1. Introduction
Ral guanine nucleotide dissociation stimulator (RalGDS), a member of the Ras-associating guanine nucleotide exchange factor (GEF) family, activates Ral GTPases by catalyzing GDP-to-GTP exchange. As a critical mediator downstream of Ras and Rap1 signaling pathways, RalGDS regulates cellular processes such as proliferation, differentiation, and apoptosis. Structurally, RalGDS family members (RalGDS, RGL, RGL2/Rlf, and RGL3) share conserved domains: an N-terminal Ras Exchange Motif (REM), a central CDC25 homology domain, and a C-terminal Ras Binding Domain (RBD) [
1,
2,
3].
The RalGDS gene was first cloned from a mouse cDNA library and has since been identified in humans, cattle, zebrafish, and
Echinococcus granulosus [
3,
4,
5,
6,
7]. Notably, extensive polymorphism has been reported in this gene across species, including five transcript variants in humans and eleven in mice. In pigs (
Sus scrofa), although the RalGDS sequence remains unannotated in public databases, in silico analysis of the porcine genome predicts at least twelve splice variants. Polymorphisms in RalGDS may lead to structural and functional divergence, potentially affecting traits such as growth, disease resistance, and metabolic efficiency. However, systematic studies on porcine RalGDS polymorphism are lacking.
Here, we amplified the full-length RalGDS open reading frame (ORF) from LLC-PK1 cells, a porcine renal proximal tubule-derived cell line, and identified seven novel polymorphisms. This work advances our understanding of RalGDS genetic diversity in pigs and its potential biological significance.
2. Materials and Methods
2.1. Cells
LLC-PK1 cells were maintained in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) (Gibco), supplemented with 7% fetal bovine serum (FBS, Invitrogen) and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator.
2.2. RT-PCR, DNA Cloning, and Sequence Assembly
Total RNA was extracted from a single layer of flattened LLC-PK1 cells using the QIAamp RNA Mini Kit (Qiagen) according to the manufacturer’s instructions and treated with DNase to remove contaminating genomic DNA. Reverse transcription was then performed using 1 µg of total RNA with the HiFiScript gDNA Removal RT MasterMix Kit (Cowin Biotech Co., Ltd., China). To determine the full RalGDS ORF, primers were first designed according to the twelve predicted RalGDS sequences (X1 (GenBank accession no. XM021070934), X2 (XM021070943), X3 (XM021070944), X4 (XM021070948), X5 (XM021070950), X6 (XM021070953), X7 (XM021070956), X8 (XM021070958), X9 (XM021070962), X10 (XR002338067), X11 (XR002338069), and X12 (XR002338070)) to obtain RT-PCR overlapping fragments. Further primers were synthesized based on newly amplified RalGDS sequences for verification (
Table 1). PCR was conducted to amplify each cDNA fragment from the RT product using 2X Taq High-Fidelity Master Mix (Tsingke Biotech Co., Ltd., China) according to the manufacturer’s protocol. The PCR reaction was performed at 94 °C for 5 minutes, followed by 30 cycles of denaturation at 94 °C for 30 seconds, annealing at 58 °C for 30 seconds, and extension at 72 °C for 30 seconds, with a final extension at 72 °C for 5 minutes. Each PCR amplicon was gel-purified, cloned into the pUC-Blunt Zero cloning vector (Sangon Biotech), transformed into DH5α E. coli, and sequenced bidirectionally. At least five clones for each PCR product were sequenced. To ensure the reliability of the sequence, each clone was sequenced three times. Sequence assembly was performed using the DNAMAN software.
2.3. Multiple Alignments and Phylogenetic Analyses
The 22 near-full-length RalGDS genome sequences, including twelve predicted pig RalGDS genes and ten sequences obtained from this study, were used in sequence alignments and phylogenetic analyses. Sequence alignments were performed using DNAMAN software. Phylogenetic trees were constructed via the neighbor-joining method in MEGA7 software, and bootstrap analysis was computed with 1000 replicates to determine percentage reliability values on each internal node of the tree.
2.4. Physicochemical Analysis of RalGDS Proteins
Physicochemical properties and signal peptide prediction analyses were performed using ExPASy (ProtParam - SIB Swiss Institute of Bioinformatics | Expasy) and the SignalP 5.0 Server (SignalP 5.0 - DTU Health Tech - Bioinformatic Services). Secondary structural analyses of the protein were performed using the online website Prabi (
https://npsa-prabi.ibcp.fr).
3. Results
3.1. Complete Genomic Characterization of RalGDS
Ten RalGDS sequences (dP1 - dP10) were identified. The results indicated that the lengths of the genomic sequences of RalGDS dP1, dP2, dP3, dP4, dP5, dP6, dP7, dP8, dP9, and dP10 were 3222, 3222, 3222, 3244, 3244, 3258, 3261, 3254, 3218, and 2792 nucleotides (nt), respectively. The 5’ untranslated regions (UTRs) of dP4 and dP5 were the same, both being 82 nt long, while the 3’ UTRs of all dP sequences were identical at 599 nt. When compared with the predicted RalGDS genomic sequences, the most significant features of the ten sequences obtained in this study were as follows: dP8 and dP9 had a contiguous 4-base deletion mutation; dP10 had a contiguous 430-nt deletion; and dP4 and dP5 had the insertion of one base (
Figure 1).
The ten complete genomic sequences of RalGDS were deposited into the GenBank database under accession numbers PV013880-PV013882, and PV137743-PV137749, respectively.
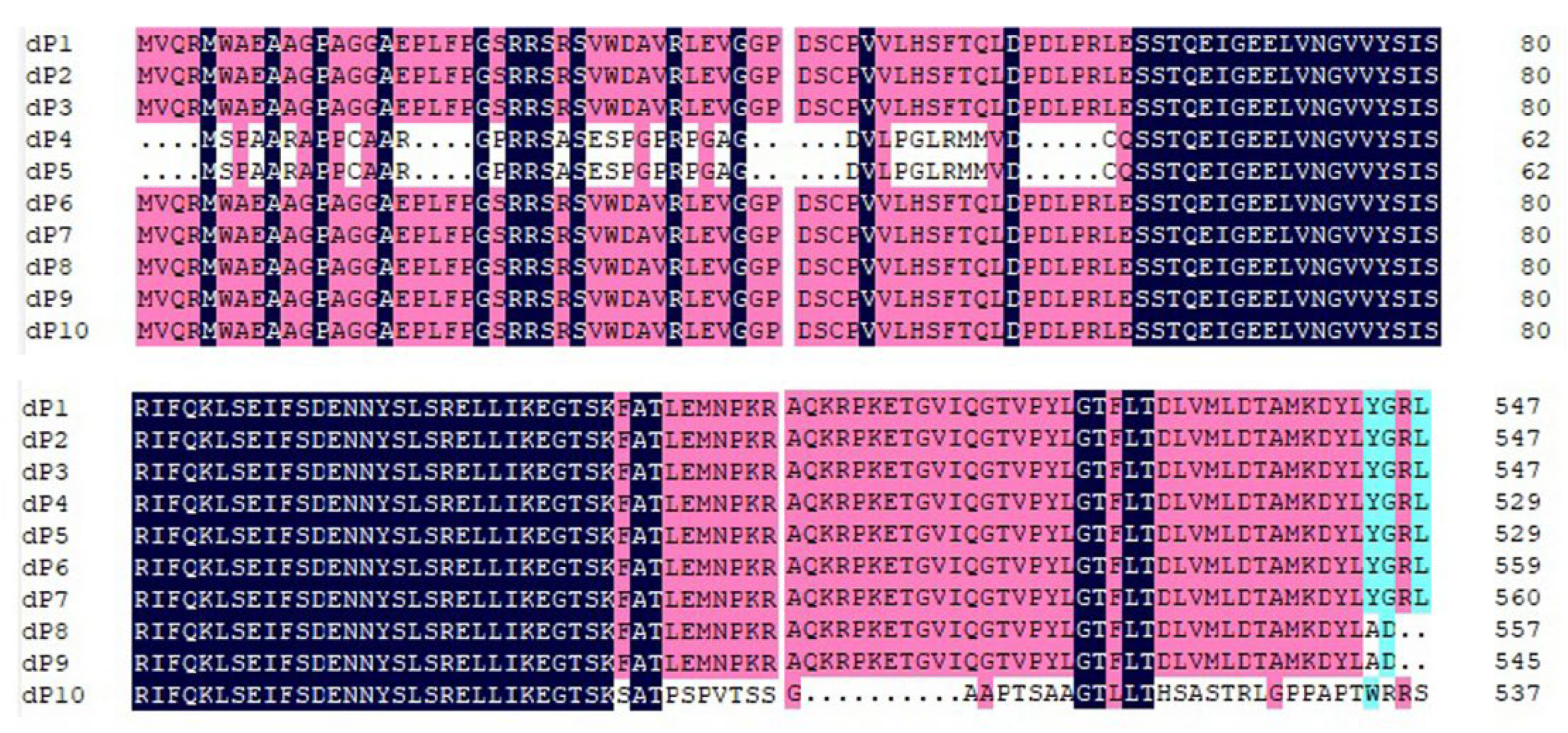
The genomes of RalGDS dP1, dP2, and dP3 possess a long open reading frame (ORF) with a length of 2616 nucleotides, encoding proteins consisting of 872 amino acids. The dP4 and dP5 genomes encode 854 amino acids. dP6 and dP7 encode 884 and 885 amino acids, respectively. However, dP8, dP9, and dP10, which have frameshift mutations in the ORF, show remarkable changes. They encode only 557, 545, and 538 amino acids, respectively. Moreover, the 38-amino acid sequence at the 3’ end of protein dP10 has extremely low homology compared to other RalGDS genomes (
Figure 2).
The ten RalGDS genes shared 82.84–99.97% identity at the nucleotide level and 51.83–100.0% identity at the amino acid level. In contrast, subsequent comparison analysis with twelve other predicted Sus scrofa RalGDS sequences showed high divergence, ranging from 33.47% similarity at the nucleotide level between dP10 and X11 to 91.42% between dP7 and X1. At the amino acid level, the similarity ranged from 50.34% between dP10 and X7 to 100.00%.
3.2. Phylogenetic Analysis
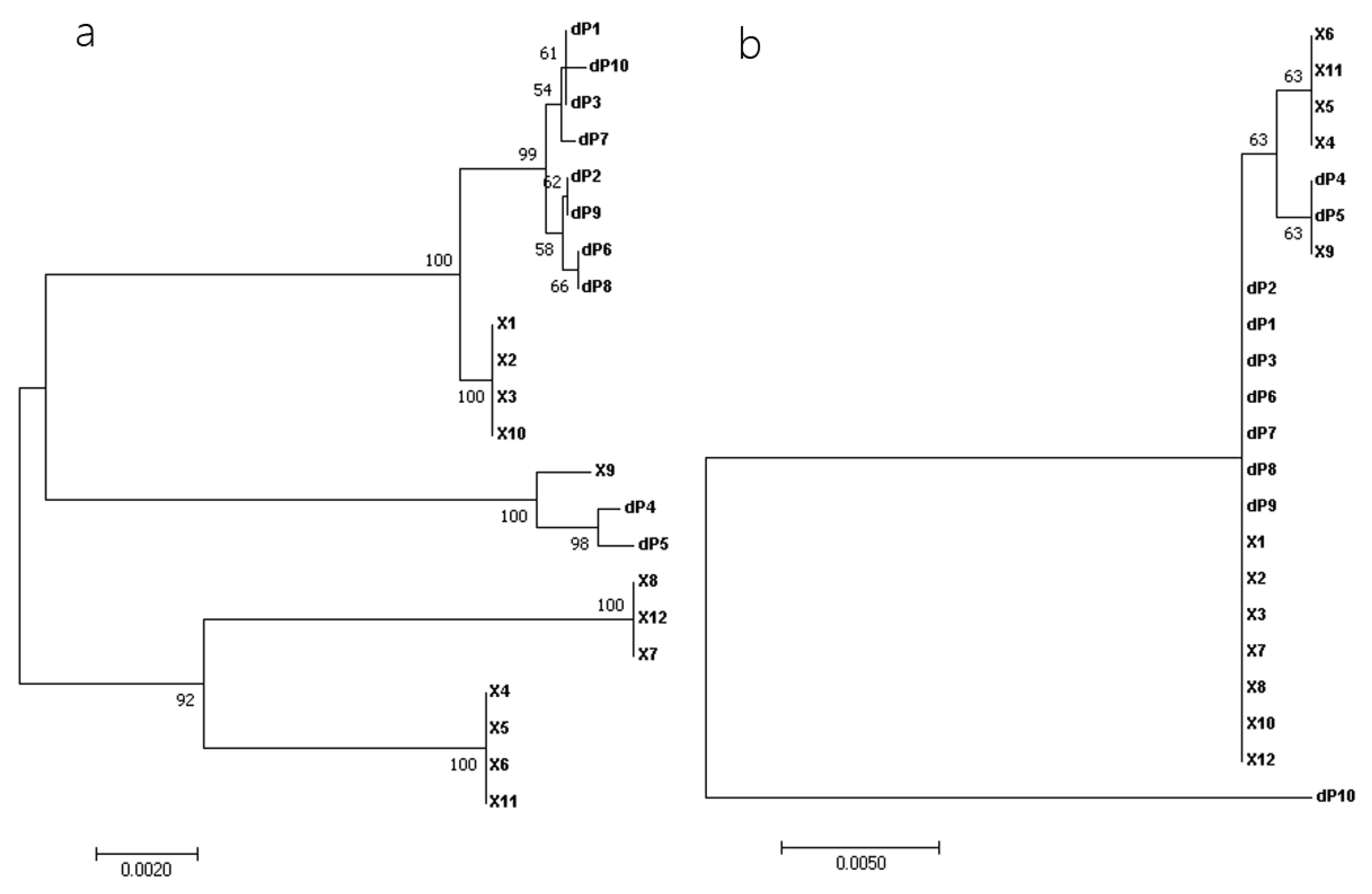
To establish genetic relationships, the phylogeny of 22 near-full-length sequenced RalGDS strains was estimated. The nucleotide sequence-based phylogenetic tree demonstrated that dP1, dP2, dP3, dP6, dP7, dP8, dP9, and dP10 were clustered together and closely related to X1, X2, X3, and X10, while dP4 and dP5 aligned with X9, distinct from two lineages: X8, X12, X7, and X4, X5, X6, X11. In contrast, the phylogenetic tree based on amino acid sequences differed from that based on nucleotide sequences, revealing that dP1, dP2, dP3, dP6, dP7, dP8, and dP9 were closely related to X1, X2, X3, X7, X8, X10, and X12, and dP4 and dP5 were closely related to X9, X4, X5, X6, and X11. Notably, dP10 formed a distinct branch, highlighting its evolutionary divergence (
Figure 3).
3.3. Physicochemical Properties and Secondary Structure Analysis of RalGDS Gene Family
The physicochemical properties and secondary structure characterization of the ten RalGDS proteins are presented in
Table 3. Their amino acid lengths vary between 538 and 884 amino acids. These proteins exhibit molecular weights ranging from 58,657.73 Da to 97,399.31 Da. Among these proteins, dP10 has the lowest relative molecular mass, and dP7 has the greatest relative molecular weight. The isoelectric points (pI) of the ten RalGDS proteins span from 5.29 to 5.96, indicating that all ten proteins are acidic. The highest aliphatic index of 89.69 was recorded for dP9, whereas dP4 and dP5 exhibited the lowest aliphatic index at 80.90. Additionally, the GRAVY (Grand Average of Hydropathicity) values for all ten RalGDS proteins are below zero, indicating that these proteins are hydrophilic. The instability index ranges from 45.95 to 53.19, suggesting that all ten RalGDS proteins are theoretically unstable.
No signal peptides (Sec/SPI) were predicted among the ten proteins. The secondary structure of the RalGDS protein was predicted, and the α-helix and random coil occupied the largest proportions at 37.47%-48.65% and 34.83%-46.60%, respectively, followed by extended strand (9.85%-12.10%) and β-turn (3.86%-5.39%). The composition of the secondary structure of dP8, dP9, and dP10 differs significantly from that of dP1, dP2, dP3, dP4, dP5, dP6, and dP7, manifested by an increase in the proportion of α-helix and β-turn and a decrease in the proportion of extended strand and random coil.
4. Discussion
This study systematically characterizes RalGDS polymorphism in porcine LLC-PK1 cells, identifying seven novel variants with potential functional implications. Based on the homology of the 5’ terminal sequences of the RalGDS gene, the twelve predicted RalGDS genes can be classified into four categories: X1, X2, X3, and X10 as one category; X7, X8, and X12 as another; X4, X5, X6, and X11 as a third; and X9 as a separate category. We designed primers for these four categories of genes, amplified them in segments, and finally designed primers based on the amplified sequences to amplify longer fragments for verification.
In this study, ten RalGDS gene family members, from dP1 to dP10, in LLC-PK1 cells were identified and obtained. According to sequence homology, the results showed that dP1, dP2, and dP3 had a close genetic relationship with the predicted RalGDS X3, dP6 had a close genetic relationship with the predicted RalGDS X2, and dP7 had a close genetic relationship with the predicted RalGDS X1. At the same time, the results showed a close genetic relationship between dP4 and dP5 and the predicted RalGDS X9. Notably, dP4 and dP5 encode extended N-terminal regions due to a single-base insertion. The observed frameshift mutations (e.g., 4-nt deletion in dP8 and dP9, and 430-nt deletion in dP10) likely disrupt RalGDS domain architecture, particularly the CDC25 and RBD regions essential for GTPase activation. Such structural alterations may impair RalGDS-mediated signaling.
The results of the phylogenetic tree based on amino acid sequences showed that RalGDS genes were divided into three categories, with dP1, dP2, dP3, dP6, dP7, dP8, and dP9 in LLC-PK1 belonging to one category, clustering with dP4 and dP5. The dP10 belongs to a separate evolutionary branch.
The physicochemical properties of proteins are critical for identifying their functions and attributes [
8]. Our results showed that most members of the RalGDS gene family, including dP1, dP2, dP3, dP4, dP5, dP6, and dP7, which exhibit high homologous conservation in their amino acid coding sequences, possess similar values for the protein instability index, aliphatic index, and secondary structure. This differs significantly from dP8, dP9, and dP10, owing to deletions resulting in frameshift mutations.
A large number of studies have shown that the polymorphism of the RALGDS gene is closely related to the occurrence and development of various diseases, such as skin cancer, breast cancer, and has also been proven to be associated with cardiovascular diseases [
9,
10,
11,
12,
13,
14,
15,
16].
The phylogenetic divergence of dP10 underscores its unique evolutionary trajectory. Its low homology to other variants (<51% amino acid identity) and distinct physicochemical properties (e.g., reduced molecular weight, altered secondary structure) suggest neofunctionalization or subfunctionalization. Further studies should explore whether dP10 retains GTPase activation capacity or acquires novel roles, such as in porcine renal physiology.
While this study focuses on a cell line, future work should validate these polymorphisms in vivo and assess their association with traits like growth efficiency or disease susceptibility.
5. Conclusions
We identified seven RalGDS polymorphisms in LLC-PK1 cells, including frameshift mutations and insertions that may alter protein function. These findings enrich the porcine genomic database and provide a framework for studying RalGDS in pig physiology and economic traits. Future research should prioritize in vivo validation and mechanistic studies to harness RalGDS diversity for genetic improvement.
Author Contributions
Conceptualization, L.W., S.Suolang. and Q.X.; Methodology, J.S., N.L., X.D., H.L. and H.Z.; Formal analysis, L.W.; data curation, L.W. and Q.X.; investigation, J.X. and K.H.; writing–the draft, L.W.; writing–review and editing, K.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 31972679).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All sequencing data are available from GenBank database on the NCBI website.
Conflicts of Interest
The authors declare no conflicts of interests.
References
- Ferro, E.; Trabalzini, L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal. 2010, 22, 1804–1810. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, R.; Umeki, N.; Yanagawa, M.; Murata, M.; Sako, Y. Single-molecule fluorescence imaging of RalGDS on cell surfaces during signal transduction from Ras to Ral. Biophys. Physicobiol. 2017, 14, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Albright, C.F.; Giddings, B.W.; Liu, J.; Vito, M.; Weinberg, R.A. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J. 1993, 12, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S.; et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009, 10, R42. [Google Scholar] [CrossRef]
- Postlethwait, J.H.; Farnsworth, D.R.; Miller, A.C. An intestinal cell type in zebrafish is the nexus for the SARS-CoV-2 receptor and the Renin-Angiotensin-Aldosterone System that contributes to COVID-19 comorbidities. bioRxiv [Preprint]. 2020, 2020.09.01.278366.
- Zheng, H.; Zhang, W.; Zhang, L.; Zhang, Z.; Li, J.; Lu, G.; Zhu, Y. ,Wang, Y., Huang, Y., Liu, J.; et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat. Genet. 2013, 45, 1168–1175. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Yang, L.; Liu, N.; Yang, J.; Zhou, X.K.; Xia, Y.C.; He, Y.; He, Y.Q.; Gong, H.J.; Ma, D.F.; et al. Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol. 2019, 19, 345. [Google Scholar] [CrossRef] [PubMed]
- González-García, A.; Pritchard, C.A.; Paterson, H.F.; Mavria, G.; Stamp, G.; Marshall, C.J. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell 2005, 7, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Lai, P.B.; Mok, T.S.; Chan, H.L.; Ding, C.; Yeung, S.W.; Lo, Y.M. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin. Chem. 2008, 54, 1528–1536. [Google Scholar] [CrossRef]
- Buhmeida, A.; Merdad, A.; Al-Maghrabi, J.; Al-Thobaiti, F.; Ata, M.; Bugis, A.; Syrjänen, K.; Abuzenadah, A.; Chaudhary, A.; Gari, M.; et al. RASSF1A methylation is predictive of poor prognosis in female breast cancer in a background of overall low methylation frequency. Anticancer Res. 2011, 31, 2975–2981. [Google Scholar]
- Miranda, E.; Bianchi, P.; Destro, A.; Morenghi, E.; Malesci, A.; Santoro, A.; Laghi, L.; Roncalli, M. Genetic and epigenetic alterations in primary colorectal cancers and related lymph node and liver metastases. Cancer 2013, 119, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Du, J.; Liu, J.; Wen, B. Identification of key target genes and pathways in laryngeal carcinoma. Oncol Lett. 2016, 12, 1279–1286. [Google Scholar] [CrossRef]
- Kawai, M.; Kawashima, S.; Sakoda, T.; Toh, R.; Kikuchi, A.; Yamauchi-Takihara, K.; Kunisada, K.; Yokoyama, M. Ral GDP dissociation stimulator and Ral GTPase are involved in myocardial hypertrophy. Hypertension 2003, 41, 956–962. [Google Scholar] [CrossRef]
- Scotland, R.L.; Allen, L.; Hennings, L.J.; Post, G.R.; Post, S.R. The ral exchange factor rgl2 promotes cardiomyocyte survival and inhibits cardiac fibrosis. PLoS ONE 2013, 8, e73599. [Google Scholar] [CrossRef] [PubMed]
- Rifki, O.F.; Bodemann, B.O.; Battiprolu, P.K.; White, M.A.; Hill, J.A. RalGDS-dependent cardiomyocyte autophagy is required for load-induced ventricular hypertrophy. J. Mol. Cell Cardiol. 2013, 59, 128–138. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).