2021, 56(3):310-324.e7. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).