Submitted:
20 June 2023
Posted:
20 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
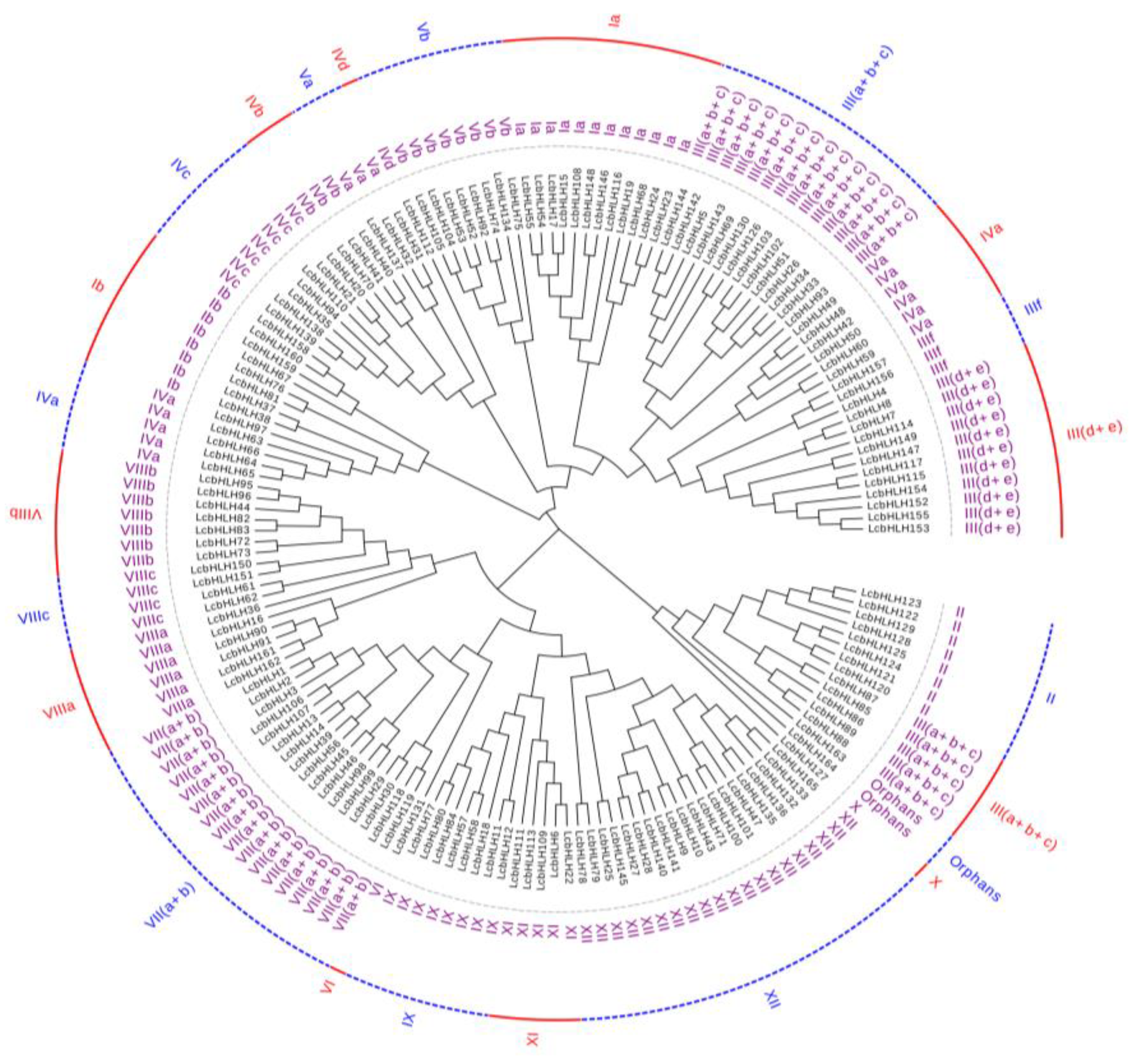
2.1. Identification of LcbHLHs gene members and Phylogenetic Analysis
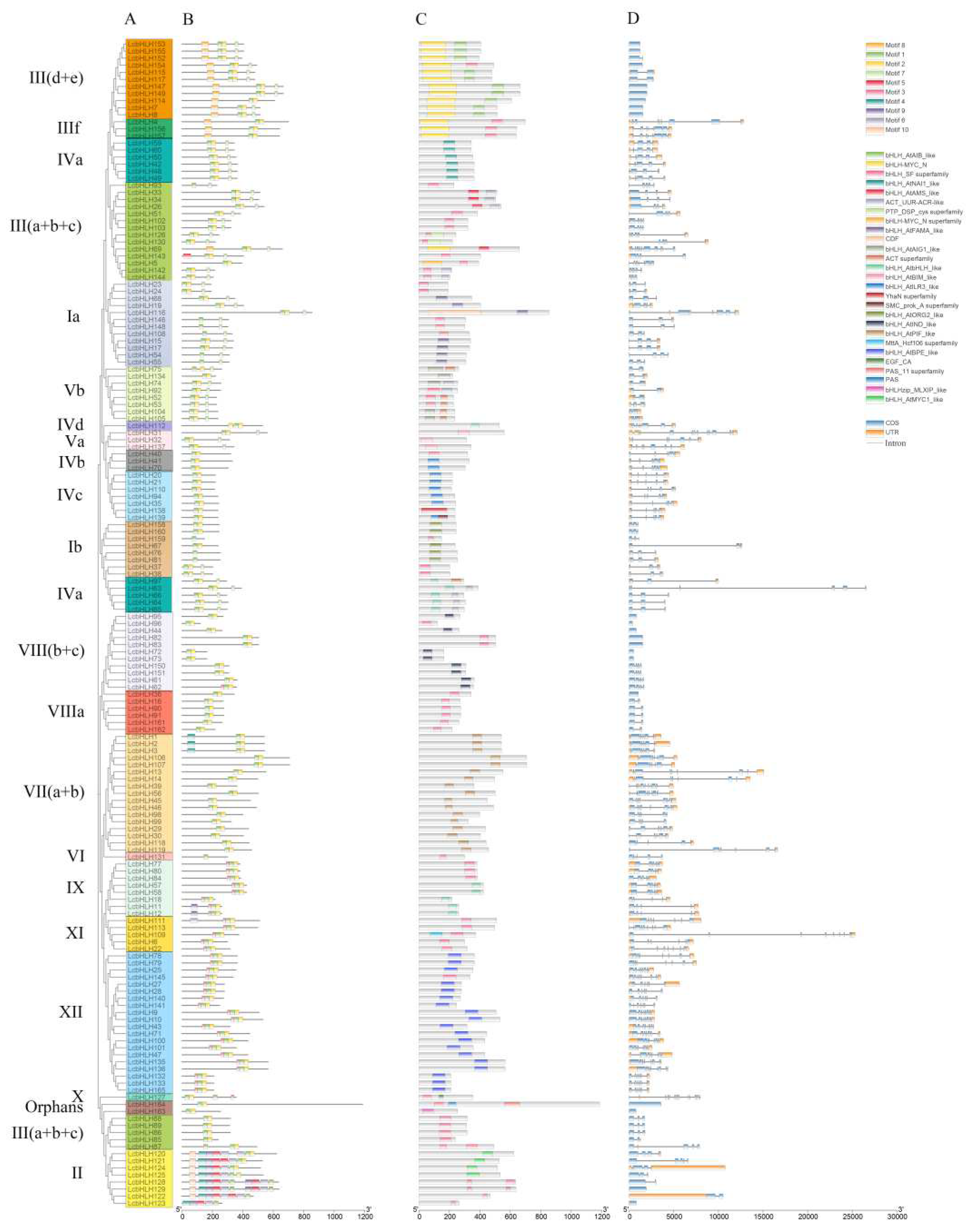
2.2. Multiple Sequence Alignment, Motif, Domain and Structure Analysis
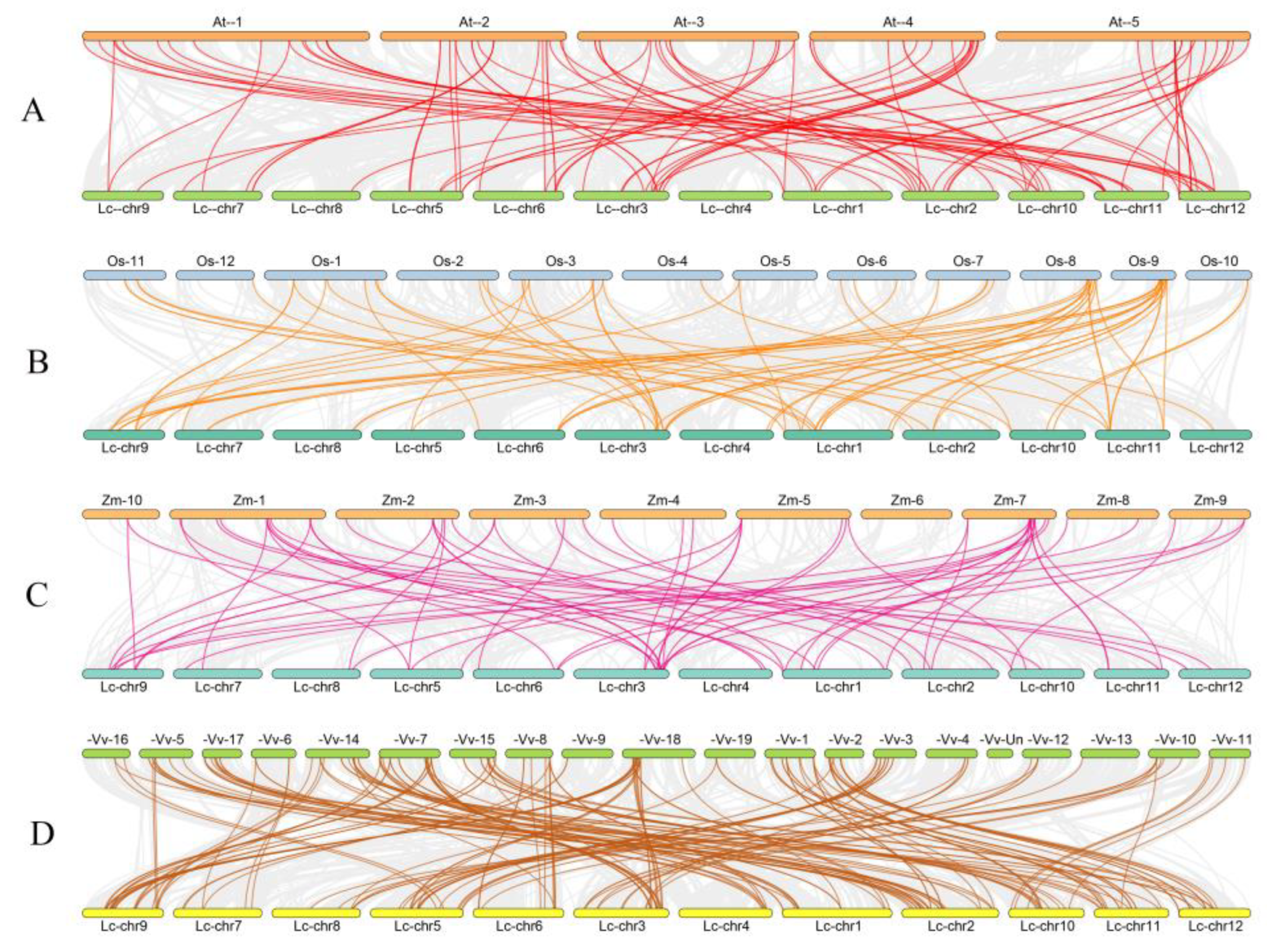
2.3. Gene Duplication and Collinear Correlation Analysis
2.4. GO annotation and analyses of cis-regulatory elements
2.5. Expression profiles of LcbHLHs and Gene Expression Analysis by qRT -PCR
2.6. The protein-protein interaction network of candidate gene.
3. Discussion
3.1. Systematic and comprehensive genome-wide detection of LcbHLHs in L. chinense
3.2. Functional prediction of LcbHLHs
4. Materials and Methods
4.1. Plant materials and data resources
4.2. Identification and Physicochemical Characterization of the bHLH gene family
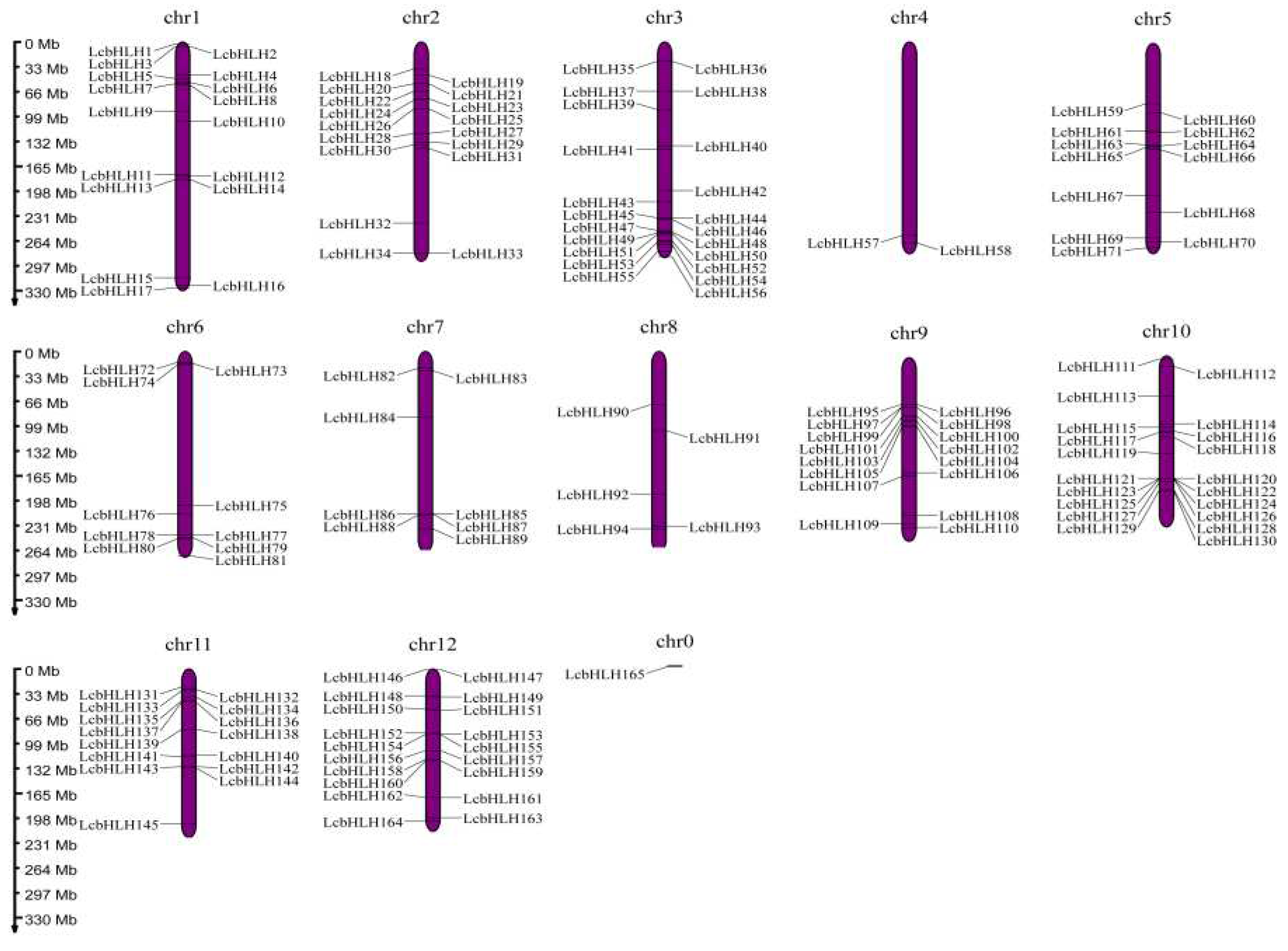
4.3. Chromosomal Localization, Tandem Duplication and Collinearity of the bHLH genes
4.4. Phylogeny and Multiple-Sequence Alignment of LcbHLHs
4.5. Analysis of Gene Structure, Conserved Motif and Family Structural Domains
4.6. GO Annotation, Analysis of Cis-acting Components and Protein-Protein Interaction of LcbHLHs
4.7. Gene expression patterns and quantitative real-time PCR analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riechmann, J.L.; Ratcliffe, O.J. A Genomic Perspective on Plant Transcription Factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.E. Built by Association: Structure and Function of Helix-Loop-Helix DNA-Binding Proteins. Structure 1994, 2, 1–4. [Google Scholar] [CrossRef]
- Atchley, W.R.; Fitch, W.M. A Natural Classification of the Basic Helix–Loop–Helix Class of Transcription Factors. Proc. Natl. Acad. Sci. 1997, 94, 5172–5176. [Google Scholar] [CrossRef]
- Massari, M.E.; Murre, C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.A. The Basic Helix-Loop-Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Ledent, V.; Vervoort, M. The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the BHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the Basic Helix-Loop-Helix Transcription Factor Gene Family in Arabidopsis Thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family[W]. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-Wide Analysis of Basic/Helix-Loop-Helix Transcription Factor Family in Rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (BHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, C.-L.; Cui, M.; Zhou, W.-J.; Wu, H.-L.; Ling, H.-Q. Four IVa BHLH Transcription Factors Are Novel Interactors of FIT and Mediate JA Inhibition of Iron Uptake in Arabidopsis. Mol. Plant 2018, 11, 1166–1183. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhang, J.; Wu, Y.-W.; Wang, Y.; Zhang, J.; Zheng, Y.; Li, Y.; Li, X.-B. BHLH Transcription Factors LP1 and LP2 Regulate Longitudinal Cell Elongation. Plant Physiol. 2021, 187, 2577–2591. [Google Scholar] [CrossRef]
- Sharma, N.; Xin, R.; Kim, D.-H.; Sung, S.; Lange, T.; Huq, E. NO FLOWERING IN SHORT DAY (NFL) Is a BHLH Transcription Factor That Promotes Flowering Specifically under Short-Day Conditions in Arabidopsis. Development 2016, 143, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yang, W.; Jia, Q.; Wang, W.; Zhang, N.; Wang, X.; Wang, S. Pleurotus Ostreatus BHLH Transcription Factors Regulate Plant Growth and Development When Expressed in Arabidopsis. J. Plant Interact. 2017, 12, 542–549. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional Regulation of BHLH during Plant Response to Stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhang, L.; Xia, C.; Fu, S.; Zhao, G.; Jia, J.; Kong, X. The Wheat Transcription Factor, TabHLH39, Improves Tolerance to Multiple Abiotic Stressors in Transgenic Plants. Biochem. Biophys. Res. Commun. 2016, 473, 1321–1327. [Google Scholar] [CrossRef]
- Ren, Y.-R.; Yang, Y.-Y.; Zhao, Q.; Zhang, T.-E.; Wang, C.-K.; Hao, Y.-J.; You, C.-X. MdCIB1, an Apple BHLH Transcription Factor, Plays a Positive Regulator in Response to Drought Stress. Environ. Exp. Bot. 2021, 188, 104523. [Google Scholar] [CrossRef]
- Zhao, R.; Song, X.; Yang, N.; Chen, L.; Xiang, L.; Liu, X.-Q.; Zhao, K. Expression of the Subgroup IIIf BHLH Transcription Factor CpbHLH1 from Chimonanthus Praecox (L.) in Transgenic Model Plants Inhibits Anthocyanin Accumulation. Plant Cell Rep. 2020, 39, 891–907. [Google Scholar] [CrossRef]
- Deng, J.; Li, J.; Su, M.; Lin, Z.; Chen, L.; Yang, P. A BHLH Gene NnTT8 of Nelumbo Nucifera Regulates Anthocyanin Biosynthesis. Plant Physiol. Biochem. 2021, 158, 518–523. [Google Scholar] [CrossRef]
- Arlotta, C.; Puglia, G.D.; Genovese, C.; Toscano, V.; Karlova, R.; Beekwilder, J.; De Vos, R.C.H.; Raccuia, S.A. MYB5-like and BHLH Influence Flavonoid Composition in Pomegranate. Plant Sci. 2020, 298, 110563. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, H.; Jiang, X.; Wang, P.; Dai, X.; Chen, W.; Gao, L.; Xia, T. A WD40 Repeat Protein from Camellia Sinensis Regulates Anthocyanin and Proanthocyanidin Accumulation through the Formation of MYB–BHLH–WD40 Ternary Complexes. Int. J. Mol. Sci. 2018, 19, 1686. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Li, Y.; Yang, S.; Gao, R.; Zhou, L.; Bao, T.; Han, T.; Wang, S.; Gao, X.; Wang, L. A Functional Homologue of Arabidopsis TTG1 from Freesia Interacts with BHLH Proteins to Regulate Anthocyanin and Proanthocyanidin Biosynthesis in Both Freesia Hybrida and Arabidopsis Thaliana. Plant Physiol. Biochem. 2019, 141, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Cai, W.; Zhang, X.; Li, W.; Zhou, Y.; Chen, Y.; Mi, Q.; Jin, L.; Xu, L.; Yu, X.; et al. Different Pruning Level Effects on Flowering Period and Chlorophyll Fluorescence Parameters of Loropetalum Chinense Var. Rubrum. PeerJ 2022, 10, e13406. [Google Scholar] [CrossRef]
- Chen, Q.; Cai, W.; Zhang, X.; Zhang, D.; Zhang, W.; Xu, L.; Yu, X.; Li, Y. The Comparative Studies on Phytochemicals of Leaf Coloration of Loropetalum chinense var. rubrum. Acta Hortic. Sin. 2021, 48, 1969–1982. [Google Scholar] [CrossRef]
- Guo, P.; Deng, S.; Zhang, Y.; Xu, L.; Yu, X.; Li, Y. Effect of Different Light Quality on Callus Growth and Flavonoids Contene of Two Loropetalum,chinense Plants. Acta Bot. Boreali Occident. Sin. 2022, 42, 118–126. [Google Scholar]
- Cai, W.; Zhang, D.; Zhang, X.; Chen, Q.; Liu, Y.; Lin, L.; Xiang, L.; Yang, Y.; Xu, L.; Yu, X.; et al. Leaf Color Change and Photosystem Function Evaluation under Heat Treatment Revealed the Stress Resistance Variation between Loropetalum Chinense and L. Chinense Var. Rubrum. PeerJ 2023, 11, e14834. [Google Scholar] [CrossRef]
- Zhang, X. Cloning, Expression and Transformation of Lc CHI and Lc ANS Genes from Loropetalum Chinense Var. Rubrum. Master, Hunan University of Technology, 2020.
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-Wide Identification and Characterization of the Potato BHLH Transcription Factor Family. Genes 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Xiong, R.; Liu, H.; Wu, M.; Chen, F.; Hanwei Yan; Xiang, Y. Basic Helix-Loop-Helix Gene Family: Genome Wide Identification, Phylogeny, and Expression in Moso Bamboo. Plant Physiol. Biochem. 2018, 132, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of BHLH Genes in Grape and Analysis of Their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Song, M.; Wang, H.; Wang, Z.; Huang, H.; Chen, S.; Ma, H. Genome-Wide Characterization and Analysis of BHLH Transcription Factors Related to Anthocyanin Biosynthesis in Fig (Ficus Carica L.). Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, R.; Ma, R.; Shen, Z.; Cai, Z.; Song, Z.; Peng, B.; Yu, M. Genome-Wide Analysis of Basic Helix-Loop-Helix Superfamily Members in Peach. PLOS ONE 2018, 13, e0195974. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 Synergistically Specify the Expression of BANYULS and Proanthocyanidin Biosynthesis in Arabidopsis Thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the Anthocyanin Biosynthetic Pathway by the TTG1/BHLH/Myb Transcriptional Complex in Arabidopsis Seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 Encodes a BHLH Protein That Regulates Trichome Development in Arabidopsis Through Interaction With GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef]
- Wen, J.; Li, Y.; Qi, T.; Gao, H.; Liu, B.; Zhang, M.; Huang, H.; Song, S. The C-Terminal Domains of Arabidopsis GL3/EGL3/TT8 Interact with JAZ Proteins and Mediate Dimeric Interactions. Plant Signal. Behav. 2018, 13, e1422460. [Google Scholar] [CrossRef]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-Interacting BHLH Transcription Factors That Regulate Jasmonate Responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-Domain Proteins Interact with the WD-Repeat/BHLH/MYB Complexes to Regulate Jasmonate-Mediated Anthocyanin Accumulation and Trichome Initiation in Arabidopsis Thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef]
- An, J.-P.; Li, H.-H.; Song, L.-Q.; Su, L.; Liu, X.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The Molecular Cloning and Functional Characterization of MdMYC2, a BHLH Transcription Factor in Apple. Plant Physiol. Biochem. 2016, 108, 24–31. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, Y.; Peng, W.; Wang, Z.; Xie, D. Molecular Mechanism for Jasmonate-Induction of Anthocyanin Accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef]
- Yu, Q.; Hua, X.; Yao, H.; Zhang, Q.; He, J.; Peng, L.; Li, D.; Yang, Y.; Li, X. Abscisic Acid Receptors Are Involves in the Jasmonate Signaling in Arabidopsis. Plant Signal. Behav. 2021, 16, 1948243. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Li, G.; Hu, P.; Zhao, X.; Li, L.; Wei, W.; Feng, J.; Zhou, H. Identification of Basic/Helix-Loop-Helix Transcription Factors Reveals Candidate Genes Involved in Anthocyanin Biosynthesis from the Strawberry White-Flesh Mutant. Sci. Rep. 2018, 8, 2721. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.-Y.; Liu, Y.-Y.; Zhu, H.-H.; Zhang, J.; Yang, J.-X.; Fu, Q.; Wang, N.; Wang, Z. Genome-Wide Analyses of the BHLH Gene Family Reveals Structural and Functional Characteristics in the Aquatic Plant Nelumbo Nucifera. PeerJ 2019, 7, e7153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L.; Li, G. Genome-Wide Analysis of the Basic Helix-Loop-Helix (BHLH) Transcription Factor Family in Maize. BMC Plant Biol. 2018, 18, 1–14. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene Duplication and Evolutionary Novelty in Plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Fujiwara, S.; Kamada, H.; Coupland, G.; Mizoguchi, T. Antisense Suppression of the Arabidopsis PIF3 Gene Does Not Affect Circadian Rhythms but Causes Early Flowering and Increases FT Expression. FEBS Lett. 2004, 557, 259–264. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.-I.; Choi, G. Light Activates the Degradation of PIL5 Protein to Promote Seed Germination through Gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.-S.; Sun, T.; Kamiya, Y.; Choi, G. PIL5, a Phytochrome-Interacting BHLH Protein, Regulates Gibberellin Responsiveness by Binding Directly to the GAI and RGA Promoters in Arabidopsis Seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef]
- Liu, X.-J.; An, X.-H.; Liu, X.; Hu, D.-G.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. MdSnRK1.1 Interacts with MdJAZ18 to Regulate Sucrose-Induced Anthocyanin and Proanthocyanidin Accumulation in Apple. J. Exp. Bot. 2017, 68, 2977–2990. [Google Scholar] [CrossRef]
- Xie, X.-B.; Li, S.; Zhang, R.-F.; Zhao, J.; Chen, Y.-C.; Zhao, Q.; Yao, Y.-X.; You, C.-X.; Zhang, X.-S.; Hao, Y.-J. The BHLH Transcription Factor MdbHLH3 Promotes Anthocyanin Accumulation and Fruit Colouration in Response to Low Temperature in Apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between BHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (BHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.; Guo, H.; Xie, D. The BHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development. PLOS Genet. 2013, 9, e1003653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Zhang, D.; Su, D.; Li, W.; Wang, X.; Chen, Q.; Cai, W.; Xu, L.; Cao, F.; et al. Comprehensive Analysis of Metabolome and Transcriptome Reveals the Mechanism of Color Formation in Different Leave of Loropetalum Chinense Var. Rubrum. BMC Plant Biol. 2023, 23, 133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Zhang, D.; Liu, Y.; Lin, L.; Xiong, X.; Zhang, D.; Sun, M.; Cai, M.; Yu, X.; et al. Transcriptomic and Metabolomic Profiling Provides Insights into Flavonoid Biosynthesis and Flower Coloring in Loropetalum Chinense and Loropetalum Chinense Var. Rubrum. Agronomy 2023, 13, 1296. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Liu, Y.; Lin, L.; Xiong, X.; Zhang, D.; Li, S.; Yu, X.; Li, Y. Genome-Wide Identification and Characterization of WRKY Transcription Factors and Their Expression Profile in Loropetalum Chinense Var. Rubrum. Plants 2023, 12, 2131. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]

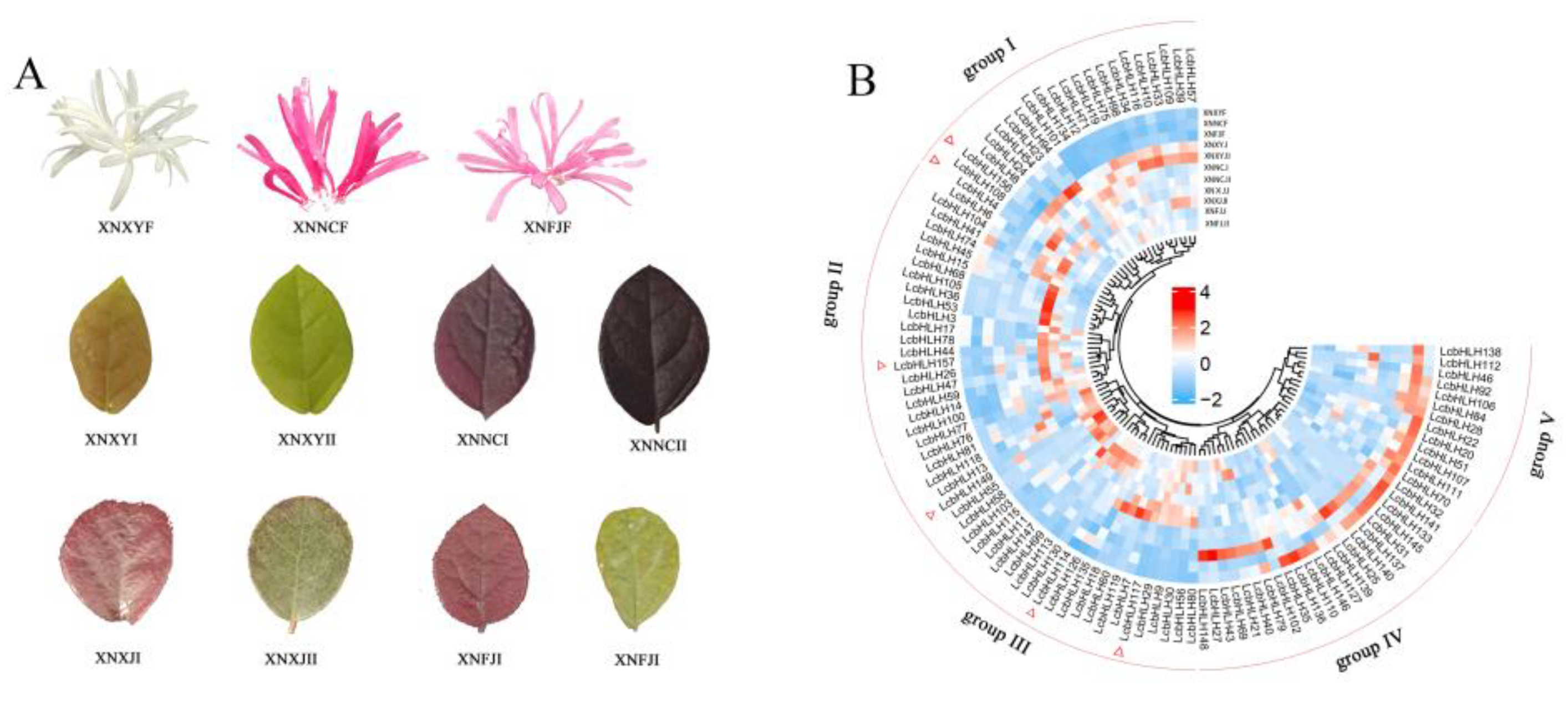
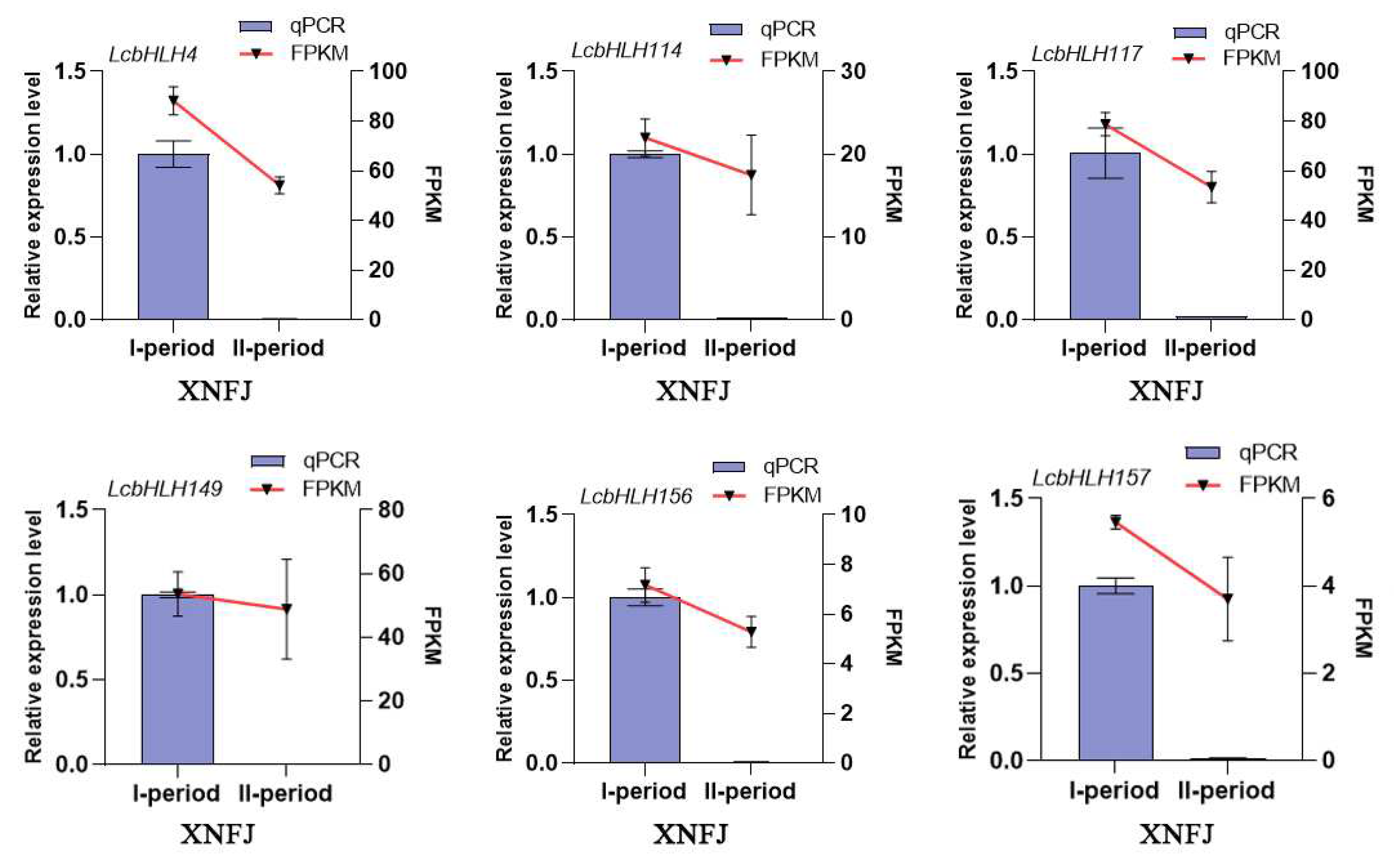
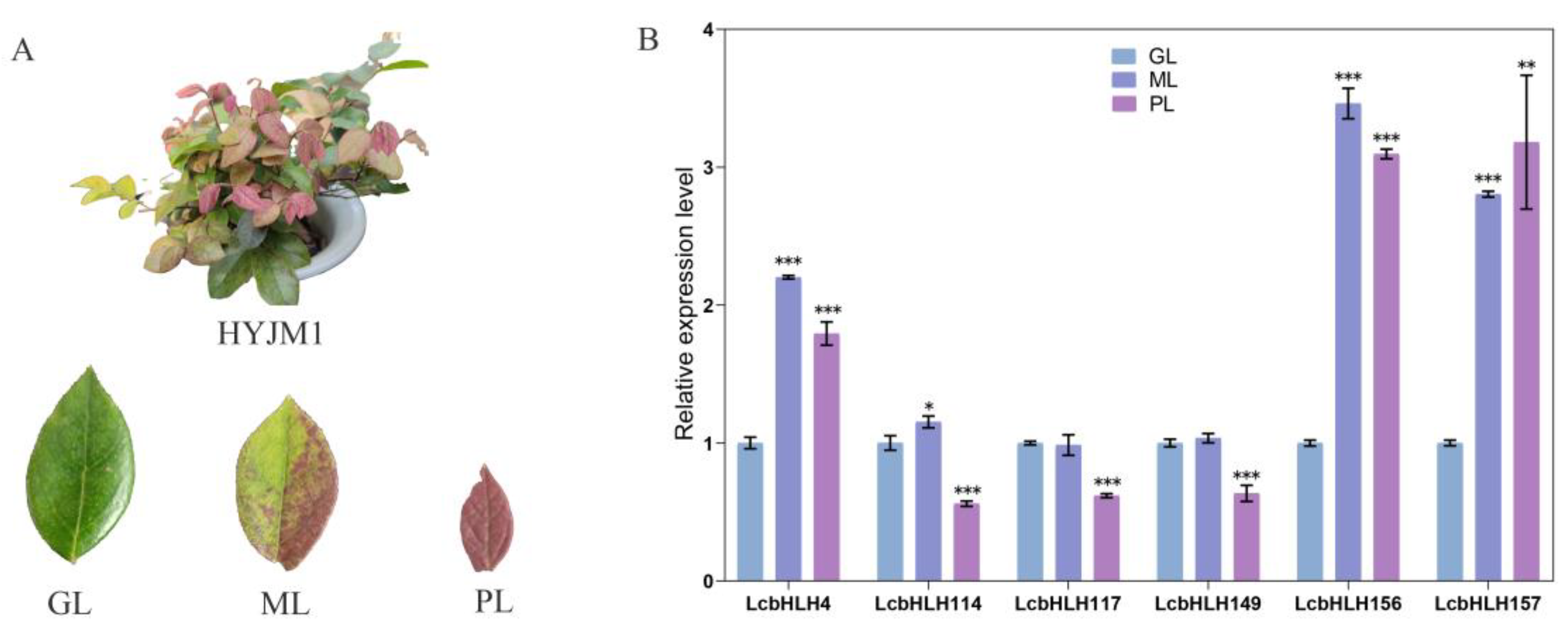
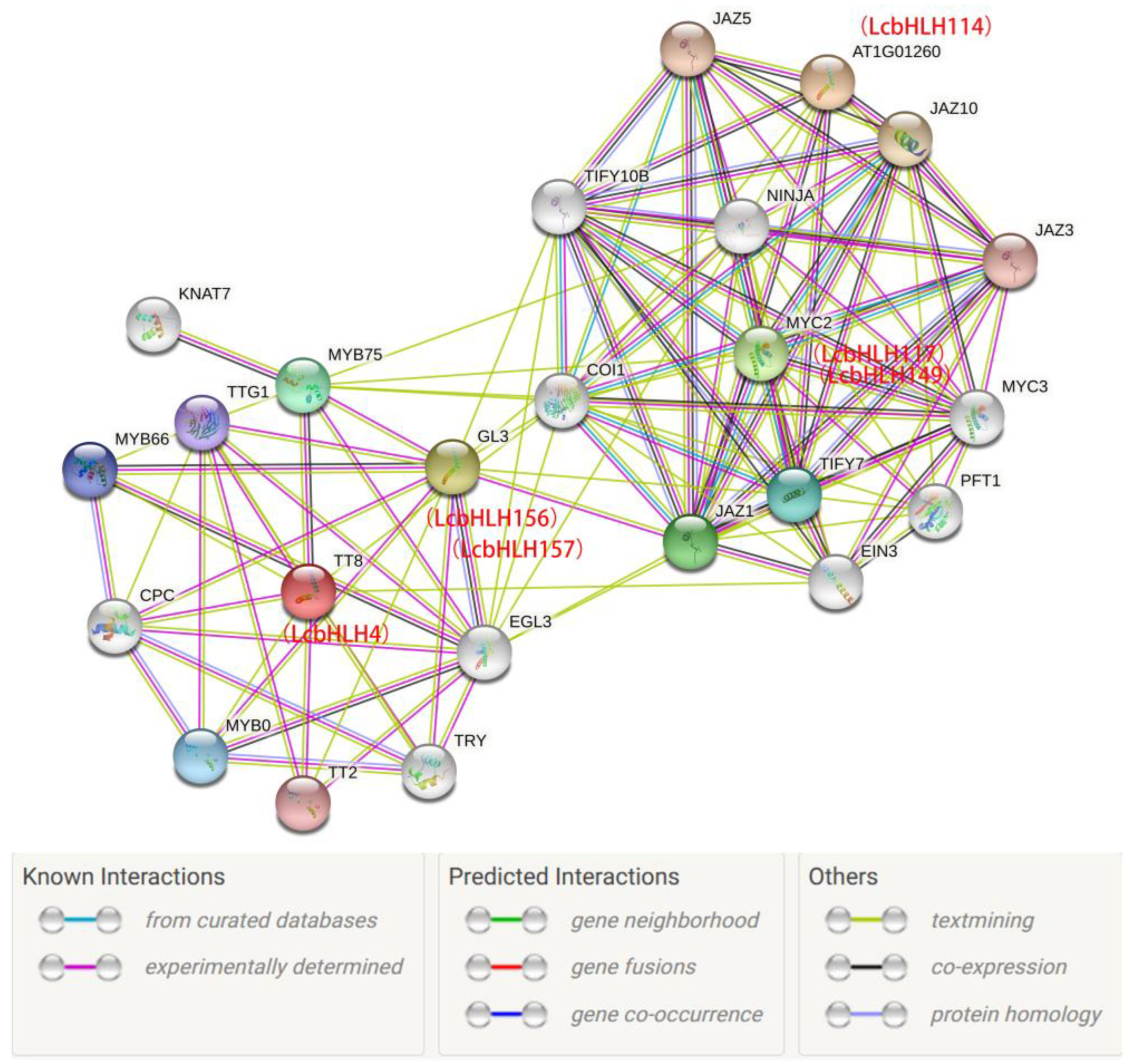
| Subfamilies | Number of LcbHLHs | Number of AtbHLHs | Number of FcbHLHs | Number of PpbHLHs |
| Ia | 12 | 10 | 8 | 8 |
| Ib | 8 | 13 | 7 | 8 |
| II | 8 | 4 | 0 | 1 |
| III(a+b+c) | 19 | 10 | 9 | 10 |
| III(d+e) | 11 | 8 | 6 | 11 |
| IIIf | 3 | 4 | 1 | 2 |
| IVa | 11 | 4 | 4 | 3 |
| IVb | 3 | 3 | 4 | 0 |
| IVc | 7 | 4 | 3 | 0 |
| IVd | 1 | 2 | 1 | 2 |
| Va | 3 | 3 | 3 | 2 |
| Vb | 8 | 5 | 6 | 6 |
| VI | 1 | 2 | 2 | 0 |
| VII(a+b) | 17 | 15 | 7 | 7 |
| VIIIa | 6 | 4 | 3 | 2 |
| VIII(b+c) | 11 | 11 | 14 | 9 |
| IX | 8 | 6 | 5 | 6 |
| X | 1 | 10 | 14 | 2 |
| XI | 5 | 5 | 4 | 4 |
| XII | 20 | 17 | 13 | 12 |
| XIII | 0 | 3 | 4 | 0 |
| XIV | 0 | 3 | 0 | 0 |
| XV | 0 | 5 | 0 | 0 |
| Orphans | 2 | 8 | 0 | 0 |
| Motif | Sequence |
|---|---|
| 1 | SHSLAERRRRERJNERFKALRSLVPNCSK |
| 2 | MDKASMLDEAIEYVKELQRQVQELSMKLE |
| 3 | EEPKSDYIHVRARRGQATD |
| 4 | QVMSFEQSNWDASVHEIQGMTSFEHPHNQDQQLHLLHEMQQNGHHHPQSF |
| 5 | FVQKPANFQTSLGFLGDLPTPDNASASSVLYDPLFHLNLPPQPPLFRDLF |
| 6 | YNLPASRTASLFGGGIDEKEGSGGVYQNGVATQFDNGVLEFTGDIGGMGK |
| 7 | RJVSALEKLGLDIVHANVSTF |
| 8 | ERAKLAKSAGIRTLVCIPTASGVVELGSTEIIKEDLGLIQLIKSLF |
| 9 | NSSTLPDTSPYINNPPTQHLLNLFHLPRCSPSSNLLPNSSI |
| 10 | DNIRLSMEELSYHQNPHQEDDAALEQHLGFDMENCYNINNN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





